Abstract
The efficacy of most marketed antimalarial drugs has been compromised by evolution of parasite resistance, underscoring an urgent need to find new drugs with new mechanisms of action. We have taken a high-throughput approach toward identifying novel antimalarial chemical inhibitors of prioritized drug targets for P. falciparum, excluding targets which are inhibited by currently used drugs. A screen of commercially available libraries identified 5,655 low molecular weight compounds that inhibit growth of P. falciparum cultures with EC50 values below 1.25 μM. These compounds were then tested in 384- or 1536-well biochemical assays for activity against nine Plasmodium enzymes: adenylosuccinate synthetase (AdSS), choline kinase (CK), deoxyuridine triphosphate nucleotidohydrolase (dUTPase), glutamate dehydrogenase (GDH), guanylate kinase (GK), N-myristoyltransferase (NMT), orotidine 5′-monophosphate decarboxylase (OMPDC), farnesyl pyrophosphate synthase (FPPS) and S-adenosylhomocysteine hydrolase (SAHH). These enzymes were selected using TDRtargets.org, and are believed to have excellent potential as drug targets based on criteria such as their likely essentiality, druggability, and amenability to high-throughput biochemical screening. Six of these targets were inhibited by one or more of the antimalarial scaffolds and may have potential use in drug development, further target validation studies and exploration of P. falciparum biochemistry and biology.
Keywords: target-based drug development, enzyme activity assays, antimalarial compounds
1. Introduction
Approximately 3 billion people, one half of the world’s population, live in at-risk regions for malaria infection. This leads to about 250 million malaria cases every year and nearly one million deaths (World Malaria Report, WHO, 2009, http://www.who.int/malaria/world_malaria_report_2009/en/index.html). The etiologic agent Plasmodium falciparum is the organism responsible for the majority of deaths due to malaria, 90% of which occur in Africa with more than 85% in children under the age of 5. The presence of widespread drug resistance is hampering the effectiveness of most of the available drug arsenal [1–6] with the notable exception of artemisinin-based derivatives. Artemisinin-based combination therapies (ACT) are the recommended first-line treatments for falciparum malaria in all countries with endemic disease [7]. Though ACT remains effective, there are recent concerns that the efficacy of artemisinin-based derivatives has declined on the Thai-Cambodian border [8–10]. Potential for clinical resistance is closely monitored in areas of Southeast Asia where artemisinin had been used as a monotherapy for decades and where antimalarial drug resistance previously developed. The potential development of artemisinin resistance by the parasite and loss of ACT would be a crisis, as ACTs have been recommended worldwide and all new agents currently in phase 3 clinical trials are based on ACT. In the battle to fight malaria, introduction of new drugs with novel mechanisms of action is essential.
Data on the essentiality of putative drug targets for Plasmodium growth are limited to a few individual genes that have been tested in gene disruption studies [11–19]. However, chemical inhibition of a target with genetic essentiality does not always translate to pharmacological efficacy against the pathogen [20], nor are genetic data on essentiality available in a comprehensive genome-wide dataset for Plasmodium. Inhibition of P. falciparum replication through chemical inhibition of potential drug targets can, instead, provide chemical validation of these targets. To this end, we undertook an effort to provide chemical tools to explore the biology of prioritized Plasmodium targets, and to provide a starting point for chemical validation of these targets. We developed biochemical assays to test the enzymatic activity of nine prioritized targets. A set of compounds which had been pre-annotated as having inhibitory activity on P. falcpiarum proliferation in erythrocytes was tested for any inhibitory activity in each of these biochemical assays. We reasoned that screening compounds already known to have cell-based activity might, at least in part, address some of the issues which have plagued biochemical approaches. These include cellular permeability, efflux and metabolism of small-molecule inhibitors.
We first screened GNF’s non-proprietary compound collection and identified a subset of 5,655 compounds which have potent inhibitory activity on P. falciparum growth in vitro as described in [21]. These compounds are non-proprietary and most are available commercially, facilitating broad exploration of their mechanism of action by the academic community. We have made the structures of these compounds available at http://www.ebi.ac.uk/chembldb/ and http://www.TDRtargets.org/. We have included in this dataset dose-response data for activity on two strains of P. falciparum, 3D7 and W2, as EC50 values, as well as cytotoxicity information on the human hepatocellular carcinoma cell line Huh7, and a measure of the promiscuity of the compounds across other historical high-throughput screens conducted at GNF. Where available, the compounds were tested as re-ordered powders from the supplier. This collection was then screened in high-throughput biochemical assays against nine Plasmodium enzymes considered to be promising drug targets based upon data compiled at TDRtargets.org [22] and other literature data. The results of these nine target-based biochemical screens are discussed below. Notably we identified chemical inhibitors of 6 of the 9 targets tested from this sub-library of only a few thousand compounds, preselected to have cellular activity.
2. Materials and methods
2.1. Identification of antimalarial compounds
A high-throughput screen of GNF’s non-proprietary compound collection was performed using an erythrocyte-based P. falciparum strain 3d7 infection assay essentially as described elsewhere [21]. Compounds were screened at 1.25 μM; those hits inhibiting growth by ≥50% were reconfirmed in dose-response format to determine EC50 values, and cellular cytotoxicity (quantified as CC50) was assessed in the Huh7 hepatocellular carcinoma cell line. Compounds with a selectivity index of <5 (CC50/EC50) were excluded from further consideration, as were compounds found to quench fluorescence. Reconfirmed hit compounds (5,655 in total) were re-ordered as powders and 3,086 compounds were available for purchase from suppliers.
2.2. Target selection
In selecting enzymes to screen (Table 1), we employed TDRtargets.org [22] and other sources to identify Plasmodium enzymes as candidates for compound screening using the following criteria: i) the enzyme is likely to be required for survival of the parasite; ii) the enzyme has a small-molecule binding pocket that might be exploited with a drug-like molecule to inhibit enzymatic activity; iii) there are exploitable differences between the parasite and human enzymes and/or the pathways to which they belong so that a drug might be developed with minimal adverse effects on the host; iv) the enzyme is amenable to recombinant expression (for which there was precedent for all selected targets [23–30]; and v) the enzymatic activity can be measured in a high-throughput format. Of these criteria we focused especially on i, iv, and v because druggability (ii) and selectivity (iii) are currently difficult to predict. (As an example of the latter, difluoromethylornithine inhibits both the human and T. brucei ornithine decarboxylases, yet selectively kills T. brucei cells, apparently because of the more rapid turnover of the human enzyme [31].) The nine enzymes chosen by these criteria were adenylosuccinate synthetase (AdSS), choline kinase (CK), deoxyuridine triphosphate nucleotidohydrolase (dUTPase), farnesyl pyrophosphate synthase (FPPS), glutamate dehydrogenase (GDH), guanylate kinase (GK), N-myristoyltransferase (NMT), orotidine 5′-monophosphate decarboxylase (OMPDC), and S-adenosylhomocysteine hydrolase (SAHH).
Table 1.
Selection of promising drug targets for biochemical screens
| Enzyme name (PlasmoDB ID) | Essentiality | Druggability score* | Genome-wide percentile ranking** |
|---|---|---|---|
| Adenylosuccinate synthase (PF13_0287) | Part of purine salvage pathway, which should be essential because Plasmodium cannot synthesize purines de novo [63]. | 0.8 | 99.5% |
| Choline kinase (PF14_0020) | Chemical validation of the Kennedy pathway for phosphatidylcholine synthesis is suggested by the work of Henri Vial [64]. | 0.6 | 97.9% |
| dUTPase (PF11_0282) | dUTPase is most likely essential because it prevents the buildup of dUTP and makes dUMP for dTTP synthesis [47]. | 0.6 | 99.8% |
| Farnesyl pyrophosphate synthase (PVX_092040) | Since protein farnesyltransferase is a valid drug target in Plasmodium [51], FPPS should be as well because it supplies the substrate for PFT. A recent paper reported an ability to predict cell-killing activity from bisphosphonates’ inhibition of FPPS [54]. | 0.8 | 99.3% |
| Glutamate dehydrogenase (PF14_0164) | Thought to be a major source of NADPH for glutathione reductase, which is thought to be essential [65]. This is based in part on the fact that Plasmodium is “exposed to multiple oxidative stress due to its high metabolic rate, the degradation of heme and reactive oxygen species imposed by the host immune system” [66]. | 0.8 | 99.7% |
| Guanylate kinase (PVX_099895) | Based on metabolic maps, guanylate kinase appeared to be the only route by which Plasmodium can convert GMP to GDP or dGMP to dGDP. (A possible bypass is noted in the text.) Also, expression during liver stage [45] suggests importance in that stage. | 0.2 | 98.4% |
| N-myristoyltransferase (PF14_0127) | As argued by Gelb et al. [49], “Genetic studies have shown that the NMT gene is essential for viability in a range of species, including Drosophila melanogaster, S. cerevisiae and Candida albicans and Cryptococcus neoformans.” | 0.8 | 98.4% |
| OMP decarboxylase (PF10_0225) | Part of the de novo pyrimidine synthesis pathway, which should be essential in Plasmodium because it does not have a pyrimidine salvage pathway [63]. Also, chemical validation of the de novo pyrimidine pathway comes from studies of dihydroorotate dehydrogenase [46]. | N/A | 98.6% |
| S-adenosylhomocysteine hydrolase (PFE1050w) | Part of a methylation cycle whose blockage should be fatal [39]. | 0.8 | 99.9% |
Druggability scores were taken from TDRtargets.org. Scores range from 0.0 to 1.0, with 1.0 signifying maximum druggability and ~0.2 signifying an average score.
TDRtargets.org was used to perform a multiparameter weighted search, with points awarded for various features considered desirable in a potential drug target. Percentiles were then computed from a rank-order list of all genes in the Plasmodium genome. Points were awarded as follows: 8 points to each gene for each life cycle stage (merozoite, early and late ring, early and late schizont, and early and late trophozoite) in which their expression ranks in the top quintile (80th to 100th percentile) for a given erythrocyte life cycle stage; 4 points to each gene for each life cycle stage in which expression was in the 60th to 80th percentile; 50 points to each gene with genetic validation (according to manual curation of the literature); 35 points to each gene associated with one or more publications (according to an automated analysis of PubMed); 50 points to each gene with one or more associated crystal structures; 30 points to each gene for which there is a 3D structural model (ModBase); 25 points to each gene lacking an ortholog in humans; 20 points to each gene lacking a predicted transmembrane domain; 20 points to each gene coding for a protein less than 100 kilodaltons; 100 points to each gene coding for an enzyme; 40 points to each gene whose orthologs are essential in at least one model organism; and 30 points to each gene for which there is an activity assay available.
2.3 Expression and purification of recombinant Plasmodium proteins
Recombinant histidine-tagged enzymes from Plasmodium falciparum and P. vivax were expressed in Escherichia coli and purified by immobilized metal affinity chromatography, essentially as described previously [32].
2.4 Biochemical assay details
Specific activities of the nine enzymes studied have been reported previously [33], along with comparisons to literature values where possible.
Enzyme activity assays were miniaturized and optimized for 1536-well plates and test compounds screened in dose-response format over 12 points in ½-log serial dilutions (0.0005 – 100 μM), the exception being SAHH, which was screened in single-point format in 384-well plates. Incubations were at room temperature unless otherwise noted. All reactions included a final [DMSO] of 1%. The initial collection of antimalarial compounds was available as a set of DMSO solutions pre-existing in the GNF compound library (available formats: 5,655 compounds, 1536-well dose-response format; 5,543 compounds, 384-well single point format), and 6 targets were screened using these collections (Table 3). As the original stocks of antimalarial compounds became depleted, the three remaining targets (FPPS, dUTPase, and NMT) were screened using DMSO solutions reconstituted from the 3,086 compounds available for repurchasing at the time of these screens. Hits were reconfirmed using re-ordered purified powders where available from the vendor and tested in dose-response format.
Table 3.
Results of biochemical screens
| Enzyme | Z′ score | Hit rate* | # Hits | Reconfirmed (powders) | Reconfirmation Enzyme (IC50 μM) | 3d7 falciparum (EC50 μM) |
|---|---|---|---|---|---|---|
| Pf Choline Kinase | 0.802 | 1.2% | 69 | 9 of 23 | 2.9 – 19.0 | 0.04 – 0.7 |
| Pf dUTPase | 0.466 | 3.1% | 95 | 0 specific to dUTPase | n/a | n/a |
| Pf OMP Decarboxylase | 0.751 | 0.5% | 26 | 4 of 11 | 2.4 – 58.0 | 0.093 – 6.0 |
| Pf S-adenosyl-Homocysteine Hydrolase | 0.400 | 0.7% | 38 | 7 of 19 | 0.1 – 9.1 | 0.9 – 7.6 |
| Pf Glutamate Dehydrogenase | 0.783 | 2.9% | 164 | 0 of 78 | n/a | n/a |
| Pv Guanylate Kinase | 0.571 | 0.3% | 17 | 4 of 7 | 2.9 – 4.0 | 0.092 – 3.4 |
| Pf Adenylosuccinate Synthase | 0.667 | 0.2% | 9 | 5 of 6 | 1.7 – 22.9 | 0.2 – 1.6 |
| Pf N-Myristoyl Transferase | 0.640 | 0.2% | 6 | 5 of 5 | 1.4 – 8.6 | 0.04 – 2.6 |
| Pf Farnesyl Pyrophosphate Synthase | 0.667 | 0.2% | 6 | 0 specific to FPPS | n/a | n/a |
Determined based on size of compound collection screened. See Methods for details.
Chemical reactions and detection strategies for each enzyme are shown in Table 2; additional details are noted below. In general, assay concentrations of most substrates were approximately 3 times their respective Km’s. Compounds, in general, were added to enzyme and buffer reagents 10 minutes prior to initiation of reaction by addition of substrate. If compounds hit in multiple target-based assays they were triaged due to lack of specificity (i.e., likely false positives) and not included in the final hit list (Supplementary Table S1).
Table 2.
Screening reactions and detection strategies
| Enzyme name (EC number) | Chemical reactions | Detection strategy | Control wells* |
|---|---|---|---|
| Adenylosuccinate synthetase (6.3.4.4) | Primary: GTP + IMP + L-aspartate → GDP + Pi + adenylosuccinate | Detect GDP using Cisbio Transcreener ADP kit. | Hadacidin [42] |
| Choline kinase (2.7.1.32) | Primary: ATP + choline → ADP + phosphocholine | Detect ADP using Cisbio Transcreener ADP kit. | No choline |
| dUTPase (3.6.1.23) | Primary: dUTP + H2O → dUMP + PPi | Detect PPi with Lonza PPiLight kit. | CAS: 14270-73-6,84472-83-3 |
| Farnesyl pyrophosphate synthase (2.5.1.−) | Primary: geranyl diphosphate + isopentenyl diphosphate → farnesyl diphosphate + PPi | Detect PPi with Lonza PPiLight kit. | compound 11 from [67] |
| Glutamate dehydrogenase (1.4.1.4) | Primary: L-glutamate + H2O + NADP+ → 2-oxoglutarate + NH3 + NADPH + H+ | Detect NADPH fluorescence. | No glutamate |
| Guanylate kinase (2.7.4.8) | Primary: ATP + GMP → ADP + GDP | Detect ADP and GDP using Cisbio Transcreener ADP kit. | Iodoacetamide |
| N-myristoyltransferase (2.3.1.97) | Primary: myristoyl-CoA + glycylpeptide → CoA + N-myristoylglycylpeptide | Detect CoA’s free –SH groups with ThioGlo. | aurintricarboxylate |
| OMP decarboxylase (4.1.1.23) | Primary: OMP → UMP + ATP CMP kinase: UMP + ATP → UDP + ADP | Detect ADP using Cisbio Transcreener ADP kit. | No OMP |
| S- adenosylhomocysteine hydrolase (3.3.1.1) | Primary: S-adenosylhomocysteine + H2O → adenosine + homocysteine Adenosine deaminase: adenosine → inosine + NH3 | Detect homocysteine’s free –SH groups with ThioGlo. | No S- adenosylhomocysteine |
Control wells either contained a known inhibitor of the enzyme or lacked a substrate so as to mimic the condition of complete inhibition of the enzyme.
AdSS was assayed in a buffer of 50 mM Tris, pH 7.0, with 5 mM DTT, 0.1% BSA, 0.001% Tween-20, and 2.5 mM MgCl2. Final concentrations were 500 ng/mL AdSS (10 nM), 30 μM GTP, 60 μM IMP, and 1 mM aspartate. Incubation time was 40 minutes, after which the GDP produced by AdSS was detected with the Transcreener GDP detection mix (Cisbio), used according to the manufacturer’s instructions. Homogeneous time-resolved fluorescence (HTRF) of the GDP produced was monitored via excitation at 337 nm and dual emission at 620 and 665 nm.
CK was assayed in a buffer of 100 mM HEPES, pH 7.5, with 150 mM NaCl and 1 mM MgCl2. Final concentrations were 312.5 ng/mL CK (5 nM), 10 μM ATP, and 280 μM choline. Incubation time was 30 minutes, after which the ADP produced by CK was detected with the Transcreener ADP detection mix (Cisbio), as for AdSS.
dUTPase was assayed in a buffer of 25 mM MOPS, pH 8.0, with 10 mM KCl, 1.25 mM MgCl2, 0.1 mg/mL BSA, 0.005% Triton X and 20% glycerol. Final concentrations were 2 ng/mL dUTPase (97 pM) and 14 μM dUTP. Incubation time was 30 minutes. Production of PPi was monitored with the PPiLight kit (Lonza) via conversion to ATP and measurement of luminescent signal corresponding to [ATP]. The PPi detection mix was diluted 1:4 with reaction buffer prior to use.
FPPS was assayed in a buffer of 50 mM HEPES, pH 7.5, with 0.625 mM MgCl2, 1 mM NaCl, 0.25 mM TCEP, 5% BSA, 10% glycerol, and 0.01% Triton X-100. Final concentrations were 1.3 μg/mL FPPS (45 nM), 25 μM geranyl diphosphate, and 25 μM isopentenyl diphosphate. Incubation time was 30 minutes. Production of PPi was monitored with the PPiLight kit (Lonza), as for dUTPase.
GDH was assayed in a buffer of 100 mM Tris, pH 8.0, with 1 mM DTT, 0.1 mg/mL BSA, and 0.005% Triton X-100. Final concentrations were 0.5 μg/mL GDH (10 nM), 400 μM NADP+, and 10 mM glutamate. Incubation time was 45 minutes. The fluorescence of the NADPH produced was monitored via excitation at 355 nm and emission at 486 nm.
GK was assayed in a buffer of 50 mM Tris, pH 7.5, with 50 mM KCl, 2 mM MgCl2 and 0.1 mg/mL BSA. Final concentrations were 1.5 ng/mL GK (130 pM), 7 μM GMP, and 10 μM ATP. Incubation time was 20 minutes, after which ADP and GDP produced by GK were detected with the Transcreener ADP detection mix (Cisbio), as for AdSS.
NMT was assayed in a buffer of 50 mM Tris, pH 7.5, with 6.25 mM NaCl, 0.625 mM MgCl2, 10% BSA, and 0.005% Triton X-100. Final concentrations were 1.5 μg/mL NMT (30 nM), 15 uM myristoyl-CoA, and 90 uM GSSYSRKNK, a synthetic peptide obtained from Peptides International or GenScript with order parameters of at least 85% purity and no N- or C-terminal modifications. This peptide was based on the N-terminal sequence of adenylate kinase 2, a substrate of N-myristoyltransferase from P. falciparum in vivo [34]. Incubation time was 45 minutes at 30 °C, after which ThioGlo [35] was added to a final concentration of 15 μM to detect the free –SH groups of CoA produced by NMT. ThioGlo fluorescence was monitored via excitation at 360 nm and emission at 540 nm.
OMPDC was assayed in a buffer of 100 mM Tris, pH 8.0, with 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 0.3 mg/mL BSA. Final concentrations were 3 ng/mL OMPDC (77 pM), 2 μM OMP, 1 μM ATP, and 1 μg/mL human CMP kinase [36], which was used to convert UMP (produced by OMPDC) + ATP to UDP + ADP. Incubation time was 180 minutes, after which the ADP was detected with the Transcreener ADP detection mix (Cisbio), as for AdSS.
SAHH was assayed in a buffer of 50 mM potassium phosphate, pH 8.0. Final concentrations were 5 μg/mL SAHH (100 nM), 4 μM S-adenosylhomocysteine, and 2.5 μg/mL adenosine deaminase from P. falciparum (added to consume the adenosine produced by SAHH, promoting continued progress of the forward reaction). Incubation time was 90 minutes, after which ThioGlo [35] was added to a final concentration of 25 μM to detect the free –SH groups of homocysteine produced by SAHH. ThioGlo fluorescence was monitored via excitation at 360 nm and emission at 540 nm. The SAHH screen was unique in that compounds were initially tested at a single concentration of 8.3 μM; therefore, initial hits were defined as compounds inhibiting SAHH by ≥25% at this concentration.
3. Results
3.1. Identification of antimalarial compounds
We identified a set of 5,655 compounds that inhibit growth of P. falciparum in erythrocytes as described in [21]. ~7,500 primary screen hits were tested in 8-point dose-response curves for reconfirmation of antimalarial activity and EC50 value determination against two drug-resistant strains: a sulfadoxine-resistant 3D7 strain and a multi-drug resistant W2 strain (resistant to chloroquine, quinine, pyrimethamine, sulfadoxine, and cycloguanil). Known inhibitors in the compound collection (e.g., artemisinin, mefloquine, dihydrofolate reductase inhibitors, quinine) were identified, providing internal validation of the assay sensitivity. Counterscreens included fluorescence quencher assessment and cytotoxicity in the Huh7 human hepatocellular carcinoma cell line. Compounds with an EC50 < 1.25 μM against P. falciparum 3d7 and W2 strains and a selectivity index (SI) over the Huh7 cell line of at least 5-fold and that did not exhibit fluorescence quenching were retained for further study.
3.2. Choline kinase
CK catalyzes a step in the synthesis of phosphatidylcholine, which is essential for intraerythrocytic growth of the Plasmodium parasite [26, 37, 38]. The Km of CK for choline was determined as ~200 μM, consistent with the value of 140 μM reported in [26]. The chemical inhibitor hexadecyltrimethylammonium bromide (HDTAB) can be used as a positive control for CK inhibition [37]; however, the physicochemical properties of HDTAB interfered with the homogeneous time resolved fluorescence (HTRF) signal detection.
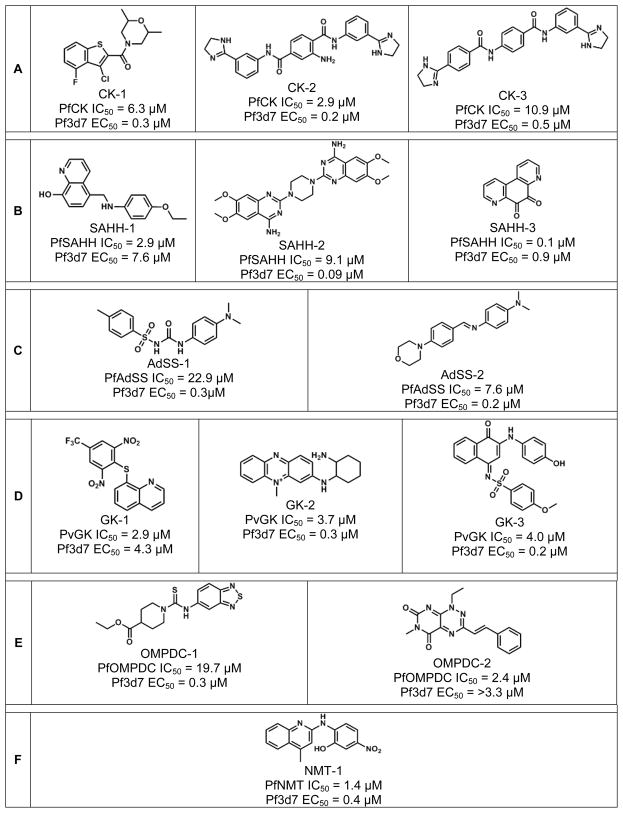
As listed in Table 2, the CK screen resulted in 69 hits for a 1.2% hit rate. IC50 values ranged from 0.06 μM to 11 μM while P. falciparum EC50 values ranged from 0.013 μM to 1.25 μM. 23 powders were available for reconfirmation as powders, nine of these reconfirmed in triplicate with IC50 values ranging from 2.9 μM to 19.0 μM. Corresponding P. falciparum EC50 values were at least 10-fold more potent. Of the nine hits identified (see Supplementary Table S1), compound CK-1 (a 3-chlorobenzothiophene amide; shown in Figure 1A) has a chemically tractable structure and may be a suitable starting point for chemical optimization. CK-2 and CK-3 (Fig. 1A) are analogs in the same scaffold. While activities on the enzyme are weaker (20-fold) than the cell-based activities, the rank orders in potency of these two compounds are the same between the cell-based and target-based assays.
Figure 1.
Chemically tractable inhibitors discovered by the biochemical screens. Abbreviations refer to the enzymes inhibited: (A) choline kinase (CK), (B) S-adenosylhomocysteine hydrolase (SAHH), (C) adenylosuccinate synthetase (AdSS), (D) guanylate kinase (GK), (E) OMP decarboxylase (OMPDC), (F) N-myristoyltransferase (NMT).
3.3. S-Adenosylhomocysteine hydrolase
SAHH, a component of the methylation cycle in which methyl groups are donated to lipids, nucleic acids, proteins, and other molecules, is almost certainly essential and is highly druggable (Table 1). The similarity of the parasite and human active sites is a concern, with only one residue of the adenosine binding site – Cys59 for PfSAHH, Thr60 for HsSAHH – differing between the two [39]. The Km of SAHH for S-adenosylhomocysteine was determined to be ~2 μM, close to the literature value of 1.2 μM [24]. The SAHH screen resulted in 38 hits and a hit rate of 0.68% (Table 2). 20 powders of the 38 hits were available, and of these, seven compounds reconfirmed in dose-response format and two were triaged based on overlap with other target screens (AdSS, below), suggesting that these were promiscuous inhibitors. IC50 values ranged from 0.1 μM to 9.1 μM and the corresponding P. falciparum EC50 values ranged from 0.9 μM to 7.6 μM. Of the five reconfirmed and specific hits identified (Supplementary Table S1), SAHH-1 (Fig. 1B) is an interesting 8-hydroxyquinoline-based hit that might serve as a potential starting point for medicinal chemistry. Other hits that may be of interest as tool compounds, though not chemically attractive for drug discovery, include SAHH-2, a quinazosin, a known vasodilator with alpha-1 adrenergic receptor as its target [40] and SAHH-3, known as phanquinone, an antimicrobial agent [41].
3.4. Enzymes involved in nucleotide metabolism (AdSS, dUTPase, GK, and OMPDC)
With AdSS, part of the purine salvage pathway, we obtained Km values of 14 μM for IMP, 105 μM for GTP, and 0.05 mM for aspartate, somewhat different from the previously reported values of 22.8 μM, 4.8 μM, and 1.4 mM, respectively [29]. The AdSS screen resulted in 9 hits and a 0.2% hit rate (Table 2). Hadacidin was used as the positive control inhibitor [42], its IC50 value being 62 μM. Of these hits, six were available as powders and five reconfirmed. Two compounds were triaged as they also inhibited SAHH activity, leaving 3 candidate AdSS inhibitors (Supplementary Table S1). However, all three of these compounds are unlikely starting points for medicinal chemistry because of chemically labile functionalities such as sulfonylureas (AdSS-1), imines derived from aldehydes (AdSS-2) and quaternary aromatic amines (AdSS-3). AdSS-1 (Fig. 1C), an N-sulfonylurea, may be useful as a tool compound; however, publicly available information cites a cytostatic mechanism of action in human cells (in PubChem: 120394059 120394064). AdSS-2 (Fig. 1C), a benzyaldehydederived imine, is relatively specific for malaria inhibition; this compound may also be useful as a tool compound for AdSS.
GK was selected as a target based on its involvement in nucleic acid synthesis. GK converts (d)GMP to (d)GDP, which in turn is used to make (d)GTP, which is essential for nucleic acid synthesis. The likely essentiality of GK is, however, in doubt, as the Plasmodium thymidylate kinase (2.7.4.9) can also convert dGMP to dGDP [43, 44]. An additional enzyme, ribonucleoside-diphosphate reductase (1.17.4.1), can convert dGDP to GDP. Thus, even if GK is blocked, (d)GDP might still be made from dGMP. Nevertheless, the expression of GK in the liver stage of the Plasmodium life cycle [45] suggests its possible importance in this stage and thus its possible usefulness as a target complementing those most prominent in the erythrocyte stages.
The Km of our P. vivax GK for GMP was 3.5 μM, lower than a value of 22 μM reported for the P. falciparum GK [28]. The GK screen resulted in 17 hits (0.3% hit rate) as shown in Table 2. The positive control iodoacetamide had an IC50 value of 36.5 μM under the assay conditions used. The range of guanylate kinase IC50 values across the 17 hits is 0.5 μM– 7.8 μM. Seven compounds were available as powders; four of these reconfirmed with 3 showing specificity for PvGK (Supplementary Table S1) and are shown in Figure 1D. GK-1 has two nitro groups and as such does not represent a chemically attractive scaffold unless analogs without these nitro groups remain active. Nevertheless, this compound is relatively specific and does not appear to be a promiscuous inhibitor in historical HTS run at GNF. Interestingly, most of the screens with overlapping activity were pathogen-based screens which may be related to the presence of the nitro groups. GK-2 and GK-3 (Figure 1D) are potentially useful as tool compounds to interrogate malaria parasite biology and to confirm PvGK is indeed the target of these compounds. However, GK-2 is an ammonium salt with low likelihood for oral bioavailability, and GK-3 is an imine of a napthoquinone which may have stability issues and trap electrophiles; thus, neither is an ideal starting point for medical chemistry efforts.
OMPDC is a pyrimidine biosynthesis enzyme downstream of the well-validated target dihydroorotate dehydrogenase [46]. The approximate Km for orotidine 5′-monophosphate (OMP) was ~0.2 μM, much lower than a previously reported value of 13.4 μM (PMID 15683248). The OMPDC screen resulted in 26 hits for an approximate hit rate of 0.5% (Table 2). IC50 values ranged from 0.22 μM to 10 μM while P. falciparum EC50 values ranged from 0.176 μM to 1.7 μM. 11 powders were available for reconfirmation and of these four reconfirmed, although two were identified to be fluorescence quenchers and therefore false positives. Unfortunately, the most attractive hit from a chemical standpoint, OMPDC-1 (Fig. 1E), is only a weak inhibitor of OMPDC (IC50 19.7 μM), although more potent in the cellular P. falciparum assay. OMPDC-1 may be useful as a tool compound for investigation of the effects on the parasite of OMPDC inhibition.
dUTPase protects DNA integrity by preventing the buildup of dUTP through conversion to dUMP, which is then converted to dTTP and used in DNA synthesis [47]. The Km of dUTPase for dUTP was measured as 4.5 μM, somewhat higher than the literature value of 1.9 μM [48]. The positive control used for dUTPase inhibition was 2′-deoxy-5′-O-(triphenylmethyl)uridine, which had an IC50 value of 2.4 μM under our assay conditions. The dUTPase screen resulted in 95 hits (3.1% hit rate) and the positive control inhibitor performed as expected. 91 compounds that were available as powders were retested in dose-response for reconfirmation of screen IC50 values. In parallel, hits were counterscreened for the potential to inhibit components of the PPi detection kit made by Lonza. The counterscreen tested putative hits for their ability to inhibit direct detection of ~5 μM PPi, using the same detection reagents as the primary dUTPase. The positive control showed the expected inhibition profile in the dUTPase assay without any inhibition in the counterscreen. However, all hits were also inhibitors in the counterscreen. In conclusion, we did not identify any hits as inhibitors of dUTPase.
3.5. Enzymes involved in pathways for adding lipid groups to proteins (NMT and FPPS)
NMT transfers myristate (saturated C14 fatty acid) groups to N-terminal glycine residues of substrate proteins, increasing their hydrophobicity and promoting associations with membranes and other proteins [49]. NMT had a Km of 7 μM for myristoyl-CoA and 21 μM for the synthetic peptide GSSYSRKNK.
The positive control inhibitor for NMT inhibition, aurintricarboxylate, had an IC50 of ~3 μM. The NMT screen resulted in 7 hits and an overall hit rate of 0.2%. The positive control inhibitor, aurintricarboxylate, performed as expected. 5 compounds were available and reconfirmed for inihibition. NMT-1 (Fig. 1F) did not score in past screens performed at GNF, suggesting a rather specific activity on NMT. While there is a single nitro group, with concomitant risk for mutagenic potential [50], this compound may be a useful tool compound to validate NMT as a viable drug target. None of the other structures are chemically attractive for drug discovery, as they contain too many structural alerts, such as nitro groups in NMT-1 and NMT-3 and ammonium salts in NMT-2, 4, and 5 (see Supplementary Table S1), but they can possibly be used as tool compounds for NMT inhibition.
FPPS is involved in the post-translational modification of proteins through protein farnesylation. FPPS produces farnesyl diphosphate (a.k.a. farnesyl pyrophosphate or FPP), which is then used by protein farnesyltransferase (PFT), a validated target in Plasmodium [51], in the attachment of farnesyl groups to proteins. Km’s of FPPS were 8 μM for geranyl diphosphate and 9 μM for isopentenyl diphosphate. The positive control inhibitor (1-hydroxy-2-phenylethylidene)bis-phosphonic acid has an IC50 of 7.7 μM against FPPS and an EC50 value of ~7.05 μM against P. falciparum in vitro. The FPPS screen resulted in 147 hits. Unfortunately, with the exception of the positive control, (1-hydroxy-2-phenylethylidene)bis-phosphonic acid, all hits that reconfirmed in the FPPS assay also were hits in the counterscreen (conducted as described above for dUTPase). In conclusion, no compounds hit with specificity against FPPS with the exception of the positive control inhibitor.
3.6. Glutamate dehydrogenase
GDH provides NADPH for biosynthesis and for glutathione reductase, an oxidative stress-fighting enzyme thought to be essential [25]. In contrast to the mammalian GDH enzyme, PfGDH is highly specific for NADP+ and NADPH, suggesting exploitable differences between the mammalian and parasite enzymes. Km’s were estimated to be 16 μM for NADP+ and 0.8 mM for glutamate, consistent with the literature values of 20 μM and 1 mM, respectively [25]. The GDH screen resulted in 164 hits, or a 2.9% hit rate; however, activity of most hits against GDH was weak. 78 powders were available and tested for reconfirmation but no hits reconfirmed under the assay conditions used. To decrease the assay stringency and increase sensitivity to inhibitors, the concentration of the substrate [NADP+] was reduced from 400 μM to 50 μM, but still no hits reconfirmed. In conclusion, no hits were identified against GDH.
4. Discussion
The availability of large compound collections and automated robotic systems for ultra high-throughput screening is now commonplace for many pharmaceutical companies, but less frequent in academia. Here we describe a collection of antimalarial chemical inhibitors identified by screening non-proprietary compound collections. In this first application, we attempted to link some of our antimalarial compounds to their possible protein targets. The cell-active compounds were tested in high-throughput assays against enzymes believed to have potential as drug targets based on considerations of essentiality, druggability, selectivity, and amenability to high-throughput screening (section 2.2 and Table 1). These enzymes included several involved in nucleotide metabolism (AdSS, dUTPase, GK, and OMPDC) and attachment of lipid groups to proteins (NMT and FPPS) as well as components of other key metabolic pathways (CK, GDH, and SAHH). While we were unable to detect inhibitors of dUTPase, FPPS and GDH, we identified inhibitors of AdSS, GK, OMPDC, NMT, CK, and SAHH as potential starting points for medicinal chemistry or as tool compounds for these Plasmodium enzymes. In our view, these results neither validate nor disprove the wisdom of our target selection strategy. We were disappointed not to find more potent inhibitors of the enzymes chosen; however, a screen of a larger chemical library might have retrieved compounds suitable for development into leads with good activity against both the enzymes and parasite cells.
IC50 values of preliminary hits against enzymes were checked with repurchased compounds (Table 3 and Supplementary Table S1). Reversible inhibitors generally act on their intracellular targets with an IC50 lower than the EC50 against the parasite. However, the compound concentrations of most of our target-based hits required to inhibit enzyme activity are higher than the concentrations needed to inhibit P. falciparum growth. There are a number of possible explanations for this cellular and enzymatic discrepancy. First, some of the targets inhibited by our antimalarial compounds may not be the only, or even the primary, target of the compound; inhibition of more than one protein (and/or non-protein targets) might contribute to cellular activity [52]. Second, it is possible that the mechanism by which a compound is affecting parasitic activity is completely unrelated to the biochemical activity reported here. Third, EC50 values would be lower relative to biochemical IC50 values if the test compound accumulates to higher concentrations inside the parasite. Lastly, comparing enzyme activity with antimalarial activity may not be particularly helpful, given unavoidable differences between intracellular and in vitro conditions (e.g. substrate concentrations), unless one is comparing structure activity relationships (SAR) among a set of chemically related analogs and relative enzyme and antimalarial inhibition. Derivation of the pharmacophore and correlation between enzyme-based and parasite-based inhibition is needed to identify meaningful SAR correlations and link target hits to the observed cell-based phenotype. Our highest priority will be to follow up on enzyme inhibitors with IC50’s < EC50’s, but investigations of other associations of high-value compounds and targets may also prove worthwhile to chemically validate targets.
Three targets (dUTPase, GDH and FPPS) lacked any hits among the compounds tested. It is possible that these enzymes are not essential to the parasite under our assay conditions. Alternatively, the broader library from which the antimalarial compounds were selected might have been lacking in chemical composition capable of inhibiting these enzymes. Regarding the latter possibility, these enzymes are predicted to have moderate to high druggability according to in silico methods (see Table 1). Our approach thus offered a test of these in silico predictions and suggested that they might benefit from further refinement.
To test whether inhibition of a particular biochemical target is causal for antimalarial activity, several approaches are possible. These include: (a) allowing cultures to evolve resistance to the compound and determining whether mutations occur in the gene for the presumed target [53]; (b) studying compounds chemically related to the original hit and looking for a correlation between IC50’s against the presumed target and EC50’s against the parasite [54]; (c) determining whether overexpression of the putative target overcomes the inhibition observed in the presence of the compound [55] (although genetic manipulation of Plasmodium can be challenging); (d) detecting a buildup of the presumed target’s substrate or a depletion of its product in extracts of compound-treated cells [56–58], and/or determining whether parasite growth can be restored via supplementation of the media with the missing product [59]; and (e) using compounds as affinity probes to isolate target proteins from cell lysates [60].
Despite the extensive screening work reported here, the mechanisms of action of our 5,655 antimalarial compounds generally remain elusive. However, modifications of the present approach might lead to greater success in identifying mechanisms of action. For example, studying compounds with EC50’s >1.25 μM might enable discovery of those with weak activity against the parasite but with EC50/IC50 data that strongly point to a particular target. Furthermore, considering that roughly 2 to 5% of an organism’s genes may have legitimate potential as drug targets [61], the number of proteins tested here was relatively small due to the labor-intensive requirement of optimizing a separate biochemical assay for each protein. A generic method for assessing compound-protein binding, such as thermal melting [32] or Surface Plasmon Resonance [62], might allow more proteins to be studied simultaneously, increasing the odds that some could be identified as the likely targets of antiparasitic compounds. Finally, it is possible that similar screening efforts might instead focus on finding new scaffolds for previously validated targets.
In conclusion, these data do not support our original proposition that target-based drug discovery can be accelerated by starting with cell-active compounds. While it is too early to say whether any of the enzyme-compound associations identified in these screens represent the compound’s true mechanism of action in inhibiting parasite growth, these compounds nevertheless have utility in exploring the biology surrounding these possible targets. Such tool compounds can be employed in basic studies of enzyme function and pathway biology as well as starting points for structure-activity relationship (SAR) and selectivity studies. Screens of the kind performed here can thus yield valuable information even when they do not result in identification of biochemically potent lead compounds for drug development.
Supplementary Material
Acknowledgments
These efforts were made possible through a combination of pharmaceutical resources (Novartis), funding by Medicines for Malaria Venture (MMV) and partnership with academia (the University of Washington). We thank Thierry Diagana of the Novartis Institute for Tropical Diseases (NITD) in Singapore for help in planning the project and critical review of the manuscript, and Christophe Bodenreider of NITD for sharing unpublished data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ekland EH, Fidock DA. In vitro evaluations of antimalarial drugs and their relevance to clinical outcomes. Int J Parasitol. 2008;38:743–7. doi: 10.1016/j.ijpara.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nkrumah LJ, Riegelhaupt PM, Moura P, et al. Probing the multifactorial basis of Plasmodium falciparum quinine resistance: evidence for a strain-specific contribution of the sodium-proton exchanger PfNHE. Mol Biochem Parasitol. 2009;165:122–31. doi: 10.1016/j.molbiopara.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–26. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 5.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–35. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–3. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eastman RT, Fidock DA. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat Rev Microbiol. 2009;7:864–74. doi: 10.1038/nrmicro2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim P, Alker AP, Khim N, et al. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White NJ. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–4. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 11.Odom AR, Van Voorhis WC. Functional genetic analysis of the Plasmodium falciparum deoxyxylulose 5-phosphate reductoisomerase gene. Mol Biochem Parasitol. 2010;170:108–11. doi: 10.1016/j.molbiopara.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slavic K, Straschil U, Reininger L, et al. Life cycle studies of the hexose transporter of Plasmodium species and genetic validation of their essentiality. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan AM, O’Neill MT, Tarun AS, et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–20. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki N, Hirai M, Maeda K, et al. The Plasmodium HU homolog, which binds the plastid DNA sequence-independent manner, is essential for the parasite’s survival. FEBS Lett. 2009;583:1446–50. doi: 10.1016/j.febslet.2009.03.071. [DOI] [PubMed] [Google Scholar]
- 15.Gunther S, Matuschewski K, Muller S. Knockout studies reveal an important role of Plasmodium lipoic acid protein ligase A1 for asexual blood stage parasite survival. PLoS One. 2009;4:e5510. doi: 10.1371/journal.pone.0005510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sijwali PS, Koo J, Singh N, Rosenthal PJ. Gene disruptions demonstrate independent roles for the four falcipain cysteine proteases of Plasmodium falciparum. Mol Biochem Parasitol. 2006;150:96–106. doi: 10.1016/j.molbiopara.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 17.El Bissati K, Zufferey R, Witola WH, Carter NS, Ullman B, Ben Mamoun C. The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc Natl Acad Sci U S A. 2006;103:9286–91. doi: 10.1073/pnas.0602590103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omara-Opyene AL, Moura PA, Sulsona CR, et al. Genetic disruption of the Plasmodium falciparum digestive vacuole plasmepsins demonstrates their functional redundancy. J Biol Chem. 2004;279:54088–96. doi: 10.1074/jbc.M409605200. [DOI] [PubMed] [Google Scholar]
- 19.Krnajski Z, Gilberger TW, Walter RD, Cowman AF, Muller S. Thioredoxin reductase is essential for the survival of Plasmodium falciparum erythrocytic stages. J Biol Chem. 2002;277:25970–5. doi: 10.1074/jbc.M203539200. [DOI] [PubMed] [Google Scholar]
- 20.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 21.Plouffe D, Brinker A, McNamara C, et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci U S A. 2008;105:9059–64. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguero F, Al-Lazikani B, Aslett M, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov. 2008;7:900–7. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menz RI, Cinquin O, Christopherson RI. The identification, cloning and functional expression of the gene encoding orotidine 5′-monophosphate (OMP) decarboxylase from Plasmodium falciparum. Ann Trop Med Parasitol. 2002;96:469–76. doi: 10.1179/000349802125001230. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi M, Iwata A, Yatome C, Kitade Y. Purification and properties of recombinant Plasmodium falciparum S-adenosyl-L-homocysteine hydrolase. J Biochem. 2001;129:101–5. doi: 10.1093/oxfordjournals.jbchem.a002819. [DOI] [PubMed] [Google Scholar]
- 25.Wagner JT, Ludemann H, Farber PM, Lottspeich F, Krauth-Siegel RL. Glutamate dehydrogenase, the marker protein of Plasmodium falciparum--cloning, expression and characterization of the malarial enzyme. Eur J Biochem. 1998;258:813–9. doi: 10.1046/j.1432-1327.1998.2580813.x. [DOI] [PubMed] [Google Scholar]
- 26.Choubey V, Guha M, Maity P, et al. Molecular characterization and localization of Plasmodium falciparum choline kinase. Biochim Biophys Acta. 2006;1760:1027–38. doi: 10.1016/j.bbagen.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Gunaratne RS, Sajid M, Ling IT, Tripathi R, Pachebat JA, Holder AA. Characterization of N-myristoyltransferase from Plasmodium falciparum. Biochem J. 2000;348(Pt 2):459–63. [PMC free article] [PubMed] [Google Scholar]
- 28.Kandeel M, Nakanishi M, Ando T, et al. Molecular cloning, expression, characterization and mutation of Plasmodium falciparum guanylate kinase. Mol Biochem Parasitol. 2008;159:130–3. doi: 10.1016/j.molbiopara.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Jayalakshmi R, Sumathy K, Balaram H. Purification and characterization of recombinant Plasmodium falciparum adenylosuccinate synthetase expressed in Escherichia coli. Protein Expr Purif. 2002;25:65–72. doi: 10.1006/prep.2001.1610. [DOI] [PubMed] [Google Scholar]
- 30.Whittingham JL, Leal I, Nguyen C, et al. dUTPase as a platform for antimalarial drug design: structural basis for the selectivity of a class of nucleoside inhibitors. Structure. 2005;13:329–38. doi: 10.1016/j.str.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Phillips MA, Coffino P, Wang CC. Cloning and sequencing of the ornithine decarboxylase gene from Trypanosoma brucei. Implications for enzyme turnover and selective difluoromethylornithine inhibition. J Biol Chem. 1987;262:8721–7. [PubMed] [Google Scholar]
- 32.Crowther GJ, Napuli AJ, Thomas AP, et al. Buffer optimization of thermal melt assays of Plasmodium proteins for detection of small-molecule ligands. Journal of Biomolecular Screening. 2009;14:700–707. doi: 10.1177/1087057109335749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crowther GJ, He P, Rodenbough PP, et al. Use of thermal melt curves to assess the quality of enzyme preparations. Anal Biochem. 2010;399:268–75. doi: 10.1016/j.ab.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahlfs S, Koncarevic S, Iozef R, et al. Myristoylated adenylate kinase-2 of Plasmodium falciparum forms a heterodimer with myristoyltransferase. Mol Biochem Parasitol. 2009;163:77–84. doi: 10.1016/j.molbiopara.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Wright SK, Viola RE. Evaluation of methods for the quantitation of cysteines in proteins. Anal Biochem. 1998;265:8–14. doi: 10.1006/abio.1998.2858. [DOI] [PubMed] [Google Scholar]
- 36.Liou JY, Dutschman GE, Lam W, Jiang Z, Cheng YC. Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue monophosphates. Cancer Res. 2002;62:1624–31. [PubMed] [Google Scholar]
- 37.Choubey V, Maity P, Guha M, et al. Inhibition of Plasmodium falciparum choline kinase by hexadecyltrimethylammonium bromide: a possible antimalarial mechanism. Antimicrob Agents Chemother. 2007;51:696–706. doi: 10.1128/AAC.00919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ancelin ML, Vial HJ. Quaternary ammonium compounds efficiently inhibit Plasmodium falciparum growth in vitro by impairment of choline transport. Antimicrob Agents Chemother. 1986;29:814–20. doi: 10.1128/aac.29.5.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka N, Nakanishi M, Kusakabe Y, et al. Crystal structure of S-adenosyl-L-homocysteine hydrolase from the human malaria parasite Plasmodium falciparum. J Mol Biol. 2004;343:1007–17. doi: 10.1016/j.jmb.2004.08.104. [DOI] [PubMed] [Google Scholar]
- 40.Greenslade FC, Scott CK, Newquist KL, Krider KM, Chasin M. Heterogeneity of biochemical actions among vasodilators. J Pharm Sci. 1982;71:94–100. doi: 10.1002/jps.2600710123. [DOI] [PubMed] [Google Scholar]
- 41.Mett H, Gyr K, Zak O, Vosbeck K. Duodeno-pancreatic secretions enhance bactericidal activity of antimicrobial drugs. Antimicrob Agents Chemother. 1984;26:35–8. doi: 10.1128/aac.26.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raman J, Mehrotra S, Anand RP, Balaram H. Unique kinetic mechanism of Plasmodium falciparum adenylosuccinate synthetase. Mol Biochem Parasitol. 2004;138:1–8. doi: 10.1016/j.molbiopara.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 43.Kandeel M, Kitade Y. Molecular characterization, heterologous expression and kinetic analysis of recombinant Plasmodium falciparum thymidylate kinase. J Biochem. 2008;144:245–50. doi: 10.1093/jb/mvn062. [DOI] [PubMed] [Google Scholar]
- 44.Kandeel M, Kitamura Y, Kitade Y. The exceptional properties of Plasmodium deoxyguanylate pathways as a potential area for metabolic and drug discovery studies. Nucleic Acids Symp Ser (Oxf) 2009:39–40. doi: 10.1093/nass/nrp020. [DOI] [PubMed] [Google Scholar]
- 45.Tarun AS, Peng X, Dumpit RF, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci U S A. 2008;105:305–10. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gujjar R, Marwaha A, El Mazouni F, et al. Identification of a metabolically stable triazolopyrimidine-based dihydroorotate dehydrogenase inhibitor with antimalarial activity in mice. J Med Chem. 2009;52:1864–72. doi: 10.1021/jm801343r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen C, Kasinathan G, Leal-Cortijo I, et al. Deoxyuridine triphosphate nucleotidohydrolase as a potential antiparasitic drug target. J Med Chem. 2005;48:5942–54. doi: 10.1021/jm050111e. [DOI] [PubMed] [Google Scholar]
- 48.Quesada-Soriano I, Leal I, Casas-Solvas JM, et al. Kinetic and thermodynamic characterization of dUTP hydrolysis by Plasmodium falciparum dUTPase. Biochim Biophys Acta. 2008;1784:1347–55. doi: 10.1016/j.bbapap.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Gelb MH, Van Voorhis WC, Buckner FS, et al. Protein farnesyl and N-myristoyl transferases: piggyback medicinal chemistry targets for the development of antitrypanosomatid and antimalarial therapeutics. Mol Biochem Parasitol. 2003;126:155–63. doi: 10.1016/s0166-6851(02)00282-7. [DOI] [PubMed] [Google Scholar]
- 50.Orsiere T, De Meo M, Rathelot P, et al. Implication of nitro group reduction in the mutagenic and chromosome damaging activities of 22 new 5-nitroisoquinolines by the Salmonella mutagenicity test and the cytokinesis-blocked micronucleus assay. Food Chem Toxicol. 2003;41:275–90. doi: 10.1016/s0278-6915(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 51.Fletcher S, Cummings CG, Rivas K, et al. Potent, Plasmodium-selective farnesyltransferase inhibitors that arrest the growth of malaria parasites: structure-activity relationships of ethylenediamine-analogue scaffolds and homology model validation. J Med Chem. 2008;51:5176–97. doi: 10.1021/jm800113p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–90. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 53.Dharia NV, Sidhu AB, Cassera MB, et al. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol. 2009;10:R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukkamala D, No JH, Cass LM, Chang TK, Oldfield E. Bisphosphonate inhibition of a Plasmodium farnesyl diphosphate synthase and a general method for predicting cell-based activity from enzyme data. J Med Chem. 2008;51:7827–33. doi: 10.1021/jm8009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luesch H, Wu TY, Ren P, Gray NS, Schultz PG, Supek F. A genome-wide overexpression screen in yeast for small-molecule target identification. Chem Biol. 2005;12:55–63. doi: 10.1016/j.chembiol.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 56.Hankins EG, Gillespie JR, Aikenhead K, Buckner FS. Upregulation of sterol C14-demethylase expression in Trypanosoma cruzi treated with sterol biosynthesis inhibitors. Mol Biochem Parasitol. 2005;144:68–75. doi: 10.1016/j.molbiopara.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Seymour KK, Lyons SD, Phillips L, Rieckmann KH, Christopherson RI. Cytotoxic effects of inhibitors of de novo pyrimidine biosynthesis upon Plasmodium falciparum. Biochemistry. 1994;33:5268–74. doi: 10.1021/bi00183a033. [DOI] [PubMed] [Google Scholar]
- 58.Vial HJ, Thuet MJ, Ancelin ML, Philippot JR, Chavis C. Phospholipid metabolism as a new target for malaria chemotherapy. Mechanism of action of D-2-amino-1-butanol. Biochem Pharmacol. 1984;33:2761–70. doi: 10.1016/0006-2952(84)90693-2. [DOI] [PubMed] [Google Scholar]
- 59.Roberts F, Roberts CW, Johnson JJ, et al. Evidence for the shikimate pathway in apicomplexan parasites. Nature. 1998;393:801–5. doi: 10.1038/31723. [DOI] [PubMed] [Google Scholar]
- 60.Kato N, Sakata T, Breton G, et al. Gene expression signatures and small-molecule compounds link a protein kinase to Plasmodium falciparum motility. Nat Chem Biol. 2008;4:347–56. doi: 10.1038/nchembio.87. [DOI] [PubMed] [Google Scholar]
- 61.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 62.Piliarik M, Vaisocherova H, Homola J. Surface plasmon resonance biosensing. Methods Mol Biol. 2009;503:65–88. doi: 10.1007/978-1-60327-567-5_5. [DOI] [PubMed] [Google Scholar]
- 63.Hyde JE. Targeting purine and pyrimidine metabolism in human apicomplexan parasites. Curr Drug Targets. 2007;8:31–47. doi: 10.2174/138945007779315524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vial HJ, Wein S, Farenc C, et al. Prodrugs of bisthiazolium salts are orally potent antimalarials. Proc Natl Acad Sci U S A. 2004;101:15458–63. doi: 10.1073/pnas.0404037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jarzyna R, Lenarcik E, Bryla J. Chloroquine is a potent inhibitor of glutamate dehydrogenase in liver and kidney-cortex of rabbit. Pharmacol Res. 1997;35:79–84. doi: 10.1006/phrs.1996.0108. [DOI] [PubMed] [Google Scholar]
- 66.Werner C, Stubbs MT, Krauth-Siegel RL, Klebe G. The crystal structure of Plasmodium falciparum glutamate dehydrogenase, a putative target for novel antimalarial drugs. J Mol Biol. 2005;349:597–607. doi: 10.1016/j.jmb.2005.03.077. [DOI] [PubMed] [Google Scholar]
- 67.Martin MB, Grimley JS, Lewis JC, et al. Bisphosphonates inhibit the growth of Trypanosoma brucei, Trypanosoma cruzi, Leishmania donovani, Toxoplasma gondii, and Plasmodium falciparum: a potential route to chemotherapy. J Med Chem. 2001;44:909–16. doi: 10.1021/jm0002578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.