Abstract
The pericentromeric regions of human chromosomes pose particular problems for both mapping and sequencing. These difficulties are due, in large part, to the presence of duplicated genomic segments that are distributed among multiple human chromosomes. To ensure contiguity of genomic sequence in these regions, we designed a sequence-based strategy to characterize different pericentromeric regions using a single (162 kb) 2p11 seed sequence as a point of reference. Molecular and cytogenetic techniques were first used to construct a paralogy map that delineated the interchromosomal distribution of duplicated segments throughout the human genome. Monochromosomal hybrid DNAs were PCR amplified by primer pairs designed to the 2p11 reference sequence. The PCR products were directly sequenced and used to develop a catalog of sequence tags for each duplicon for each chromosome. A total of 685 paralogous sequence variants were generated by sequencing 34.7 kb of paralogous pericentromeric sequence. Using PCR products as hybridization probes, we were able to identify 702 human BAC clones, of which a subset, 107 clones, were analyzed at the sequence level. We used diagnostic paralogous sequence variants to assign 65 of these BACs to at least 9 chromosomal pericentromeric regions: 1q12, 2p11, 9p11/q12, 10p11, 14q11, 15q11, 16p11, 17p11, and 22q11. Comparisons with existing sequence and physical maps for the human genome suggest that many of these BACs map to regions of the genome with sequence gaps. Our analysis indicates that large portions of pericentromeric DNA are virtually devoid of unique sequences. Instead, they consist of a mosaic of different genomic segments that have had different propensities for duplication. These biologic properties may be exploited for the rapid characterization of, not only pericentromeric DNA, but also other complex paralogous regions of the human genome.
[The sequence data described in this paper have been submitted to the GenBank data library under accession numbers AC002038, AC002307, AF182004-AF182009, AF183323-AF183331, AF183333-AF183337, AF183339-AF183350, AF183352-AF183356, AF183358-AF183362, AF183366-AF183369, AF183371-AF183375, and AF262624–AF262695.]
The human genome contains several different classes of repetitive elements that are categorized based largely on their copy number and their mode of propagation (Gardiner 1996; Vogt 1990). Two broad classes of repeats are generally recognized: interspersed and tandem repeat elements (Brown 1999). Tandemly repeated DNA, such as centromeric α-satellite and microsatellite DNA, is believed to expand and contract by mechanisms involving unequal crossing-over or replication slippage. In contrast, interspersed repetitive elements such as LINEs and SINEs, which comprise more than one-third of the total genome (Smit and Riggs 1996), are propagated via mechanisms of retrotransposition. Both classes of repeats are easily recognized as repetitive because of both their high copy number and their defined sequence characteristics. As more of the human genome is sequenced, it is becoming apparent that yet another class of repetitive DNA exists. Low copy repeat sequences are being discovered as many unique regions of the genome are found to have duplicate counterparts. Portions of some genes and even entire gene segments have been duplicated and exist at multiple, discrete locations within the genome (Eichler et al. 1996, 1997; van Deutekom et al. 1996; Regnier et al. 1997; Zimonjic et al. 1997; Trask et al. 1998; Horvath et al. 2000).
Mapping and sequencing of the human genome indicates that a large number of these duplicated segments lie within pericentromeric and subtelomeric regions (Eichler 1998). These duplicated sequences, or paralogs, are nonprocessed. This suggests an underlying DNA transposition mechanism for their duplication and dispersal. Partial or complete paralogous genomic segments have been identified for several gene loci including ALD, SLC6A8, NF1, HERC2, KGF, FRG1, olfactory receptor, immunoglobulin variable κ-chain and immunoglobulin variable heavy chain segments. A large number of pericentromeric and subtelomeric regions (1p12, 1q12, 2p11, 3p13, 3qter, 15qter, 19pter, 9p11, 10p11, 13q11, 14q11, 15q11, 16p11, 17p11, 18p11, 18q11, 20p, 20q, 21p11, 21q11, 22p11, and 22q11) have been shown to be sites of these recent duplications (Zachau 1993; Arnold et al. 1996; Eichler et al. 1996, 1997; van Deutekom et al. 1996; Regnier et al. 1997; Reiter et al. 1997; Zimonjic et al. 1997; Potier et al. 1998; Ritchie et al. 1998; Trask et al. 1998; Amos-Landgraf et al. 1999; Brand-Arpon et al. 1999; Christian et al. 1999). These data suggest that the process of pericentromeric/subtelomeric duplication may be a general property of the human genome. The reason for this apparent location bias is unclear, although we have suggested that GC-rich repeat elements may play a role in the accumulation of duplicated segments within pericentromeric regions (Eichler et al. 1999).
Paralogous regions can be very large (>150 kb) and can exhibit a high degree of sequence similarity (>98%) (Horvath et al. 2000; Orti et al. 1998). These properties make such segments particularly problematic for both mapping and sequencing of the human genome (Eichler 1998). Where the degree of similarity approaches levels observed for allelic variation, it is anticipated that it will be difficult to disentangle assembled sequence contigs constructed using whole shotgun sequencing approaches (Eichler 1998; Green 1997). Even high-throughput sequencing centers using traditional STS mapping and sequencing strategies have encountered difficulties building physical maps across such duplicated regions. Since large insert clones are linked into contiguous sets of overlapping clones based on short STS PCR products, the absence of unique STSs and/or the presence of a large number of clones from paralogous loci within a single contig can create significant ambiguities. This translates into either gaps in the sequence/physical map (DeSilva et al. 1999; Dunham et al. 1999) or the construction of physical maps in which BAC clones have been misassigned due to the presence of highly similar paralogous blocks (Cao et al. 1999). Indeed, in the recently reported DNA sequence of human chromosome 22 (Dunham et al. 1999), many of the gaps and map inconsistencies are biased toward the pericentromeric region at sites of large inter- and intrachromosomal repeats. The importance of such repeat elements in mediating recurrent chromosomal structural rearrangements (Ji et al. 1999; Mazzarella and Schlessinger 1998) and their overall abundance (∼10% based on chromosome 22 data) necessitate the development of specialized mapping and sequencing strategies to provide a comprehensive view of human genome organization.
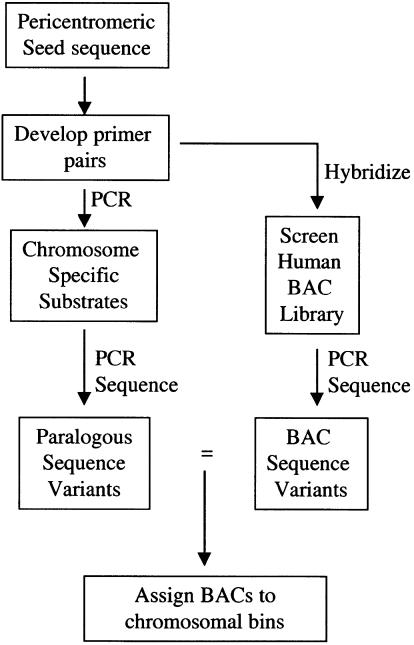
To circumvent some of the problems of paralogy, we developed a sequence-based strategy that exploits the paralogous nature of these complex regions. Our strategy is outlined in Figure 1. A completely sequenced clone from the pericentromeric region of 2p11 was chosen as a reference seed sequence. FISH analysis and database sequence similarity searches were used to provide a preliminary overview of the duplicative nature of this sequence. Next, a series of STS primer pairs were developed from the reference sequence and were used to screen a monochromosomal hybrid DNA panel by PCR. The STS product from each hybrid was directly sequenced, effectively generating a catalog of paralogous sequence variants that could be used to distinguish each chromosomal copy. The paralogous sequence variant is analogous to the single nucleotide polymorphism with the exception that sequence variation accumulates after a duplication event as opposed to descent from a common founding allele. Finally, the same STSs were used as probes to screen a human genomic BAC library. The BACs that positively hybridized were PCR amplified with the STS primer pairs and the products directly sequenced. Comparisons between the BAC sequence variants and monochromosomal paralogous sequence variants allowed us to unambiguously assign highly paralogous BAC clones to different chromosomal bins. This transchromosomal approach not only allows the rapid identification and characterization of other pericentromeric DNA but also provides insight into the unique structure and biology of these complex regions of the genome.
Figure 1.
Flowchart of pericentromeric characterization strategy.
RESULTS
Characterization of a Pericentromeric Reference Sequence
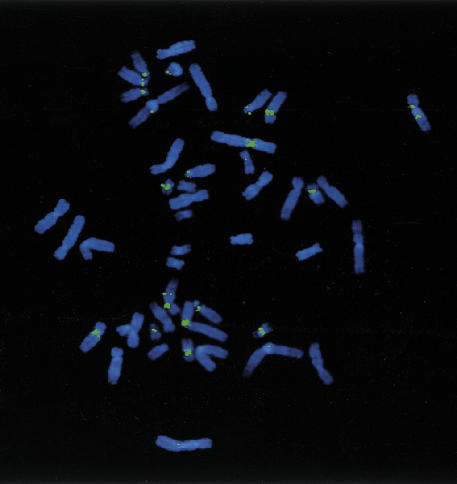
A completely sequenced BAC clone CIT978SK-A-101B6 (GenBank accession no. AC002038) was first assigned to 16p11 by STS D16S2816 (Cao et al. 1999). FISH analysis using 101B6 as a probe indicated multiple pericentromeric signals observed on 1p12, 2p11/q11, 4q24, 7, 9p12/q12–13, 10p11, 15q11/q13, 16p11/q11, 22q11, and Y (Fig. 2). More extensive analysis of 101B6 in relation to the centromere of chromosome 2 was performed using two-color FISH with101B6 and higher order α-satellite DNA from chromosome 2 (data not shown). This analysis placed 101B6 within 1–3 Mb (the limit of resolution of FISH) of the centromere on 2p11. Sequence analysis of the clone revealed the presence of a previously characterized 9.7 kb ALD segment which had been duplicated (∼5–10 million years ago) from Xq28 to the pericentromeric regions of 2p11, 10p11, 16p11, and 22q11. Sequence variants within this segment were identified that were specific to chromosome 2p11. Additional STS analysis confirmed a 2p11 rather than 16p11 origin of the sequence. Furthermore, the presence of FISH signals within the pericentromeric regions of chromosomes 1, 7, 9, 13, 14, 15, and 21 (Fig. 2) that had not been observed during the characterization of the ALD duplication (Eichler et al. 1997; Horvath et al. 2000) suggested the presence of additional duplications within this sequence. Therefore, 101B6 was chosen for further analysis because it was a completely sequenced pericentromeric BAC with a complex paralogous organization that had proven difficult to map based on traditional STS techniques.
Figure 2.
FISH of 101B6. Hybridization of the entire insert of BAC clone, A-101B6, shows consistent fluorescent signals on 1q12, 2p11/q11, 9p12/q12–13, 10p11, 15q11/q13, 16p11/q11, and 22q11. Less intense signals are observed for 4q24 and the centromeric regions of chromosomes 7 and Y. Note the difference in size and intensity of signals on some chromosomes (compare 2 and 16), which may suggest copy number differences.
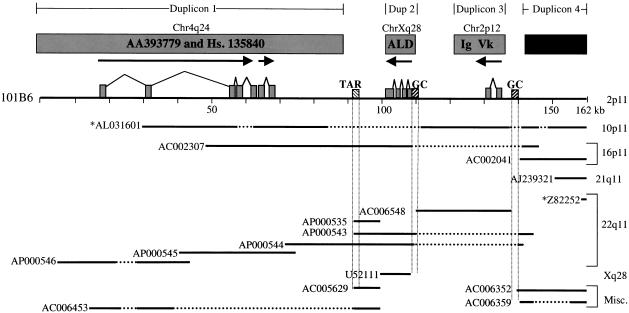
A series of database searches were initially used to characterize duplicons (blocks of duplicated sequence) within 101B6. These searches identified at least three genic duplicons (Fig. 3, Table 1). The first duplicon is approximately 85 kb in length and is composed of a duplicated segment with conserved exon–intron structure. Duplicated exonic sequences within this segment are, on average, 95.4% similar to cDNA sequences AA393779 and those identified within Unigene cluster Hs.135840 (see Methods). The expressed copies of both AA393779 and cDNAs in Hs.135840 have been mapped to 4q24 and are contained on one contiguous genomic segment (Horvath et al. 2000). The second duplicon (9.7 kb), which has been previously described (Eichler et al. 1997), contains four paralogous ALD exons, which are 94.1% similar to the expressed Xq28 adrenoleukodystrophy gene. The third duplicon contains a segment (∼15 kb) with conserved exon–intron structure paralogous to the expressed 2p12 immunoglobulin variable κ-locus that is 97.2% similar over two exons. Sequences within a third paralogous exon have similarity to other immunoglobulin sequences on chromosomes 1 and 22. A fourth segment was identified that appeared highly duplicated based on database searches, but showed no evidence of paralogous intron–exon structure. Sequence between and outside of paralogous exons is composed of a mixture of highly repetitive elements and nongenic sequences. Interestingly, this pericentromeric BAC contains one telomeric associated repeat (TAR) located at position 92 kb within the sequence. There are also two interspersed GC-rich repetitive sequences located at positions 109 kb and 139 kb within the reference sequence. Both the TAR and GC-rich repeat elements lie in close proximity to the points of transition between the genic duplicons (Fig. 3).
Figure 3.
Database sequence similarity searches. The diagram depicts the extent of overlap between the (101B6) reference sequence (top solid line) and a subset (as of 12–99) of other highly paralogous (>90%) GenBank sequences (lower solid lines). Sequences with an * before them denote clones in htgs phase of GenBank. These overlaps are placed in the context of ancestral duplications from 4q24, Xq28, and 2p12 (see text). Horizontal broken lines indicate a gap in the target sequence, whereas vertical broken lines indicate the positions of repeat sequences. The paralogous nonprocessed pseudogene fragments of the adrenoleukodystrophy, AA393779 and Unigene cluster Hs. 135840, and the immunoglobulin κ-variable chain segment are shown as filled boxes. The direction of transcription (arrows) and the exon–intron structure with respect to the ancestral (expressed) sequence are indicated. GC-rich repeat elements such as the telomeric associated repeat (TAR) and GC-rich interspersed repeats are indicated by hatched boxes.
Table 1.
cDNA Sequence Similarity Searches
| cDNA Duplicons | ||||
| target | map | bp aligned | % sim | 101 B6 position |
|---|---|---|---|---|
| AA393779 | 4q24 | 451 | 94.3 | 19.5–57.5 |
| AI740992 | 4q24 | 537 | 94.8 | 57.5–61.3 |
| AI963884 | 4q24 | 345 | 95.3 | 66.8–66.6 |
| AW135265 | 4q24 | 380 | 94.8 | 66.8–66.6 |
| AI027746 | 4q24 | 394 | 95.5 | 57.5–66.8 |
| AI797613 | 4q24 | 478 | 96 | 67.0–66.6 |
| AA581067 | 4q24 | 203 | 97 | 66.8–67.0 |
| AI654903 | 4q24 | 408 | 97 | 66.8–35.7 |
| 4557300 | Xq28 | 3616 | 94.1 | 99.7–109.4 |
| x64641 | 2p12 | 359 | 97.2 | 131.7–132.2 |
In addition to these duplicated segments, database searches revealed that 101B6 shares large blocks of sequence with multiple clones from other chromosomes (Fig. 3, Table 2). The boundaries of these shared duplicated segments do not always correspond precisely to the duplicons described above. A 63 kb region of 101B6 sequence is 96.9% similar to clone AC002307 from 16p11 and spans both duplicons 1 and 2. There is also a considerable amount of overlap with sequences Z82252, AP000535, AP000543–546, and AC006548 from chromosome 22 and clone AL031601 from chromosome 10. A schematic overview of all duplicons identified upon database searches (Fig. 3) indicates that virtually none of the sequence within clone 101B6 is unique. Instead, the sequence is composed of a variety of different duplicated segments of varying lengths distributed over at least five chromosomes. This absence of unique sequence and the high degree of sequence similarity among various clones from different chromosomes may help to explain the difficulties in mapping these regions of the genome.
Table 2.
Genomic Segment Sequence Similarity Searches
| Paralogous Genomic Sequence | |||||
| accession | map location | bp aligned | % sim w/indels | position in 101B6 | position in accession |
|---|---|---|---|---|---|
| AL031601* | 10p11 | 28279 | 97.0 ± .1% | 29030–57363 | 201797–230218 |
| " | 10p11 | 21985 | 97.3 ± .1% | 62773–84828 | 230254–252655 |
| " | 10p11 | 26042 | 97.3 ± .1% | 110154–139591 | 161707–135464 |
| " | 10p11 | 4907 | 96.1 ± .3% | 140110–145045 | 134605–129689 |
| " | 10p11 | 13180 | 96.9 ± .2% | 148036–161973 | 77319–93720 |
| AC002307 | 16p11 | 60268 | 96.9 ± .1% | 47856–109695 | 1–61389 |
| " | 16p11 | 2940 | 96.9 ± .1% | 140119–143065 | 61582–64521 |
| AC002041 | 16p11 | 21528 | 96.2 ± .1% | 140099–161973 | 123955–146849 |
| " | 16p11 | 9387 | 96.3 ± .2% | 152567–161973 | 89735–99146 |
| AJ239321 | 21q11.1 | 11394 | 95.0 ± .2% | 150277–161973 | 32583–47845 |
| Z82252* | 22 | 4714 | 95.0 ± .3% | 157235–161973 | 40814–36075 |
| AC006548 | 22q11 | 29200 | 97.5 ± .1% | 110153–139533 | 59792–89142 |
| AP000535 | 22q11 | 1805 | 93.3 ± .6% | 91389–93314 | 17826–19788 |
| " | 22q11 | 5795 | 94.9 ± .3% | 92479–99554 | 28981–23067 |
| AP000543 | 22q11 | 5430 | 96.1 ± .1% | 91830–110155 | 36955–19791 |
| " | 22q11 | 17002 | 94.5 ± .3% | 139529–145044 | 20213–14626 |
| AP000544 | 22q11 | 36530 | 97.0 ± .1% | 71773–109772 | 38993–2260 |
| " | 22q11 | 2228 | 92.2 ± .6% | 139529–141827 | 2284–1 |
| AP000545 | 22q11 | 37069 | 97.1 ± .1% | 38918–76099 | 37247–1 |
| AP000546 | 22q11 | 12863 | 97.4 ± .1% | 3299–16187 | 39984–24657 |
| " | 22q11 | 6078 | 97.0 ± .2% | 16179–22409 | 23603–17335 |
| " | 22q11 | 14312 | 97.2 ± .2% | 28789–43152 | 14351–1 |
| U52111 | Xq28 | 9680 | 95.4 ± .2% | 99671–109435 | 63289–53533 |
| AC005629 | ? | 6158 | 95.7 ± .3% | 92082–99552 | 64544–70776 |
| AC006352 | ? | 14998 | 96.5 ± .2% | 140099–155195 | 81100–64760 |
| " | ? | 9391 | 96.2 ± .2% | 152562–161973 | 114915–105500 |
| AC006359 | ? | 4074 | 93.8 ± .4% | 139629–143774 | 31664–27484 |
| " | ? | 7133 | 95.4 ± .2% | 154823–161973 | 63949–56793 |
| AC006453 | ? | 7822 | 96.5 ± .2% | 14554–22404 | 133259–142417 |
| " | ? | 9879 | 96.7 ± .2% | 28788–38702 | 145388–155314 |
| " | ? | 6152 | 95.8 ± .3% | 92082–99552 | 55134–61357 |
Denotes unordered clone in htgs phase.
All alignments as of December 1999.
Generation of a Paralogy Map
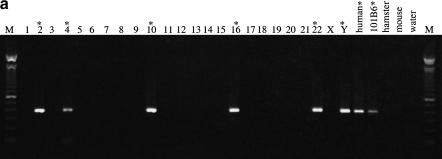
Because database searches suggested that 101B6 shares a patchwork of sequences from other chromosomes, we used PCR assays with chromosome-specific reagents to delineate specifically the genomic distribution of duplicons on different chromosomes. Twenty-four PCR primer pairs (see Methods) were developed in unique regions (as determined by RepeatMasker) of the 101B6 reference sequence at a density of approximately one pair every 8 kb. Multiple primer pairs were developed within each duplicon to eliminate failure of cross-amplification from potential sequence differences located within the primer binding site. These primer pairs amplified PCR products ranging in size from 303–1124 bp and were used to screen a monochromosomal somatic cell hybrid DNA panel to determine the interchromosomal distribution of each pair. As expected, the vast majority of primer pairs (23/24 = 96%) amplified products of nearly identical length from several different monochromosomal hybrids indicating a multicopy distribution for these particular genomic segments. These primer pairs amplified paralogous sites and are referred to as paralogous sequence tagged sites, or pSTSs. A typical PCR amplification is shown in Fig. 4a.
Figure 4.
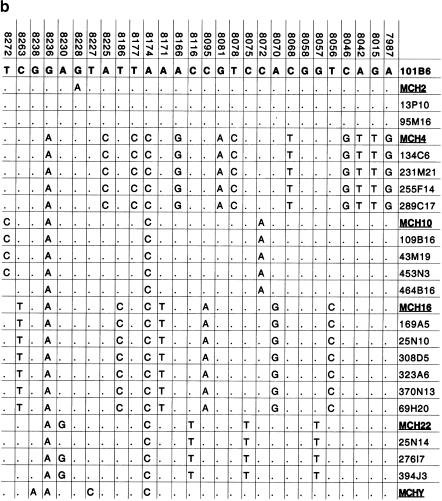
Paralogous STS and sequence variants. (a) A typical PCR amplification of a paralogous STS against a panel of monochromosomal somatic cell hybrid DNAs. pSTS1 was designed to 101B6 (chromosome 2) sequence (see Methods) yet amplified a ∼383 bp product from chromosomes 2, 4, 10, 16, 22, and Y (marked with asterisks). (b) The PCR products from pSTS 1 were bidirectionally sequenced and aligned (Consed). Basepairs in bold represent 101B6 basepairs, whereas the numbers above each bp represent its location in 101B6. Only the paralogous sequence variants (PSVs) that distinguish each chromosome are shown; a period represents the same bp as 101B6. Along the right are the sequences of the monochromosomal hybrid sequence (MCH). Below each chromosomal sequence signature, a subset of RPCI-11 BAC clones corresponding to each PSV is indicated. The numbers correspond to pSTSs developed to the 101B6 reference sequence. Similar analyses were performed for 16 other pSTS.
Seventeen pSTSs pairs were chosen for sequence analysis. PCR was performed against a panel of monochromosomal hybrid DNAs and the PCR products were directly sequenced. Sequences derived from the monochromosomal hybrids were aligned, using Consed, to identify sequence variants that were specific to each chromosome (Fig. 4b). Paralogous sequence variants (PSVs) were found that uniquely identified the different monochromosomal hybrid sequences for a given primer pair. Monochromosomal hybrid sequences for a single primer pair are very similar at the sequence level, ranging from 95.4% to 97.6% (data not shown). Consequently, on average we expected ∼17 sequence variants in 500 bp of sequence from a pSTS to distinguish two chromosomal paralogs. During the course of our monochromosomal hybrid analysis we generated a total of 35.7 kb of paralogous sequence corresponding to 5.9 kb of original 101B6 sample sequence. Comparisons of the monochromosomal hybrid sequence signatures yielded a total of 685 paralogous sequence variants distributed among 14 different human chromosomes. In some cases, sequence analysis of several pSTSs from the hybrids showed the presence of heterozygous signals. Because of the monochromosomal origin of these chromosomes, the heterozygous signals likely represent intrachromosomal duplications.
Cytogenetic analysis was used to confirm the distribution of duplicons and to identify cytogenetic band positions for each duplicated segment. A series of six nonoverlapping DNA fragments, which effectively represent different portions of the 101B6 sequence, were used as probes in FISH metaphase assays. The locations of these six probes are schematically shown within Fig. 5a. FISH using paralogous chromosome 16 cosmid 308A5 had previously been used to confirm the ancestral 4q24 locus and the pericentromeric localizations of duplicon 1 (Horvath et al. 2000). Similarly, a 9.7 kb long range PCR probe spanning exons 7–10 of the ALD gene confirmed the expressed Xq28 locus as well as the pericentromeric localizations of duplicon 2 (Eichler et al. 1997). Four additional long-range PCR probes were designed within duplicated segments 3 and 4 of 101B6 (Fig 5a). Analysis of these probes revealed localizations that could not be detected by whole BAC hybridizations (data not shown). For example, probe LR-3 hybridizes to chromosomes 13, 14, 17, 18, and 21 in addition to chromosomes 1, 2, 7, 9, 10, 15, 16, 17, and 22 as seen with the entire 101B6 insert used as probe. Because these probes are smaller and less complex in comparison to the complete BAC insert, we believe that the FISH signals obtained when using long-range PCR probes are more representative of the true distribution of these duplicons within the genome. The cytogenetic analyses indicate, with the exception of the signals of the ancestral loci within 4q24 and Xq28, that the other 12 chromosomal locations are exclusively pericentromeric.
Figure 5.
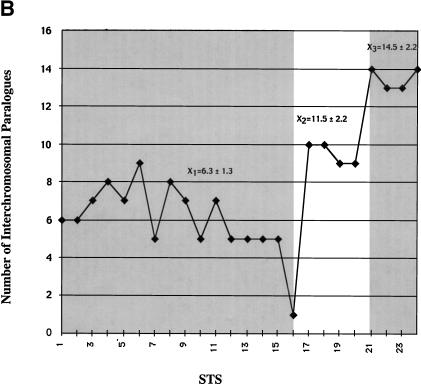
Paralogy map. (a) Summary of PCR and FISH analysis of 101B6. Each column describes the PCR results of one primer pair tested against a panel of 24 monochromosomal somatic cell hybrid DNAs. A total of 24 paralogous STS (pSTS 1–24) primer pairs were developed based on the 101B6 reference sequence. Dots along the top line indicate the approximate position of each primer pair in 101B6 (see Table 3 for the exact location of each primer). The filled gray boxes indicate chromosomal hybrids tested positive by PCR and, therefore, represent the extent of paralogy of each chromosome with respect to the 2p11 reference sequence. As expected, only chromosome 2 tested positive for all pSTS. A schematic of the duplication organization (see Fig.3) of the 2p11 sequence is provided. The positions of long-range PCR (LR-ALD, LR-1 to 4) and the cosmid (c308a5) probes used in FISH assays are indicated. FISH localizations are summarized on the right side of the figure. These confirm the interchromosomal distribution and cytogenetic position of each pSTS. (b) The number of observed interchromosomal duplications is plotted (y axis) against the position of each paralogous STS. The mean number of duplications is calculated for three groups (X1=duplicon 1 and 2, X2=duplicon 3, and X3=duplicon 4). A significant difference is observed for each pairwise comparison of the means (P < 0.001; two-tailed test; unequal variances).
Data from database searches, pSTS hybrid, and FISH analyses reveal a complex, highly paralogous organization of the 101B6 sequence. The pericentromeric regions of chromosomes 10, 16, and 22 share the largest blocks of sequence similarity with the 2p11 pericentromeric segment 101B6 (Table 2, Figure 5a). With one exception, all pSTS tested coamplify from chromosomes 2p11, 10p11, and 22q11. pSTS pair 16, which was developed across one of the GC-rich repeats at position 109 kb, is the only site that amplifies solely from chromosome 2. Based on our monochromosomal analysis with 24 pSTSs, an average of 7.7 different chromosomal loci are detected for each pSTS. Interestingly, some groups of pSTS are distributed among more chromosomes (pSTSs 17–24, Fig 5a) than others (pSTSs 1–16, Fig. 5a). If the number of chromosomes scored positive for a given pSTS is plotted against the position of the pSTS within the 101B6 reference sequence, a distinct pattern emerges (Fig. 5b). Three statistically significant (p<0.001, two-tailed t test, unequal variances) blocks can be discerned. pSTSs within duplicons 1 and 2 show a similar number of interchromosomal duplicons (6.3 +/− 1.3 chromosomes). In contrast, the number of interchromosomal signals more than doubles for pSTSs designed within duplicon 4 (mean=14.5 +/− 2.2 chromosomes). An intermediate number of chromosomes cross amplify for pSTSs developed within the third duplication segment (11.5 +/− 2.2 chromosomes). These data indicate that different regions of the pericentromeric sequence have had markedly different propensities to duplicate.
Identification of Other Complex Regions of the Human Genome
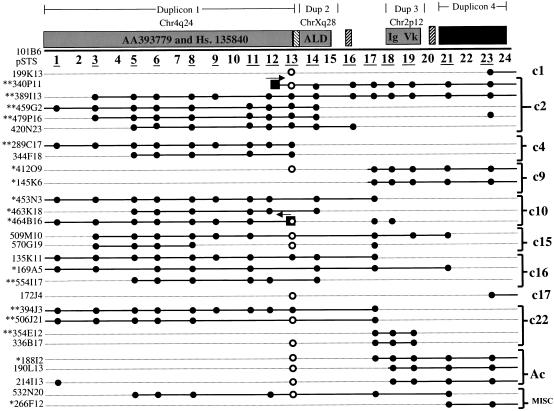
To confirm the contiguous genomic organization of 101B6, and to identify paralogous clones from other chromosomes, a human BAC library (RPCI-11), was screened by radioactive hybridization with four 101B6-derived probes (PCR products from pSTS 1, pSTS 14–15, pSTS18, pSTS23) and a cDNA insert (AA393779 representing duplicon 1). A total of 702 BAC clones were identified. Approximately 10% of these clones (65/702) cohybridized with two or more probes. As expected, hybridization with pSTS23 (duplicon 4) identified the largest number of clones (397 BACs). Based on the library redundancy (12-fold coverage), we estimate that this single paralogous locus may be represented more than 30 times throughout the human genome. From our initial BAC screen, a subset of BACs from each pSTS hybridization to the RPCI-11 library was randomly selected for further analysis. A total of 107 (∼15%) BAC clones were selected for subsequent sequence analysis. Each of these BACs was amplified with a series of pSTS primers and the products were directly sequenced and compared to the chromosomal signatures. This analysis served two purposes. Firstly, it provided an assessment of the organization and continuity of the pSTSs on other chromosomes. Secondly, it allowed us to unambiguously assign the chromosomal origin of each BAC. A BAC was assigned to a chromosome after at least five sequence variants were identified that were concordant with the previously determined chromosome pSTS signature. Comparison of the paralogous sequence variants allowed us to unambiguously assign 65 of the 107 BACs to specific chromosomal pericentromeric regions. A total of 42 BACs, representing 26 distinct sequence signatures, could not be identified; these were placed into a miscellaneous bin for later analysis. Figure 6 summarizes BACs representative of each chromosome that share large segments of sequence with 101B6 as determined by PCR and sequence analysis. Using this sequence-based approach we have begun to construct 11 different BAC contigs in complex pericentromeric regions of the human genome. Analysis of sequence data collected from chromosome 22 as well as other chromosomes indicates that these BACs map to regions either near or within the gaps of existing maps (see below).
Figure 6.
Identification of pericentromeric BAC clones. A total of 702 individual BAC clones were identified upon hybridization of the RPCI-11 BAC library (segments 1 and 2) with 101B6-derived probes. 107 of these clones were characterized at the sequence level with 16 of the paralogous STSs indicated by an underline. 65/107 BACs could be assigned to a chromosomal bin based on at least five diagnostic paralogous sequence variants between the BAC and monochromosomal hybrid signature. A representative subset of paralogous BACs are depicted. Filled circles show the representative STS content of each BAC based on amplification with 101B6-derived pSTSs. Open circles indicate that a product larger than expected was amplified. Asterisks indicate BACs for which one (*) or both (**) end sequences were generated. Boxes show the position of the BAC-end sequence with respect to the 101B6 reference sequence. Eleven different contig bins were created corresponding to BACs from chromosome 1, 2, 4, 9, 10, 15, 16, 17, 22, acrocentric bin (13, 14, 15, 21, 22), as well as a miscellaneous bin, which includes BACs that have not yet been assigned to a chromosome but possess a distinct paralogous sequence signature.
DISCUSSION
Our analysis reveals several interesting features of human pericentromeric DNA. First, these data support the observation that pericentromeric regions of our genome have been subject to an unprecedented level of genomic duplication among nonhomologous chromosomes. Certain pericentromeric regions are composed almost exclusively of duplicated sequence. These segments are arranged as a patchwork or mosaic of different duplicons that correspond to ancestral, originally gene encoding, DNA that has been transposed to pericentromeric regions (Eichler et al. 1996, 1997; Jackson et al. 1999; Horvath et al. 2000). Sequence analysis indicates that the TAR and interspersed GC-rich repetitive elements (CAAAAAGCGGG) demarcate the transition from one duplicon to another in 101B6. Within the limits of resolution of this study (<5 kb), the transition between the 4q24, Xq28, and 2p12 duplicons each occurred across one of these GC-rich repeats (Fig. 3), implicating these elements as potential transpositional integration signals. The duplicons themselves may be arranged into larger units that are distributed to multiple pericentromeric regions (Compare 2p11, 10p11, 16p11, and 22q11 for duplicons 1 and 2; Table 2 and Fig. 3). Interestingly, estimates of copy number for the different duplicons suggest a difference in the degree of duplication. For example, the most proximal region analyzed, (duplicon 4 pSTS 20–24) is shared among more than half of all human chromosomes, indicating that either these sequences were more evolutionarily mobile or that the sequence defines a canonical sequence motif of many human pericentromeric regions. In contrast, other segments show a significantly reduced genomic distribution. The molecular basis for this difference is unclear, although it is possible that differences in proximity to centromeric α-satellite DNA may influence the spread of duplicated material among pericentromeric regions, as has been previously proposed (Regnier et al. 1997).
The degree of sequence similarity (Table 2) among the pericentromeric regions ranged over a narrow interval (93.3–97.5%). This we have proposed is a consequence of a pericentromeric swapping or exchange event that occurred among nonhomologous chromosomes within a very narrow window of human evolution. Previous comparative and phylogenetic analyses indicated that these duplication events occurred approximately 5 million years ago (Horvath et al. 2000). While nonhomologous exchange of DNA is not thought to be common, studies of acrocentric chromosomes indicate that the short arms of these chromosomes share α-satellite DNA subsets and rDNA gene sequence polymorphisms (Arnheim et al. 1980; Krystal et al. 1981; Choo et al. 1988; Greig et al. 1993). Because the acrocentric chromosomes are associated with the cell nucleoli during cell division, it has been postulated that this physical proximity may promote nonhomologous recombination or conversion events leading to an evolutionary homogenization of α-satellite as well as other pericentromeric DNA (Choo et al. 1988; Greig et al. 1993). Our data may suggest that the pericentromeric regions of many other nonacrocentric chromosomes are also capable of undergoing similar types of nonhomologous exchange events. Such a model could help explain the high degree of sequence similarity among localized patches of genomic sequence on chromosomes 1, 2, 7, 9, 10, 16, 17, 18, and Y.
Another interesting observation is the considerable variation in colinearity among different pericentromeric regions. Our BAC sequence analysis (Fig. 6) indicates that some regions contain many contiguous STSs, whereas other regions are much more fragmented with respect to 101B6. These observations are confirmed by both monochromosomal hybrid analysis (Fig. 5a) as well as database sequence similarity searches (Table 2, Fig. 3). In some cases, it appears that these fragment transitions occur, once again, near or within GC-rich repeats. For example, the extent of chromosome 9 and 21 paralogy to chromosome 2p11 (Fig. 5a) begins within pSTS16, which spans a CAAAAAGCGGG repeat. Similarly, 1q12, 7p11, 16p11, 17p11, and 18p11 paralogy all begin immediately after another such repeat (pSTS21). It is possible that these elements, in addition to serving as transposition integration signals, may also represent focal points for the transfer of genomic material among pericentromeric regions. In other cases, the variation in the extent of paralogy, particularly among BACs, appears to be the result of secondary events that rearranged these large pericentromeric blocks after duplication. For example, the recently published sequence organization of 22q11 compared to the sequence within 2p11 is complex. Instead of a single-step duplication of material between these chromosomes, at least two additional rearrangement events must be invoked to account for this organization (Fig. 7). All of these observations are consistent with a rapid evolutionary turnover in the pericentromeric region of human and other primate chromosomes (Eichler et al. 1999; Jackson et al. 1999).
Figure 7.
2p11 vs. 22q11 pericentromeric organization. Miropeat analysis was performed using the 162 kb of 2p11 reference sequence and 600 kb of finished chromosome 22 sequence contig 3. Miropeats identifies regions of sequence similarity and displays this similarity information graphically in the positional context of the sequence (vertical line) as black bars delineated by joining lines between the two sequences (http://www.genome.ou.edu/miropeats.html). Comparisons were performed using repeat-masked versions (RepeatMasker v. 3.0) of the sequences (consequently small breaks in the sequence similarity are indicated). Note the colinearity of duplicons 1 and 2 (see Fig. 3 for a detailed description of duplication content). Duplicon 3 is located 300 kb distal to the first sequence overlap in an inverted orientation. At least two rearrangement events must be invoked to account for this comparative organization. Duplicon 4, although present by monochromosomal hybrid analysis within chromosome 22 (Fig. 5a) could not be identified in any of the current finished sequence. This duplicated segment presumably lies within one of the remaining sequence gaps of 22q11.
Traditional methods of physical mapping (i.e., fingerprinting, STS-content mapping, BAC-end sequence characterization) focus on the identification of overlapping sets of contiguous clones. Implicit in this methodology is the presence of unique sequence characteristics that allow such overlaps to be detected. Although these strategies have been effective in the generation of large overlapping contigs within euchromatic DNA, regions near or within heterochromatic DNA have proven much more difficult (Green 1997; Dunham et al. 1999; Peterson et al. 1999). Instead of a cis-based approach for characterization of such regions, we have employed a transchromosomal assay to characterize these regions. The approach exploits the highly paralogous nature of these areas to identify other complex regions of the genome that have been linked evolutionarily by recent duplication events. Sequence similarity provides the specificity to detect clones, whereas sequence differences (paralogous sequence variants) provide the sensitivity needed to distinguish between closely related copies. Application of this approach to 162 kb of sequence from chromosome 2p11 has allowed us to effectively identify 702 BAC clones and assign more than 65 BACs to 9 contigs located within different pericentromeric regions across the genome. Based on the total number of BAC clones and distinct sequence signatures identified, we estimate that our analysis of this single 162 kb reference sequence facilitated the identification and characterization of 6–8 Mb of pericentromeric DNA (see Methods). Such BAC clones may now be selected as templates to complement sequencing efforts on these chromosomes and to facilitate closure in particularly problematic regions. Because of the paralogous nature of this and other pericentromeric regions, it is likely that a relatively limited number of reference sequences must be analyzed in this fashion to obtain clones representing a significant fraction of human pericentromeric DNA.
One of the limitations of our approach has been the inability to assign a portion (39%) of our BAC clones to specific chromosomal bins. Some of this variability could be attributed to human polymorphism. Because the number of bp differences between any two variant sequences exceeds the level of human polymorphism (1 difference in 1200–2000 bp) (Cargill et al. 1999; Halushka et al. 1999), this is likely not the sole explanation for our inability to assign all BAC clones to a chromosome. The ability to discern all possible paralogous signatures using hybrids as templates begins to diminish as the number of paralogous copies distributed throughout the genome increases. Many of these high-copy paralogous segments often showed the presence of heterozygous sequence signatures or an unusually high level of background when PCR products were directly sequenced from specific monochromosomal hybrids. These data suggest that multiple copies of duplicons exist on single chromosomes and that the sequencing of a single PCR product from a monochromosomal hybrid is not sufficient to resolve this intrachromosomal duplicity. In such cases, these problems may be eliminated by selection of a more refined set of chromosome-specific substrates (i.e., monochromosomal deletion hybrids or chromosome-specific cosmid libraries that have been derived from a single chromosomal haplotype). Indeed, the frequent occurrence of highly paralogous intrachromosomal duplicated segments in the genome near gaps in sequence or clonal continuity (Dunham et al. 1999; Loftus et al. 1999) may require the use of such resources to achieve final sequence closure.
Database sequence similarity searches (Table 2, Fig.3) reveal that GenBank entries that are paralogous to 101B6 belong to one of two categories: sequences that have been not been assigned to a specific chromosome or those that have been assigned largely to one of three specific pericentromeric regions (10p11, 22q11, and 16p11). Sequences from the latter group have been generated for the most part as a consequence of specialized sequencing efforts. For example, the paralogous segments within 10p11 are the product of a random shotgun library construction and sequence assembly of overlapping large-insert YAC clones that previously had been mapped near the chromosome 10 centromere (Jackson et al. 1996, 1999). Similarly, the corresponding paralogous segments from 22q11 have been derived almost exclusively from overlapping cosmid (40 kb insert) clones, which were isolated from a chromosome 22-specific library (Dunham et al. 1999). Due to the clonal instability and/or size limitations of YAC and cosmid clones, it is unlikely that such methods will become widely adopted by sequencing centers.
Our approach obviates the need for specialized sequencing templates by focusing specifically on these problematic regions, exploiting their highly paralogous nature and using the sensitivity of sequence data to specify BAC location. Our analysis, for example, has identified a series of chromosome 22 paralogous sequence signatures (from duplicon 4) that are not currently represented in the finished sequence of chromosome 22 (Dunham et al. 1999). These sequences presumably lie within one of the remaining gap regions of this chromosome. Using these paralogous STSs as probes, it should now be possible to identify and sequence clones within these regions to provide a complete sequence representation of this chromosome. Similarly, until recently, chromosome 10 sequencing efforts were unable to incorporate the chromosome 10 ALD paralog, a known pericentromeric marker, into existing YAC maps (Jackson et al. 1999). Our approach identified two BACs (Fig. 6) belonging to chromosome 10 that contained the ALD paralog and one of these (accession number AL133173) has been placed into the sequencing queue at the Sanger Center to facilitate closure in this region. These results suggest that these types of analyses may not only be useful for de novo sequencing of complex regions but also may complement existing sequencing efforts within the genomic community. Although such an approach requires more upfront effort, its pangenomic application should help to provide a more balanced representation of both the heterochromatic and euchromatic portions of our genome.
METHODS
Sequence Analysis
Interspersed repeat sequences were masked using RepeatMasker version 3.0 software (A.F.A. Smit and P. Green, http://ftp.genome.washington.edu/RM/ RepeatMasker.html) BLASTN (v.2.0.10) sequence similarity searches were performed using repeatmasked 101B6 (AC002038) sequence as query against both htgs and nr divisions of GenBank. (Horvath et al. 2000). Only genomic sequence greater than 1 kb, and unprocessed cDNA sequences with a minimum of 90% identity to query were considered (Tables 1 and 2). A combination of BLAST, sim4 (Florea et al. 1998) and Miropeats (Parsons 1995) software delineated the extent of duplication. Global pairwise genomic sequence alignments were performed with ALIGN software (http://genome.cs.mitu/edu/align /align.html). Percent similarity was calculated as {[(number of matched bases)]/ [L (number of bases aligned) + (number of indels)]} × 100%. Standard error was estimated as the square root of the binomial distribution. Sequence manipulations and alignment calculations were performed using Alignscorer software (Jeff Bailey, unpubl.). Paralogous sequence variants generated from BAC and chromosomal hybrid sequencing were identified using Javascripts that manipulate Phrap-generated ace files (Ewing and Green 1998). Only sequence variants with a phrap value >20 for forward or reverse strands were considered.
Hybridization
The RPCI-11 human BAC library (segments 1 and 2) was hybridized with PCR-generated probes representing pSTS 1, pSTS 18, pSTS 23, and the gel-purified insert of cDNA clone AA393779 (Table 2, Table 3). The pSTS probes were designed to reference sequence A101B6 (CIT978SK, from the California Institute of Technology library). High-density arrayed BAC filters (Roswell Park Cancer Institute, Buffalo, NY) were hybridized and washed as described previously (Eichler et al. 1997) with the exception that nylon membranes were blocked using 1 mg of sonicated salmon sperm DNA (Stratagene). PCR probes were purified (Qiagen QiaQuick kit) and 25–50 ng of product was random-hexamer labeled (MegaPrime) using [α-32P] dCTP and 2 U Klenow (Amersham, manufacturer's specifications). A total of 702 strongly hybridizing BAC clones were identified using probes derived from the 162 kb 101B6 clone. Based on the depth of library coverage (11.8 ×) and an average insert size of 166 kb (http://bacpac.med.buffalo.edu), we estimate the BAC clones represent ∼9.9 Mb of pericentromeric sequence. A similar estimate is obtained if the total number of distinct BAC paralogous signatures is considered (37 different patterns × 166 kb insert = 6.1 Mb).
Table 3.
Paralogous STR Primers
| Primer pair | OLIGO | Sequence (5′ to 3′) | Location | TA °C |
|---|---|---|---|---|
| 1 | 101b6-1 | GAGAAGGTTCTGGTGGCAGATGCTG | 7926–7950 | 65 |
| 101B6-2 | TTACCCAGAGTTTGCCAACCCAGAC | 8309–8285 | ||
| 2 | 101B6-3 | CGAGTGACAGTTAACTGGCTACAG | 13953–13676 | 65 |
| 101B6-4 | CCAAATAGCAATCTAGACAAAGCTG | 14475–14451 | ||
| 3 | 101b6-65 | CAGATTGGCTATAGGTCCATGCC | 19536–19558 | 55 |
| 101b6-66 | GGAGTTAGGATCTAGAGACAGTAG | 20149–20126 | ||
| 4 | 101B6-5 | GTGCACTCATGTGCTGCTGGAGAC | 32227–32250 | 55 |
| 101b6-36 | CAAGTGAACGGTGTTGTGTATTGGC | 33075–33051 | ||
| 5 | 101b6-67 | TGACCATTCTTACAGTGGTACTCC | 35201–35224 | 55 |
| 101b6-68 | ATGTCATCCATACTGCTAGCAGCC | 35862–35839 | ||
| 6 | 101B6-7 | CGGGTAGGACATGATATTGTGGC | 37778–37756 | 55 |
| 101b6-37 | GTGCTGCATCATGATTACTTATCCTG | 37233–37258 | ||
| 7 | 101B6-74 | CTGTATCAATCACTGCTGTGCTCAG | 56741–56717 | 55 |
| 101B6-109 | GAGCTAAGTGTTTTCATACATGTC | 56308–56331 | ||
| 8 | 101B6-78 | CTAGTATCAGAGATGTGGCAGAAG | 57193–57216 | 55 |
| 101B6-79 | CAACCAGAATGAGGGGATTTCCTA | 57654–57631 | ||
| 9 | 101B6-10 | TATCAAGCTGGTTCCAGGAACTGG | 64911–64934 | 55 |
| 101b6-38 | GTACTGAACATGATCCAGTGTGCTG | 65590–65566 | ||
| 10 | 101B6-116 | AACTCCTGGTGTTATGAGGGCAAC | 66346–66369 | 55 |
| 101B6-118 | AAGAAGTAGGCAGATGATGACAGG | 66894–66871 | ||
| 11 | 101B6-11 | CACTTGGTACAATCACCAATGCAAAG | 70731–70706 | 65 |
| 101b6-39 | GGAAGCTGTGAAGAAGCTGGTCTC | 70252–70275 | ||
| 12 | 101B6-14 | TGGCTGATCTGTCTGACAACAGTG | 85255–85278 | 60 |
| 101b6-41 | CAACACCTAGTTGGCCATATAGTCC | 86050–86026 | ||
| 13 | 101B6-81 | AGTTTCCTGCCTGGGATGGTTCAC | 90575–90552 | 55 |
| 101b6-42 | CAAACAGCTTTGGATCCATAGCCAC | 90201–90225 | ||
| 14 | 83192 | TCACAGGCTAGTGGACATGGCAGAC | 100389–100366 | 55 |
| 83191 | CACCCGCAGCACCTGGATGTCAGC | 100165–100189 | ||
| 15 | 101b6-85 | CCTTGTGTGACCAGGTGATCTACC | 109336–109313 | 55 |
| 101b6-86 | ACAGTAGCCATCACTGCACACATG | 108700–108723 | ||
| 16 | 101B6-27 | GGTAGATCACCTGGTCACACAAGG | 109313–109336 | 55 |
| 101b6-51 | CCAAGAAGTTAGATTCTGTCTTTGG | 110437–110413 | ||
| 17 | 101b6-55 | GGTGACATGATGCTCTCATCTGGC | 115316–115339 | 55 |
| 101b6-56 | CCTTTGGTAGGGATCCAGGGATTG | 115740–115717 | ||
| 18 | 101B6-20 | TGTAACATTCTCATAGCCATCTGG | 124982–124959 | 55 |
| 101b6-44 | GAAACTTTTGGTTACCTGAGATTGC | 124701–124725 | ||
| 19 | 101B6-22 | CACATGCAGTTAGGTGTGGACTGG | 131151–131174 | 55 |
| 101b6-45 | ACGTGACAATGCCTGTCCTGACTG | 131537–131514 | ||
| 20 | 101B6-24 | CTGCTGTGAAGTGAATGGTGTCTTC | 138788–138764 | 55 |
| 101b6-46 | CGTGGTAGACAGAGCTTCATTCAAC | 138308–138332 | ||
| 21 | 101B6-29 | GGAGATCTGGGATGGAATAGGGTTC | 148181–148205 | 55 |
| 101b6-47 | GAGAGATCATAGTGGGTTTGTGGAG | 148514–148490 | ||
| 22 | 101B6-31 | CGCCAGTCACCTCTAAACCGTATTG | 153385–153361 | 55 |
| 101b6-48 | GCCTATCTGTGTAATTGACTGGTTAG | 152747–152772 | ||
| 23 | 101B6-32 | CAGTATCTTCACATTCTCTCCCTGTCC | 155471–155497 | 55 |
| 101b6-49 | GAAAGAAGCAAGAGTGCGCTAAAC | 155774–155751 | ||
| 24 | 101B6-34 | CACACCTGCGAGGTGGATGGAAGAG | 160943–160919 | 55 |
| 101b6-50 | GGTAGCACCTACTTTTCAAATAGCG | 160514–160538 |
PCR and Sequencing
PCR amplifications of somatic cell hybrid templates (NIGMS, Human Genetic Mutant Cell Repository Mapping Panel 2) were performed as previously described (Horvath et al. 2000). BAC DNA templates were isolated from 5 ml overnight cultures (Qiagen Qiawell DNA isolation kit), resuspended in TE or water, and 1/25 of the total volume (∼15 ng) was used for PCR. Table 3 summarizes the oligonucleotide sequence, its position within the 101B6 reference sequence, and the PCR annealing temperature for each of the 24 PCR assays. All PCR products were directly sequenced (both forward and reverse strands) using a modified dye-terminator sequencing protocol (Horvath et al. 2000). BAC end cycle sequencing reactions using T7.29 and SP6.22 primers and a modified Big-Dye terminator sequencing protocol (http://bacpac.med.buffalo.edu) consisting of 2 μg DNA, 8μl Big Dye terminator mix (Perkin Elmer Applied Biosystems, Norwalk, CT) and 1μl (20 pmoles) primer. BAC DNA templates for end-sequencing were prepared using a Nucleobond DNA purification kit (Clontech, Palo Alto, CA). All fluorescent traces were analyzed using the Applied Biosystems Model 377 DNA Sequencing System (Perkin-Elmer Applied Biosystems, Norwalk, CT) and the quality of sequence data assessed with PHRED/PHRAP/CONSED software (http://genome.wustl.edu). A total of 37.5 kb and 41.2 kb of paralogous pericentromeric sequence was generated from monochromosomal and BAC templates, respectively. All sequences have been deposited into GenBank.
Fluorescent In Situ Hybridization
Metaphase and prometaphase chromosomes were prepared from phytohemagglutinin (PHA)-stimulated leukocyte cultures from a normal male donor using standard procedures. Fluorescence in situ hybridization was performed on prometaphase cells on unstained slides as described elsewhere (Sullivan et al. 1996). The BAC probes were labeled with biotin 14-dATP by nick translation (BioNick Labeling System18247–015, Gibco BRL, Gaithersburg, MD). And the chromosomes were counterstained with DAPI. Twenty metaphases were analyzed for the presence of probes and the localization of the signals determined from the DAPI counterstain. Digital images were collected using a Leitz DMRB microscope controlled by CytoVision ChromoFluor software manufactured and distributed by Applied Imaging Corporation (Santa Clara, CA).
Acknowledgments
We thank Carl Kashuk, Jeff Bailey, Theresa Depinet, and Laurie Christ for computational and technical assistance, and Drs. Aravinda Chakravarti, Christine O'Keefe, and Rob Nicholls for helpful comments in the preparation of this manuscript. This work was supported by grants NIH GM58815, NIH HG01847, DOE ER62862–1013741–0005006, and a Basil O'Connor Scholar award (FY99–0519) to E.E.E. J.E.H. was supported in part by NIH GM08613–04, Genetics training grant. The financial support of the W.M. Keck Foundation and a Howard Hughes Medical Institute grant to Case Western Reserve University, School of Medicine are also gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL eee@po.cwru.edu.
REFERENCES
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci USA. 1980;77:7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold N, Stanyon R, Jauch A, O'Brien P, Wienberg J. Identification of complex chromosome rearrangements in the gibbon by fluorescent in situ hybridization (FISH) of a human chromosome 2q specific microlibrary, yeast artificial chromosomes, and reciprocal chromosome painting. Cytogenet Cell Genet. 1996;74:80–85. doi: 10.1159/000134387. [DOI] [PubMed] [Google Scholar]
- Brand-Arpon V, Rouquier S, Massa H, de Jong PJ, Ferraz C, Ioannou PA, Demaille JG, Trask BJ, Giorgi D. A genomic region encompassing a cluster of olfactory receptor genes and a myosin light chain kinase (MYLK) gene is duplicated on human chromosome regions 3q13–q21 and 3p13. Genomics. 1999;56:98–110. doi: 10.1006/geno.1998.5690. [DOI] [PubMed] [Google Scholar]
- Brown TA. Genomes. New York: Bios Scientific Publishers: Wiley-Liss; 1999. [Google Scholar]
- Cao Y, Kang HL, Xu X, Wang M, Dho SH, Huh JR, Lee BJ, Kalush F, Bocskai D, Ding Y, et al. A 12-Mb complete coverage BAC contig map in human chromosome 16p13.1– p11.2. Genome Res. 1999;9:763–774. [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalayanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [published erratum appears in Nov. 1999. Nat. Genet. 23: 373]. [DOI] [PubMed] [Google Scholar]
- Choo KH, Vissel B, Brown R, Filby RG, Earle E. Homologous alpha satellite sequences on human acrocentric chromosomes with selectivity for chromosomes 13, 14 and 21: Implications for recombination between nonhomologues and Robertsonian translocations. Nucleic Acids Res. 1988;16:1273–1284. doi: 10.1093/nar/16.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader–Willi/Angelman syndrome chromosome region (15q11–q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- DeSilva U, Massa H, Trask BJ, Green ED. Comparative mapping of the region of human chromosome 7 deleted in Williams syndrome. Genome Res. 1999;9:428–436. [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Masquerading repeats: Paralogous pitfalls of the human genome. Genome Res. 1998;8:758–762. doi: 10.1101/gr.8.8.758. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Archidiacono N, Rocchi M. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 1999;9:1048–1058. doi: 10.1101/gr.9.11.1048. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW. Interchromosomal duplications of the adrenoleukodystrophy locus: a phenomenon of pericentromeric plasticity. Hum Mol Genet. 1997;6:991–1002. doi: 10.1093/hmg/6.7.991. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet. 1996;5:899–912. doi: 10.1093/hmg/5.7.899. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner K. Base composition and gene distribution: critical patterns in mammalian genome organization. Trends Genet. 1996;12:519–524. doi: 10.1016/s0168-9525(97)81400-x. [DOI] [PubMed] [Google Scholar]
- Green P. Against a whole-genome shotgun. Genome Res. 1997;7:410–417. doi: 10.1101/gr.7.5.410. [DOI] [PubMed] [Google Scholar]
- Greig GM, Warburton PE, Willard HF. Organization and evolution of an alpha satellite DNA subset shared by human chromosomes 13 and 21. J Mol Evol. 1993;37:464–475. doi: 10.1007/BF00160427. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Horvath JE, Viggiano L, Loftus BJ, Adams MD, Archidiacono N, Rocchi M, Eichler EE. Molecular structure and evolution of an alpha satellite/non-alpha satellite junction at 16p11. Hum Mol Genet. 2000;9:113–123. doi: 10.1093/hmg/9.1.113. [DOI] [PubMed] [Google Scholar]
- Jackson MS, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Ricco A, Piccininni S, et al. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications, and unstable sequences with homologies to telomeric and other centromeric locations. Hum Mol Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- Jackson MS, See CG, Mulligan LM, Lauffart BF. A 9.75-Mb map across the centromere of human chromosome 10. Genomics. 1996;33:258–270. doi: 10.1006/geno.1996.0190. [DOI] [PubMed] [Google Scholar]
- Ji Y, Walkowicz MJ, Buiting K, Johnson DK, Tarvin RE, Rinchik EM, Horsthemke B, Stubbs L, Nicholls RD. The ancestral gene for transcribed, low-copy repeats in the Prader–Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- Krystal M, D'Eustachio P, Ruddle FH, Arnheim N. Human nucleolus organizers on nonhomologous chromosomes can share the same ribosomal gene variants. Proc Natl Acad Sci USA. 1981;78:5744–5748. doi: 10.1073/pnas.78.9.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Kim UJ, Sneddon VP, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby ML, Barnstead M, Cronin L, et al. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- Mazzarella R, Schlessinger D. Pathological consequences of sequence duplications in the human genome. Genome Res. 1998;8:1007–1021. doi: 10.1101/gr.8.10.1007. [DOI] [PubMed] [Google Scholar]
- Orti R, Potier MC, Maunoury C, Prieur M, Creau N, Delabar JM. Conservation of pericentromeric duplications of a 200-kb part of the human 21q22.1 region in primates. Cytogenet Cell Genet. 1998;83:262–265. doi: 10.1159/000015201. [DOI] [PubMed] [Google Scholar]
- Parsons J. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Peterson ET, Sutherland R, Robinson DL, Chasteen L, Gersh M, Overhauser J, Deaven LL, Moyzis RK, Grady DL. An integrated physical map for the short arm of human chromosome 5. Genome Res. 1999;9:1250–1267. doi: 10.1101/gr.9.12.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier M, Dutriaux A, Orti R, Groet J, Gibelin N, Karadima G, Lutfalla G, Lynn A, Van Broeckhoven C, Chakravarti A, et al. Two sequence-ready contigs spanning the two copies of a 200-kb duplication on human 21q: Partial sequence and polymorphisms. Genomics. 1998;51:417–426. doi: 10.1006/geno.1998.5389. [DOI] [PubMed] [Google Scholar]
- Regnier V, Meddeb M, Lecointre G, Richard F, Duverger A, Nguyen VC, Dutrillaux B, Bernheim A, Danglot G. Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition. Hum Mol Genet. 1997;6:9–16. doi: 10.1093/hmg/6.1.9. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR. The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet. 1997;6:1595–1603. doi: 10.1093/hmg/6.9.1595. [DOI] [PubMed] [Google Scholar]
- Ritchie RJ, Mattei MG, Lalande M. A large polymorphic repeat in the pericentromeric region of human chromosome 15q contains three partial gene duplications. Hum Mol Genet. 1998;7:1253–1260. doi: 10.1093/hmg/7.8.1253. [DOI] [PubMed] [Google Scholar]
- Smit A, Riggs A. Tiggers and other DNA transposon fossils in the human genome. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Jenkins LS, Karson EM, Leana-Cox J, Schwartz S. Evidence for structural heterogeneity from molecular cytogenetic analysis of dicentric Robertsonian translocations. Am J Hum Genet. 1996;59:167–175. [PMC free article] [PubMed] [Google Scholar]
- Trask B, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen OT, Eichler EE, van den Engh G, Rouquier S, Shizuya H, et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum Mol Genet. 1998;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Lemmers RJ, Grewal PK, van Geel M, Romberg S, Dauwerse HG, Wright TJ, Padberg GW, Hofker MH, Hewitt JE, et al. Identification of the first gene (FRG1) from the FSHD region on human chromosome 4q35. Hum Mol Genet. 1996;5:581–590. doi: 10.1093/hmg/5.5.581. [DOI] [PubMed] [Google Scholar]
- Vogt P. Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved “chromatin folding code.”. Hum Genet. 1990;84:301–336. doi: 10.1007/BF00196228. [DOI] [PubMed] [Google Scholar]
- Zachau H. The immunoglobulin k locus — or what has been learned from looking closely at one-tenth of a percent of the human genome. Gene. 1993;135:167–173. doi: 10.1016/0378-1119(93)90062-8. [DOI] [PubMed] [Google Scholar]
- Zimonjic D, Kelley M, Rubin J, Aaronson S, Popescu N. Fluorescence in situ hybridization analysis of keratinocyte growth factor gene amplification and dispersion in evolution of great apes and humans. Proc Natl Acad Sci USA. 1997;94:11461–11465. doi: 10.1073/pnas.94.21.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]