Abstract
Purpose
Prominin is a family of pentaspan transmembrane glycoproteins. They are expressed in various types of cells, including many stem/progenitor cells. Prominin-1 plays an important role in generating and maintaining the structure of the photoreceptors. In this study, we identified three prominin homologs in Xenopus laevis, a model animal widely used in vision research, and characterized their messenger RNA (mRNA) expression in selected tissues of this frog.
Methods
Reverse-transcription PCR (RT–PCR) and rapid amplification of cDNA ends (RACE) were used to isolate cDNAs of prominin homologs. Semiquantitative RT–PCR was used to measure the relative expression levels of mRNAs of the three prominin homologs in four X. laevis tissues, specifically those of the retina, brain, testis, and kidney. Sequences of prominin homologs were analyzed with bioinformatic software.
Results
We isolated cDNAs of three prominin homologs from X. laevis tissues and compared their sequences with previously described prominin-1, 2, and 3 sequences from other species using phylogenetic analysis. Two of these homologs are likely to be the X. laevis orthologs of mammalian prominin-1 and 2, respectively, while the third homolog is likely to be the X. laevis ortholog of prominin-3, which was only found in nonmammalian vertebrates and the platypus. We identified alternatively spliced exons in mRNAs of all three prominin homologs. Similar to mammalian prominin-1, we found that exons 26b, 27, and 28a of the X. laevis prominin-1 gene are alternatively spliced, and that the splice isoforms of mRNA show tissue-specific expression profiles. We found that prominin-1 was the most abundant homolog expressed in the retina, brain, and testis, while prominin-3 was the most abundant homolog in the kidney. The expression level of prominin-2 was the lowest of the three prominin homologs in all four examined tissues of this frog.
Conclusions
Our results suggest that the mRNAs of prominin homologs are expressed in many tissues of X. laevis, but differ in their expression levels and mRNA splicing. Prominin-1 is the most abundant of the three prominin homologs expressed in the frog retina.
Introduction
The prominins are a family of pentaspan transmembrane glycoproteins [1-5]. The first member, identified as prominin-1, was originally cloned from murine kidney epithelial cells [6]. It was later characterized as an immunological determinant of hematopoietic and neuroepithelial stem/progenitor cells [6-8], referred to as CD133 or AC133 [2,7]. Prominin-1 is also expressed in embryonic stem cells [9], cancer cells [10-14], some epithelial cells [15-17], glial cells [18], and rod photoreceptors [19,20]. Prominin-1 is specifically associated with membrane protrusions such as microvilli, filopodia, and lamellipodia [17,21]. Maw et al. [19] reported that a frameshift mutation (PROML11878, now designated as c.1841delG) in prominin-1 that results in the premature termination of translation causes autosomal recessive retinal degeneration in a consanguineous Indian pedigree. Another mutation, R373C, of human prominin-1 was reported to cause autosomal dominant macular degeneration in patients [22,23]. Permanyer et al. identified a third mutation, c.869delG, of human prominin-1 that causes autosomal recessive retinitis pigmentosa [24]. Zacchigna et al. found that knockout of prominin-1 in mice causes slow retinal degeneration [20]. The specific localization of prominin-1 in murine rod photoreceptors and the pathological effect caused by its mutations in humans and mice suggest that it may be involved in the generation or maintenance of photoreceptor outer segment structure.
A second member of the prominin family, prominin-2, shares many features with prominin-1, including a predicted pentatransmembrane topology [3], preferential subcellular localization to membrane protrusions [25], binding to cholesterol, and association with vesicles released into the body fluids [16,25-27]. However, the expression profile of prominin-2 is different from that of prominin-1 in many tissues [3,28,29], and prominin-2 is not detected in the mouse eye [3].
The prominin-1 gene is alternatively spliced in human and mouse tissues, producing multiple splice isoforms of mRNA [2,18,30-32]. Kemper et al. showed that the expression of these splice isoforms is regulated in mouse tissues, based on the fact that these splice isoforms differ in their expression profiles or levels in analyzed tissues [30]. A region that encodes the C-terminus of the human and mouse prominin-1 is extensively spliced [2,31,32]. The significance of the alternative splicing of prominin-1, especially at the C-terminus, is unknown. PDZ (Postsynaptic density protein, PSD-95; Discs large tumor suppressor, DlgA; Zonula occludens-1 protein, ZO-1)-binding motifs have been identified in certain splice isoforms of human and mouse prominin-1 [31]. It was postulated that these splice isoforms interact with different partners and therefore participate in different cellular functions [31-33].
Our goal is to study the function of prominin homologs in the retina of X. laevis, in which photoreceptor outer segment membrane synthesis and structure has been investigated by retinal cell biologists for decades [34,35], and for which genetic manipulation methods are available [36,37]. Here, we present the results of the isolation of cDNAs of three prominin homologs in X. laevis, comparisons with prominin homologs from other species by phylogenetic analysis, characterization of alternative splicing of prominin-1, and analysis of mRNA expression levels of three prominin homologs by semiquantitative RT–PCR.
Methods
Animals
Adult X. laevis and X. tropicalis were purchased from Xenopus Express, Brooksville, FL. The animals were kept in an environment of artificially controlled 12 h:12 h light-dark cycle and constant temperature of 18 °C. All experiments were conducted in accordance with the Animal Care and Use Committee of the University of Connecticut Health Center and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Cloning of Xenopus laevis prominin-1 cDNA
A protein versus translated nucleotide BLAST search was performed against the Sanger Institute X. laevis expressed sequence tag (EST) database, using the protein sequence of human prominin-1 (GenBank accession number: NP_006008) as a query. Nested reverse-transcription (RT)-PCR was performed to isolate a partial cDNA from an X. laevis (adult) retinal cDNA library (a gift from Dr. Joseph C. Besharse, Medical College of Wisconsin, WI) [38]. Full-length cDNAs were subsequently isolated by performing 5′ and 3′ rapid amplification of cDNA ends (RACE) using cDNA synthesized from X. laevis retina total RNA as a template [39]. Sequences of primers used in nested RT–PCR and RACE are provided in Table 1. The isolated cDNAs were cloned into pCR 2.1 TOPO cloning vector (Invitrogen, Carlsbad, CA) and sequenced using the dye-termination method [40].
Table 1. Primers used in cloning of xlProminin-1, 2 and 3 cDNAs.
| Targets | Sequences (5′-3′) | Purpose |
|---|---|---|
| xlProminin-1 | CGGGGTACCGTAGATCTCTTGGCATTCGCTAATG | RT–PCR |
| CGCGGATCCTTATGATAACCATTATTACCATTTTCCA | RT–PCR | |
| ATTGACTGCTCCAGTGGCAC | 5′RACE outer primer | |
| CATTAGCGAATGCCAAGAGATCTAC | 5′RACE inner primer | |
| GTTTGGTTTGGGAGGAGCAACTGTG | 3′RACE outer primer | |
| ATGGACACCGAAGATGTTTATGATG | 3′RACE inner primer | |
| xlProminin-2 | CAGAAGGTCCAAGAGGAGTTTGC | RT–PCR |
| CCAGGGATTAGTTATATTGTCACACA | RT–PCR | |
| ATTTGCATCGACTACAGCCTGC | 5′RACE outer primer | |
| TTTTCAAGCACCAGGGGAAGGC | 5′RACE inner primer | |
| TTGGAAAACCTGGCAACCGTGC | 3′RACE outer primer | |
| AACTCCTTGGCTGAACTGGTCC | 3′RACE inner primer | |
| xlProminin-3 | CATGTTGGCTGGAAATATCTGCATGTT | RT–PCR |
| TTGCCGAGATGTTGTCATCTCCAT | RT–PCR | |
| TAGTAGCCACCAGGAGCTTAGC | 5′RACE outer primer | |
| TCACGGCATTTACGGTGTTCAC | 5′RACE inner primer | |
| AATTTTGCTCGCTGTGGACCAGTTGCCAGA | 3′RACE outer primer | |
| TGCAAAGCACTACAGGAGGATGAAGACTTCT | 3′RACE inner primer | |
| Adaptor or anchor primers for RACE | GGCCGCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | 5′RACE adaptor primer |
| GGCCGCACGCGTCGACTAGTAC | 5′RACE anchor primer | |
| CGACTCACTATAGGGCGATTTTTTTTTTTTTTTTT | 3′RACE adaptor primer | |
| CGGGGTACCGTAATACGACTCACTATAGGGCGA | 3′RACE anchor primer |
Cloning of Xenopus laevis prominin-2 and 3 cDNAs
We performed BLAST searches with sequences of known prominin homologs, including human prominin-1 (NP_006008) and 2 (NP_653308), and mouse prominin-1 (NP_032961) and 2 (NP_620089), against the Ensembl genomic database of Xenopus tropicalis [41]. Partial sequences resulting from this search were used to search the X. laevis EST databases at the Sanger Institute and to design primers to amplify prominin homologs from X. laevis by RT–PCR and RACE, using cDNA synthesized from X. laevis retina or testis total RNA as templates. Sequences of primers used in RT–PCR and RACE are provided in Table 1.
Sequence analysis of prominin homologs
Maximum likelihood phylogeny of prominin homologs from different species was generated using PhyML 3.0 software [42]. cDNAs of prominin homologs of the human, chimpanzee, rhesus monkey, rat, mouse, horse, cattle, dog, zebrafish, nematode, and fly were retrieved from GenBank. Prominin homologs of X. tropicalis were cloned by methods similar to those used for X. laevis; cDNAs synthesized from X. tropicalis retina or testis total RNA were used as templates for PCR and RACE. Bovine and platypus prominin homologs were predicted from genomic sequences by BLAST search and by aligning them with cDNA sequences of known prominin homologs and searching for the consensus exon/intron boundaries [43,44]. Platypus genomic sequences were retrieved from Ensembl Ornithorhynchus anatinus version 60.1n (OANA5) Chromosome 4. Bovine genomic sequences were retrieved from Ensembl Bos taurus version 60.4i (Btau_4.0) Chromosome 6. Other software used in sequence analysis included ClustalW for sequence alignments [45], SignalIP 3.0 for signal peptide prediction [46], NetOGlyc 3.1 for glycosylation site prediction [47], TMHMM Server, version 2.0 for prediction of transmembrane domains [48,49], and ScanProsite software for identification of PDZ-binding motifs [50].
RNA isolation and cDNA synthesis
RNA was extracted from the adult X. laevis retina, brain, testis, and kidney using TRIzol reagent (Invitrogen) and used to synthesize first-strand cDNAs with SuperScript II reverse transcriptase (Invitrogen) and oligo-dT primers (Amersham, Piscataway, NJ) according to protocols supplied by the manufacturers.
Analysis of alternative splicing
Alternatively spliced exons of xlProminin-1, 2, and 3 were identified by sequencing and alignment of multiple cDNA clones. Exon/intron boundaries were determined by comparing cDNA sequences with X. tropicalis genomic DNA sequences. The reference genomic sequences used were: X. tropicalis unplaced genomic scaffold, v4.2 XENTRscaffold_393, whole genome shotgun sequence, containing the prominin-1 gene, GenBank accession NW_003163719; X. tropicalis unplaced genomic scaffold, v4.2 XENTRscaffold_30, whole genome shotgun sequence, containing the prominin-2 gene, GenBank accession NW_003163356; X. tropicalis unplaced genomic scaffold, v4.2 XENTRscaffold_640 and 3098, whole genome shotgun sequence, containing the prominin-3 gene, GenBank accession NW_003163966 and NW_003166412, respectively. Alternative splicing at the C-terminus of xlProminin-1 in the retina, brain, testis, and kidney was examined by amplifying a cDNA fragment containing this region by RT–PCR. Primers used in this analysis were: 5′-ATA AGA ATG CGG CCG CCT GCA GAG AAC ATC CTT TGA TAT CGA G-3′ and 5′-CGC GGA TCC CAG ATT ATT TTC TGA AAG TCA TTT CTC C-3′. (Note that the Not I and BamH I sites were incorporated into the primer sequences to facilitate cloning.) The profiles and relative abundances of differentially spliced transcripts were analyzed by comparing the sizes and intensities of RT–PCR products resolved by electrophoresis on a 1% agarose gel. Optical densities were measured with ImageJ software [51] for quantification of different splicing isoforms of xlProminin-1. Individual bands were extracted from the gel using the GeneClean® III Kit (Qbiogene, Irvine, CA), cloned into the pCR 2.1 TOPO cloning vector (Invitrogen) and sequenced.
Semiquantitative reverse-transcription PCR
Two primer pairs were designed for specific amplification of each prominin homolog (Table 2 for sequences of primers). Reverse-transcriptase PCR amplification of a fragment of β-actin was used as a loading control to establish that an equal amount of starting materials was used (Table 2). PCR reactions were assembled using an Advantage® 2 PCR Kit (Clontech, Mountain View, CA) in a final volume of 100 μl containing 4 μl of cDNA. Aliquots (20 μl) taken at the end of the 27th, 31st, 35th, and 39th cycles were visualized on a 1% agarose gel by ethidium bromide staining, and digital images were analyzed using Image J software. The total amount of DNA in selected areas was represented with integrated optical densities. Results obtained with two different primer pairs were averaged and plotted.
Table 2. Primers used in semiquantitative reverse-transcription (RT)-PCR of xlProminin homologs.
| Targets | Primer pairs | Sequences (5′-3′) | Expected MWs |
|---|---|---|---|
| xlProminin-1 | 1 | GAAATTGGCTTTATCATTGCGGCAGTA | 1939-1993a bps |
| AGTAGCCAATGACTTCATCCATTAACTT | |||
| 2 | AACCTTACCAACCAGATTAGAGGATCA | 1601-1655a bps | |
| ACCTTCCCCTCGATATCAAAGGATG | |||
| xlProminin-2 | 1 | CAGAAGGTCCAAGAGGAGTTTGC | 1800 bps |
| CCAGGGATTAGTTATATTGTCACACA | |||
| 2 | GTGCTTGAAAAGATTGTATCTTCTTTC | 1370 bps | |
| AAGGCATTGGCTTCACTCTGTAGC | |||
| xlProminin-3 | 1 | GAAGTTGACTTAATAATTGGCCAAAGT | 1881 or 1902b bps |
| GAGAGCAATGAGAGAGACATTTCAG | |||
| 2 | CATGTTGGCTGGAAATATCTGCATGTT | 1436 bps | |
| AACTGTCTTTCGCCAGCGTAGC | |||
| X. laevis β-actin (as control) | GGCTATGCTCTACCACATGCCA | 532 bps | |
| TGGAGCCACCAATCCAGACA |
aThe expected MWs of these fragments varies with the presence or absence of the alternatively spliced exons 8a (24 bps), 11a (12 bps) and 19 (18 bps). The differences in sizes of these PCR products are too small to be resolved on a 1% agarose gel.
bThe expected MWs of these fragments varies with the presence or absence of the alternatively spliced exons 24a (21 bps). The differences in sizes of the two PCR products are too small to be resolved on a 1% agarose gel.
Results
Identification of three prominin homologs in Xenopus laevis
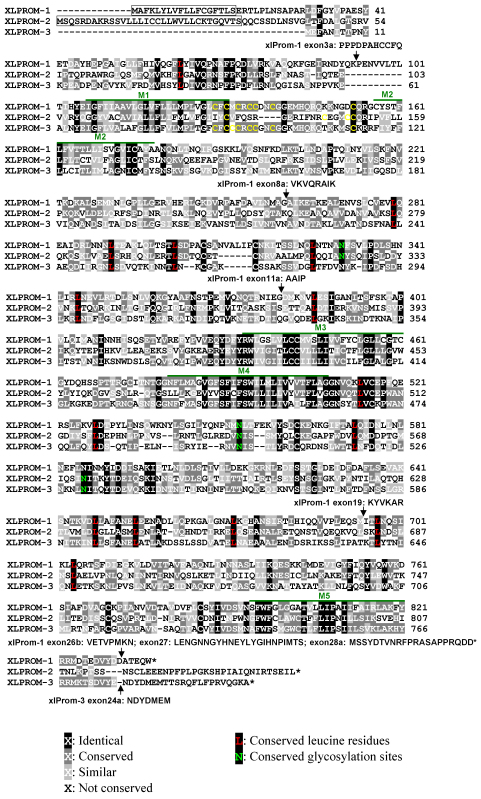
By performing a BLAST search of the X. laevis EST database, we retrieved a sequence (GenBank accession: BJ061435) with 62% similarity to human prominin-1. BLAST search of the NCBI nucleotide database and the Ensembl X. tropicalis genomic database using the available sequences of prominin homologs from different species as query sequences revealed three different prominin homologs in X. tropicalis. These sequences have the assigned GenBank accession numbers of BC127277, XM_002932561, and XM_002940570. Sequence BC127277 showed a high degree of protein sequence identity (95%) to the above mentioned X. laevis sequence BJ061435. However, X. laevis homologs of sequences XM_002932561 and XM_002940570 could not be retrieved from any public cDNA or genomic database by BLAST search, perhaps due to incomplete coverage of the X. laevis sequences by those databases. They were instead cloned by RT–PCR using specific primer pairs based on X. tropicalis prominin homolog sequences, employing cDNA synthesized from total RNA isolated from X. laevis tissues as templates. Multiple clones were obtained and sequenced. Full-length cDNA sequences of the three X. laevis prominin homologs were obtained by performing 5′ and 3′ RACE. Predicted protein sequences are aligned in Figure 1.
Figure 1.
Alignment of the predicted protein sequences of three X. laevis prominin homologs, showing characteristic features of prominins, including the pentaspan transmembrane topology, a conserved cysteine rich domain, conserved leucine residues and N-glycosylation sites. We designated the three X. laevis prominin homologs as xlProminin-1, 2, and 3, respectively. Identical and conserved residues are indicated by differentially shaded boxes. Predicted transmembrane domains are marked with M#. The predicted signal peptides of xlProminin-1 and 2 are boxed. No signal peptide was predicted for xlProminin-3. Positions of alternatively spliced exons are indicated by arrows and the sequences of the alternatively spliced exons are given. Note that the N- and C-termini of the three X. laevis prominin homologs are less similar than other regions. Cysteine residues at the junction of M1 and the first intracellular domain (I1) are marked with yellow. Conserved leucine residues of the three xlProminin homologs are marked with red. Conserved glycosylation sites are marked with green.
Phylogenetic analysis of prominin homologs
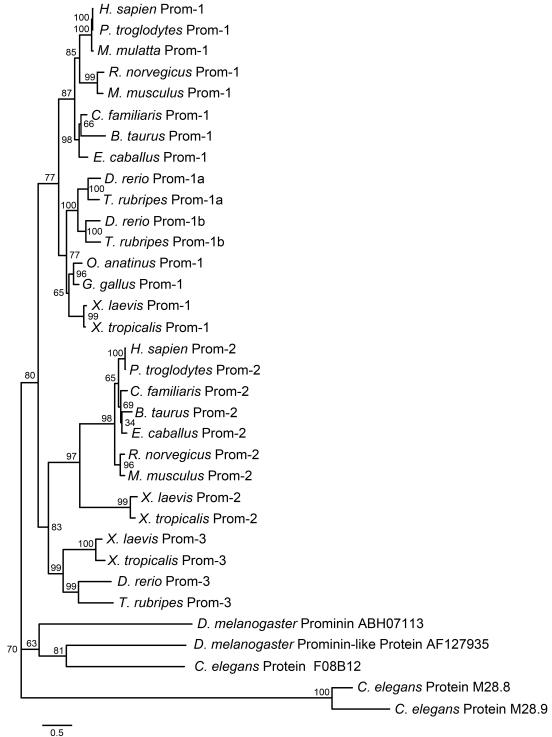
Previous taxonomic studies of prominins have divided prominin homologs into three groups. These are prominin-1 [2,31] and prominin-2 in mammals [3], and prominin-3 in zebrafish [52]. Two variants of prominin-1, namely prominin-1a and 1b, were identified in zebrafish [52]. We performed protein phylogenetic analysis using the PhyML 3.0 software to compare X. laevis prominin homologs with available prominin sequences; the results are presented in Figure 2. The sequences used to construct the phylogenetic tree were from online public databases, newly cloned cDNAs, or predicted mRNAs from genomic sequences (Methods). Since the N- and C-termini of prominin homologs were incomplete in many of the retrieved sequences, only the sequences from exon 4 to exon 24 of prominin-1 and homologous sequences of other prominins were used for the phylogenetic analysis. The analysis revealed that the first cloned prominin homolog is the X. laevis ortholog of prominin-1, and is therefore designated as xlProminin-1. The other two prominin-like cDNAs found in X. laevis were named xlProminin-2 and xlProminin-3 according to their positions on the phylogenetic tree.
Figure 2.
Phylogenic analysis of prominin homologs from several species. This phylogenetic tree was constructed to show the existence of multiple prominin homologs in metazoan animals from fly to human and their evolutionary relationship. Three X. laevis prominin homologs, designated as xlProminin-1, 2, and 3, are placed in branches of the consensus phylogenetic tree with prominin-1, 2, and 3 from other species. The numbers at each node are the percentage bootstrapping values of 100 replicates. The evolutionary distance of any two sequences is represented by the length of the branches. Sequences used in the phylogenetic analysis are excerpted from: H. sapien Prom-1 (NP_006008), P. troglodytes Prom-1 (XP_517115), M. mulatta Prom-1 (NP_001070888), R. norvegicus Prom-1 (NP_068519), M. musculus Prom-1 (NP_032961), C. familiaris Prom-1 (XP_850831), B. taurus Prom-1 (HQ159409), E. caballus Prom-1 (XM_001498729), D. rerio Prom-1a (NP_001108615), T. rubripes Prom-1a (HQ159405), D. rerio Prom-1b (NP_932337), T. rubripes Prom-1b (HQ159406), O. anatinus Prom-1 (HQ159408), G. gallus Prom-1 (XP_001232165), X. laevis Prom-1 (XM_001163920), X. tropicalis Prom-1 (HQ159400), H. sapien Prom-2 (NP_653308), P. troglodytes Prom-2 (XP_001143498), C. familiaris Prom-2 (XP_854455), B. taurus Prom-2 (XP_599188), E. caballus Prom-2 (XP_001494215), R. norvegicus Prom-2 (AAN63818), M. musculus Prom-2 (NP_620089), X. laevis Prom-2 (NP_001163922), X. tropicalis Prom-2 (HQ159402), X. laevis Prom-3 (NP_001163921), X. tropicalis Prom-3 (HQ159403), D. rerio Prom-3 (XP_684527), T. rubripes Prom-3 (HQ159407), D. melanogaster Prominin (ABH07113), D. melanogaster Prominin-like protein AF127935 (NP_647770), C. elegans protein F08B12 (NP_509907), C. elegans protein M28.8 (NP_496294), C. elegans protein M28.9 (NP_496296).
All newly cloned and predicted prominin homologs were also named according to their positions on the phylogenetic tree, and full sequences (including certain splice variants) were deposited in GenBank with assigned accession numbers (see Table 3 and Table 4), including sequences from: X. tropicalis, fugu, platypus, and cattle. We made new predictions for bovine prominin-1 from genomic sequences, because previously predicted bovine prominin-1 sequences (XP_875477 and XP_002688513) contain obvious errors and cannot be used for an accurate phylogenetic analysis. Our newly predicted bovine prominin-1 was assigned with the GenBank accession HQ159409. Sequences used in the phylogenetic analysis and the alignment of these sequences are presented in Appendix 1.
Table 3. Evidence for existence of full-length or partial sequences of prominin homologs in non-mammalian vertebrates and platypus.
| Species | Prominin-1 | Prominin-2 | Prominin-3 | |
|---|---|---|---|---|
| Prominin-1a | Prominin-1b | |||
| Zebrafish (D.rerio) | NP_001108615 | NP_932337 | Not found | XP_684527 |
| Fugu (T. rubripes) | HQ159405 | HQ159406 | Not found | HQ159407 |
| Frog (X. laevis) | NP_001163920 | NP_001163922 | NP_001163921 | |
| Frog (X. tropicalis) | HQ159400, HQ159401, BC127277 | HQ159402 | HQ159403 | |
| Chicken (G. gallus) | XP_001232165 | XP_425759 | AF406812 | |
| Platypus (O. anatinus) | HQ159408 | XP_001509143 | XP_001511144 | |
Table 4. Presence or absence of alternatively spliced exons in full-length cDNAs of the xlProminin-1 and xtProminin-1 splice variants.
| Sources | Splice variants | 3a | 8a | 11a | 19 | 26b | 27 | 28a | X. laevis (xlProminin-1) | X. tropicalis (xtProminin-1) |
|---|---|---|---|---|---|---|---|---|---|---|
| Retina | s1 | - | - | - | + | - | - | - | JF274976 | HQ159400 |
| s2 | - | - | + | + | - | - | - | JF274977 | ||
| s3 | + | - | + | + | - | - | - | JF274978 | ||
| s4 | - | - | + | + | + | - | - | JF274979 | ||
| s5 | - | - | - | - | + | + | + | JF274980 | HQ159401 | |
| Kidney | s6 | + | - | - | + | + | + | + | FJ358518 | |
| s7 | - | + | + | + | - | - | - | JF274981 |
We compared the results of our phylogenetic analysis of prominin homologs with previous findings and found that X. laevis and X. tropicalis are the only species verified so far to possess all three prominin orthologs. Only two prominin homologs, prominin-1 and 2, were found in human and mouse. We conducted BLAST searches of the NCBI nucleotide database and the Ensembl database for prominin homologs in more species of mammals, including the human, chimpanzee, rhesus monkey, cattle, horse, dog, and rat. All retrieved prominin homolog sequences from these mammals were better grouped with prominin-1 or 2 than with prominin-3. A total of three prominin homologs were found in zebrafish (Danio rerio) and fugu (Takifugu rubripes). However, these fish do not have a prominin-2 homolog; they instead have two prominin-1 homologs, prominin-1a and 1b, and a prominin-3 homolog [52]. Incomplete sequences of prominin homologs were retrieved from EST or genomic databases of the chicken (Gallus gallus) and platypus (Ornithorhynchus anatinus, a primitive egg-laying mammal). GenBank accession numbers of these sequences are given in Table 3. They seem to group with prominin-1, 2, or 3 by sequence alignment. However, further taxonomic studies are needed to precisely position them in the phylogeny of prominins. Three C. elegans prominin homologs and two D. melanogaster prominin homologs were used in constructing the prominin phylogeny. These sequences did not belong to any of the three groups of prominins in our analysis, suggesting the possibility of other prominin homologs.
All three prominin homologs are predicted to be pentatransmembrane glycoproteins
Similar to mammalian prominins, hydropathy analysis using TMHMM software predicted that all X. laevis homologs have five hydrophobic domains, which likely constitute membrane-spanning domains that divide the sequence into two small intracellular loops and two large extracellular domains. However, the last hydrophobic sequence in xlProminin-2 did not reach the set threshold of TMHMM for prediction of a transmembrane domain (Appendix 2). Alignment of translated protein sequences from three cloned X. laevis prominin homolog cDNAs reveals that the most conserved regions are the transmembrane domains, as illustrated in Figure 1. A cysteine-rich region at the junction of the first transmembrane domain and intracellular loop (M1 and I1) is conserved in xlProminin-1 and 3, but is less conserved in xlProminin-2. Multiple leucine residues on the extracellular domains (E2 and E3) are conserved in all three xlProminin homologs. The conserved cysteine-rich region and leucine residues were all previously identified in mammalian prominin-1 and 2 [3,6,53,54]. The extracellular domains of all three xlProminins contain consensus sequences for N-linked glycosylation. In contrast to mouse and human prominin-1, no PDZ-binding motifs were identified in the C-terminal splice variants of xlProminin-1, 2, or 3 using ScanProsite software. PDZ-binding motifs were identified on the C-termini of certain splice variants of prominin-1 in the rodent (s1, s2, s7, s8, and s11) and human (s3, s4, s5, and s9) models, [31] and could potentially modulate interactions of prominin-1 with other proteins.
Three prominin homologs are differentially expressed in Xenopus laevis tissues
Although prominin-1 is broadly expressed, the principle phenotype observed in patients is retinal degeneration [19,20,22-24]. It was proposed that there may be functional and spatial overlap of prominin homologs in most tissues, such that they compensate for one another if a loss of function mutation occurs [3]. We therefore analyzed expression of the three xlProminin homologs in different X. laevis tissues to assess their relative expression levels in those tissues. We were particularly interested in determining whether any prominin homolog is predominantly expressed or absent in particular tissues among the four we studied.
We performed semiquantitative RT–PCR with mRNAs isolated from the X. laevis retina, brain, testis, and kidney to analyze the expression profiles of xlProminin-1, 2, and 3, because these four tissues are mRNA rich and differ significantly in their function and metabolism. The results are presented in Figure 3. Precautions were taken to ensure specificity of the PCR reaction and accuracy of the analysis, including ensuring that measurements were consistent when using two completely different primer pairs for each prominin homolog. We also used relatively large PCR products (>1.4 kb), such that small nonspecific products could easily be recognized and disregarded. For the primer design, we carefully chose primers that only complement the desired homolog. We found that mRNAs of three xlProminin homologs were differentially expressed in these tissues: xlProminin-1 was the predominant form found in the retina, brain, and testis, while x1Promnin-3 was the predominant form found in the kidney. The expression level of xlProminin-2 was the lowest of the three prominin homologs in all examined tissues in this frog.
Figure 3.
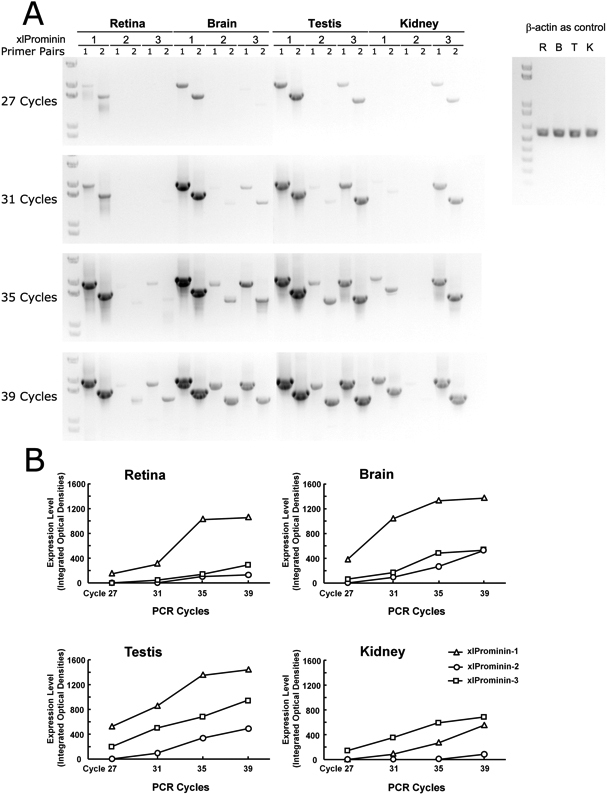
Semiquantitative reverse-transcription PCR analysis of xlProminin-1, 2, and 3 performed on the retina, brain, testis, and kidney. A: Aliquots (20 μl) of the PCR reaction were taken at the end of the 27th, 31st, 35th, and 39th cycles, separated and visualized on a 1% agarose gel. Two independent primer pairs (see Table 2) were used to amplify each xlProminin homolog in each tissue to ensure the specificity and efficiency of the PCR reaction. A fragment of Xenopus laevis β-actin cDNA was also amplified from equivalent starting material of total RNA to serve as a control. The Invitrogen 1KB PLUS DNA (Cat. No. 10787–018) ladder was used for size standards. B: Integrated signals in areas occupied by individual DNA bands were quantified and plotted to compare the relative expression level of xlProminin-1, 2, and 3 in four X. laevis tissues: the retina, brain, testis, and kidney. xlProminin-1, 2 and 3 are expressed in all four tissues. Among the three prominin homologs, prominin-1 is predominantly expressed in the retina, brain, and testis, while prominin-3 is predominantly expressed in the kidney. The expression level of prominin-2 is the lowest of the three prominin homologs in all examined tissues, and is barely detectable in the kidney.
Splice variants of Xenopus laevis prominin-1
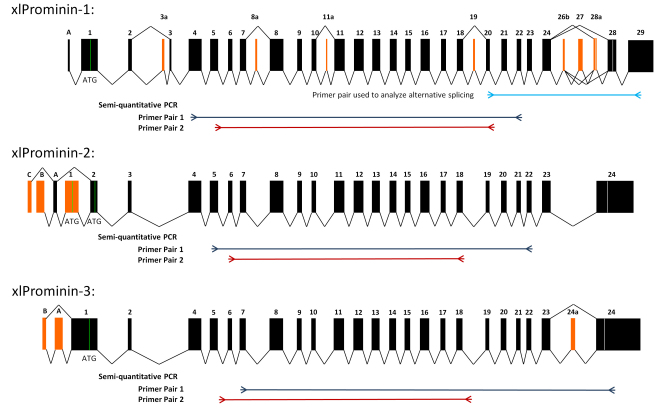
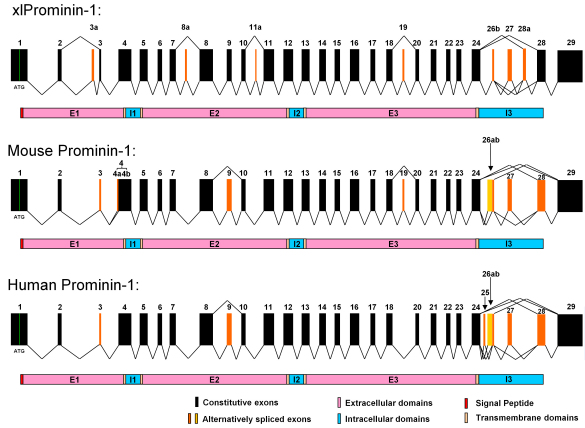
A total of seven full-length splice variants (s1–s7) of xlProminin-1 were cloned and sequenced, and the presence or absence of certain alternatively spliced exons is summarized in Table 4. These splice variants were not named following the nomenclature for splice variants of human and mouse prominin-1 [31], because some alternative exons in xlProminin-1 do not have corresponding exons in human and mouse, and vice versa. Comparison of exon organization of X. laevis, mouse, and human prominin-1 is described in detail below. Alignment of these cloned cDNAs reveals that there are at least seven alternatively spliced exons in the xlProminin-1 gene (Figure 4). We numbered exons following the established nomenclature [31]. Exons 3a (“a” for “alternative”), 8a, 11a, and 28a are newly discovered alternative exons found in X. laevis. They were numbered based on their positions relative to the constitutive exons. Exons 19, 26b, and 27 are alternative exons in the human and mouse that were alternatively spliced in X. laevis. Alternatively spliced exons 26b to 28a of xlProminin-1 reside in a region encoding the C-terminus of the protein. Exons 3 and 28 of xlProminin-1 were always retained, despite the fact that they appear to be alternative in the mouse and human [2,31]. We noticed that the inclusion or deletion of these alternatively spliced exons does not change the downstream reading frame, except for exon 28a, within which an in-frame stop codon resides. Inclusion of exon 28a results in a longer but different protein sequence for the C-terminus of xlProminin-1 relative to other isoforms.
Figure 4.
Exon organization of xlProminin-1, 2, and 3. Homologous exons of xlProminin-1, 2, and 3 are aligned. Genes of all three xlProminins are alternatively spliced and their exon organization is evolutionarily conserved. Constitutive exons are marked in black. Alternatively spliced exons are marked orange. Spliced forms identified in cDNA clones are indicated by joining lines. Translation start sites (ATG) are marked with green lines and the positions of translational stop codons are marked with white lines. Note the complex splicing of exons 26b, 27, and 28a, which generates several distinct isoforms of xlProminin-1. Positions of primers used in reverse-transcription (RT)-PCR for semiquantitative analysis of mRNA expression and for alternative splicing are indicated on the diagram.
Xenopus laevis prominin-2 and 3 are also alternatively spliced
Multiple variants arising from alternative splicing of all three prominin homologs were identified among mRNAs isolated from different tissues (Figure 4). A region near the 5′ end of xlProminin-2 transcript was alternatively spliced. Exon 1 of xlProminin-2 was predicted to contain the translation initiation site (ATG). Surprisingly, exon 1 was skipped in one of the sequenced clones, implying an alternative start codon in exon 2. This proposed alternative start codon was associated with a suboptimal Kozak consensus sequence (−3 CAAATGG +4). The sequence of one full-length xlProminin-2 was obtained; this was assigned GenBank accession FJ358519. The 5′ end of xlProminin-3 transcript and its exon 24a were found to be alternatively spliced. Two full-length splice variants (s1 and s2) of xlProminin-3 were obtained, and assigned GenBank accession numbers FJ358520 and JF274982, respectively. xlProminin-3.s1 contains an alternatively spliced exon 24a, while xlProminin-3.s2 does not. Details of exon organization, alternative 5′ untranslated region (UTR) or alternative polyA sites at 3′ UTR of xlProminin-1, 2, and 3 are provided in Appendix 3. The functional significance of the alternatively spliced variants is unknown.
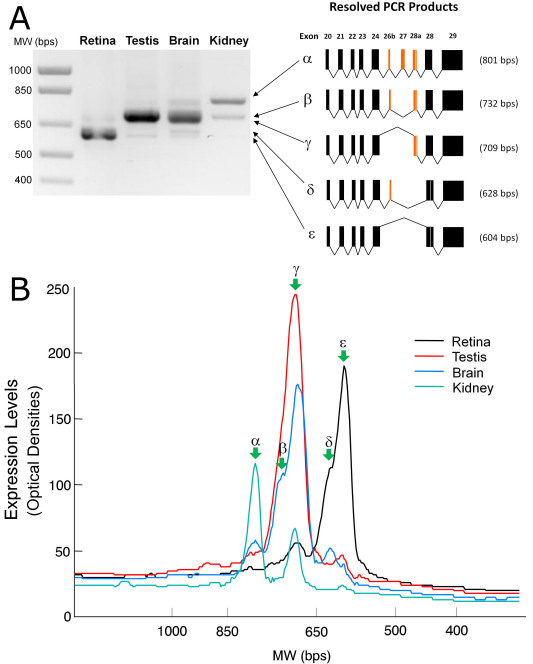
Characterization of alternative splicing of Xenopus laevis prominin-1 in exons 20 to 28
Fargeas et al. previously reported that splicing of the prominin-1 gene in mice results in great variations in the sequences of alternative C-termini [32]. We therefore examined the splicing profile of xlProminin-1 within a region from exon 20 to 28 by RT–PCR (Figure 5). We found that the two most abundant isoforms of xlProminin-1 in the X. laevis retina did not contain exon 27; their exon compositions were: (20–24)+26b-27-28a-28+ (ε in Figure 5) or (20–24)+26b+27-28a-28+ (δ in Figure 5). Isoforms containing exon 28a were also detected in the retina; their exon compositions were: (20–24)+26b-27-28a+28+ (γ in Figure 5), (20–24)+26b+27-28a+28+ (β in Figure 5) and (20–24)+26b+27+28a+28+ (α in Figure 5). However, isoforms containing exon 28a only accounted for a very small proportion of all xlProminin-1 in the retina (Figure 5). This result was unlikely to be biased by preferential amplification of smaller PCR products, since profiling of xlProminin-1 alternative splicing in this region gave very different results in the kidney, testis, and brain: Those tissues preferentially express the isoforms with exon 28a (Figure 5).
Figure 5.
Analysis of the xlProminin-1 alternatively spliced isoforms in four tissues: the retina, brain, testis, and kidney. We found that profiles of the xlProminin-1 alternative splicing are different in these tissues. A: Products of reverse-transcription PCR (RT–PCR) from a region of extensive alternative splicing on the xlProminin-1 gene (exon 20 to 28) were separated on a 1% agarose gel and visualized with ethidium bromide staining. Five discrete PCR products were excised from the gel, cloned, and sequenced. Their exon compositions were determined and each product was designated with a Greek letter (α, β, γ, δ, and ε) for identification when used in the quantitative analysis shown in panel B. The predominant isoforms of xlProminin-1 expressed in the retina lack the alternatively spliced exons 26b, 27, and 28a, whereas the predominant isoform of xlProminin-1 expressed in the kidney retains these exons. The predominant isoforms of xlProminin-1 expressed in the brain and kidney retain exon 28a. Exon 27 was retained only when exon 26b was retained as well. B: Quantification of the xlProminin-1 alternatively spliced isoforms in four tissues: the retina, brain, testis, and kidney. The resolved RT–PCR products were linearly scanned and the optical densities were plotted. Green arrows indicate peaks that represent isoforms resulting from alternative splicing. The major isoform of xlProminin-1 expressed in the retina, ε, lacks the alternatively spliced exons 26b, 27, and 28a, whereas the major form of xlProminin-1 expressed in the kidney, α, retains all possible exons. The majority of xlProminin-1 expressed in the brain and kidney retains exon 28a.
Comparison of the exon organization of Xenopus laevis, mouse, and human prominin-1
The organization and splicing of prominin-1 exons in the mouse and human have been well characterized [30-32]. To determine whether the gene structure is conserved in xlProminin-1, we aligned all identified exons of prominin-1 from X. laevis, mice, and humans (Figure 6). Some exons found in one species were absent in another, but these were always alternatively spliced exons (Figure 6). We also correlated the positions of all alternatively spliced exons of xlProminin-1 to its membrane topology and found that all alternatively spliced exons represent sequences coding for the extracellular domains (E1, E2, and E3) or the C-terminus of the protein, as was previously observed in prominin-1 from mouse and human [3,30-32].
Figure 6.
Comparison of exon organization of prominin-1 gene from X. laevis, mouse, and human. It appears that the gene structure of prominin-1 is evolutionarily conserved in these animals. Alternative splicing of the prominin-1 gene is seen in all three animals, however, with considerable differences in the choices of alternative exons and the splicing patterns. Homologous exons of xlProminin-1 and mouse and human prominin-1 are aligned for comparison of their exon organization. Constitutive exons are marked in black. Alternatively spliced exons are marked in orange. Spliced forms identified in cDNA clones are indicated by jointed lines. Note that homologous exons are assigned with the same number to maintain consistency with preexisting nomenclature [2,31]. Exons 26b and 27 are conserved and alternatively spliced in all three species. Alternative exons 3a, 8a, 11a, and 28a of xlProminin-1 are not found in mouse or human prominin-1. Splicing of alternative exons 4a results from using an alternative 3′ splice site in exon 4, and is only observed in mouse prominin-1 [31]. Alternative exon 19 of xlProminin-1 is alternatively spliced in mouse prominin-1, but no evidence has been found that this exon is alternatively spliced in human prominin-1. Alternative exon 25 of human prominin-1 is not found in prominin-1 from the mouse or X. laevis. The alternative exon 26a of mouse prominin-1 and the alternative exons 25 and 26a of human prominin-1 are not found in xlProminin-1. Exons 3, 9, and 28 of mouse prominin-1 are alternative, but appear to be constitutive in X. laevis. The region encompassing exons 24 to 28 of xlProminin-1 and homologous sequences of the prominin-1 gene from the mouse and human are regions of extensive alternative splicing. Diagrams of translated proteins are aligned with their coding sequences. Divisions of proteins by predicted transmembrane domains are marked.
Discussion
All members of the prominin family are predicted to be pentaspan transmembrane proteins with extracellular N-termini and intracellular C-termini [1,3,7,55]. They have two large extracellular domains (around 250 amino acids each) and two small intracellular domains (around 30 amino acids each). The two extracellular domains are predicted to be N-glycosylated and have conserved leucine residues. The first intracellular domain is unusually rich in cysteine (there are 7, 4, and 8 cysteine residues in the I1 domain of xlProminin-1, 2, and 3, respectively). These features are highly conserved in all identified prominin homologs, including xlProminin-1, 2, and 3 (Figure 1), suggesting that they are functionally important. However, the cysteine-rich domain of xlProminin-2 is not as well conserved as that of xlProminin-1 and 3.
Only two prominin homologs, namely prominin-1 and 2, were found in the human and mouse in previous studies [3,6,8]. Prominins in other mammals are less studied, but consistent with the findings in the human and mouse, we did not find prominin-3 homologs in the chimpanzee, rhesus monkey, horse, cattle, dog, or rat. By contrast, three prominin homologs were identified or their existence was inferred in some nonmammalian vertebrates (Table 3 and Table 4), including fish (D. rerio [52] and T. rubripes), frogs (X. laevis and X. tropicalis), and birds (G. gallus). However, phylogenetic analysis revealed that zebrafish and fugu lack an ortholog of prominin-2. These fish instead have two prominin-1 genes, namely prominin-1a and 1b [52]. In contrast, we determined that both X. laevis and X. tropicalis have all three prominin homologs that are orthologs of prominin-1, 2, and 3 (Figure 2). It appears that the third homolog was lost during mammalian evolution. Interestingly, genomic sequences encoding three prominin homologs are retrievable from the platypus (O. anatinus; Table 3). Studies of platypus prominin may provide clues for how prominin homologs have evolved. It may be that prominin-1 and 2 have extensive functional overlap, since there is shared expression of prominin-1 and 2 in most mouse tissues except the eye [3]. Our results showed that mRNAs of all three xlProminin homologs are expressed in the four examined tissues of X. laevis at various levels (Figure 3). The localization and functions of prominin homologs may overlap, and thus the loss of one homolog through mutation may be compensated for in most tissues by other prominins.
Several prominin homologs have been identified or predicted in C. elegans (e.g., NP_509907, NP_496294, and NP_496296) and D. melanogaster (e.g., NP_647770 and ABH07113) [1,6,56,57], while no prominin homologs have been found in bacteria, fungi, or plants, suggesting that the prominins appeared during metazoan evolution, increased to three genes in nonmammalian vertebrates, and subsequently reduced to two in mammals. Since prominins are not present in any unicellular species whose genome has been sequenced, we postulate that prominin-like proteins are important for intercellular interactions, such as cell-cell adhesion, migration, and communication, or in developmental processes. For example, Zelhof et al. demonstrated that in the Drosophila retina, prominin interacts with an extracellular matrix molecule, spacemaker, to control the apposition of photoreceptor rhabdomeres [57]. Similar to those described in Bardet-Biedl syndrome [58], polydactyly phenotypes observed in PROML11878 (now designated as c.1841delG) patients [19] suggest a role in development: Mutations may directly or indirectly interfere with ciliary signaling pathways such as those mediated by smoothened [59].
Our study specifically addresses the mRNA expression of prominin homologs in X. laevis. The processes of mRNA and protein expression are often not well correlated. mRNAs may be regulated independently of the encoded proteins, and some mRNAs are not translated at all. Such genes are categorized as “pseudogenes” [60]. In fact, nontranslated mRNAs can serve important roles in regulating the expression and degradation of other homologous mRNAs [61-64]. We have verified that the protein encoded by the xlProminin-1 gene is detectable in X. laevis tissues by both immunoblotting and immunohistochemistry (data not shown). To date, we have been unable to determine whether proteins of xlProminin-2 and 3 are synthesized in frog tissues. Nevertheless, the presence of long, uninterrupted, evolutionarily conserved reading frames (Figure 1) suggests that these do not represent pseudogenes.
We observed alternative splicing of all prominin homologs in X. laevis (Figure 4). Alternative splicing sites were not found in exons coding for the two small intracellular loops, but only in exons coding for the three extracellular domains and the C-terminus. A region encompassing exons 26b to 28a of the X. laevis prominin-1 gene is a region of extensive alternative splicing (Figure 5). In extreme cases, alternative splicing of prominin-1 genes can yield proteins with completely different C-termini, which can also vary greatly in length. The significance of these isoforms is unknown. Alternative splicing is regarded as a mechanism to diversify the mRNA products from a single gene, and can confer multifunctionality to gene products by generating isoforms that have novel protein interaction domains or localization signals. We found that certain xlProminin-1 splice variants are preferred in certain tissues; splicing variants with exon composition of 24+26b-27-28a-28+ and 24+26b+27-28a-28+ that encode proteins of shorter C-terminus length are the preferred isoforms in the X. laevis retina (Figure 5). It may therefore prove informative to examine the localization of alternatively spliced forms, or to search for novel interaction partners by techniques such as crosslinking, coimmunoprecipitation, or two-hybrid analysis. Tissue-specific distributions of certain isoforms could also explain the tissue-specific phenotypes (retinal degeneration) observed with mutations in this otherwise broadly expressed gene [19,20,22-24]. Comparison of exon organization of prominin-1 in the human, mouse, and frog reveals that although there are considerable differences in both the existence and composition of certain alternatively spliced exons, all three prominin-1 homologs share similar forms of alternative splicing, especially at the region encoding for the C-termini of the proteins (Figure 6). A recent study found that isoforms s11 and s12 that lack exon 27 are the predominant alternative splicing form in the human retina [24]. These findings support the use of X. laevis as a suitable model system for investigating prominin-1 function in the retina.
Our findings of three prominin homologs in X. laevis and characterization of their mRNA expression and alternative splicing paves the way for future studies of the function of these prominins in photoreceptors in this frog. There are many advantages of studying X. laevis photoreceptors in comparison to mammalian photoreceptors: The photoreceptors of X. laevis are much larger than mammalian photoreceptors, and are easily imaged in detail by confocal microscopy. This frog’s retina contains nearly equal numbers of rods and cones, allowing for comparative investigation of both cell types. Furthermore, studies of photoreceptor outer segment biosynthesis have been conducted in frogs because of their large size and high rates of membrane turnover [34,35]. Finally, transgenic manipulation of X. laevis is comparatively easy and allows for the study of the transgene effects in relatively large numbers of animals in a timely manner [36,37].
Future directions for studying the function of prominins in photoreceptors may be focused on the turnover, transport, subcellular localization, and interacting partners of these molecules in X. laevis. Disease-causing mutations of human prominin-1 could be introduced to xlProminin-1, if they reside in conserved domains, and used to develop useful transgenic X. laevis disease models. This approach has proven successful in many studies, including investigations of the pathogenesis of the rhodopsin P23H mutation [65-67] and T22N mutation of rab8 [68]. None of the currently identified mutations of prominin-1 that cause human disease are located in alternative exons. It is unclear if any of the alternatively spliced isoforms is associated with disease pathogenesis. It would be informative to elucidate the function of each specific splice isoform by the specific knockdown of its mRNA with the RNA interference technique [69,70], which has been successfully applied in X. laevis [71-73].
Acknowledgments
This work was supported by NIH grant R01EY006891 and the John A. and Florence Mattern Solomon Chair in Vision Research and Eye Diseases to D.S.P. We thank Dr. Joseph C. Besharse (Medical College of Wisconsin) for providing the X. laevis retina cDNA library. We thank Dr. Orson L. Moritz (University of British Columbia) for helpful discussion of the manuscript.
Appendix 1. Sequences used in phylogenetic analysis.
To access the file, click or select the words “Appendix 1.” This will initiate the download of a pdf archive that contains the file.
Appendix 2. Prediction of membrane topology of three xlProminin homologs.
To access the file, click or select the words “Appendix 2.” This will initiate the download of a pdf archive that contains the file.
Appendix 3. Exon organization of the xlProminin genes.
To access the file, click or select the words “Appendix 3.” This will initiate the download of a pdf archive that contains the file.
References
- 1.Corbeil D, Roper K, Fargeas CA, Joester A, Huttner WB. Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic. 2001;2:82–91. doi: 10.1034/j.1600-0854.2001.020202.x. [DOI] [PubMed] [Google Scholar]
- 2.Fargeas CA, Corbeil D, Huttner WB. AC133 antigen, CD133, prominin-1, prominin-2, etc.: prominin family gene products in need of a rational nomenclature. Stem Cells. 2003;21:506–8. doi: 10.1634/stemcells.21-4-506. [DOI] [PubMed] [Google Scholar]
- 3.Fargeas CA, Florek M, Huttner WB, Corbeil D. Characterization of prominin-2, a new member of the prominin family of pentaspan membrane glycoproteins. J Biol Chem. 2003;278:8586–96. doi: 10.1074/jbc.M210640200. [DOI] [PubMed] [Google Scholar]
- 4.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 5.Jászai J, Fargeas CA, Florek M, Huttner WB, Corbeil D. Focus on molecules: prominin-1 (CD133). Exp Eye Res. 2007;85:585–6. doi: 10.1016/j.exer.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Weigmann A, Corbeil D, Hellwig A, Huttner WB. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci USA. 1997;94:12425–30. doi: 10.1073/pnas.94.23.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–12. [PubMed] [Google Scholar]
- 8.Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, Bray RA, Waller EK, Buck DW. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013–21. [PubMed] [Google Scholar]
- 9.Kania G, Corbeil D, Fuchs J, Tarasov KV, Blyszczuk P, Huttner WB, Boheler KR, Wobus AM. Somatic stem cell marker prominin-1/CD133 is expressed in embryonic stem cellderived progenitors. Stem Cells. 2005;23:791–804. doi: 10.1634/stemcells.2004-0232. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L, Wei X, Cheng L, Tian J, Jiang JJ. CD133, one of the markers of cancer stem cells in Hep-2 cell line. Laryngoscope. 2007;117:455–60. doi: 10.1097/01.mlg.0000251586.15299.35. [DOI] [PubMed] [Google Scholar]
- 11.Suetsugu A, Nagaki M, Aoki H, Motohashi T, Kunisada T, Moriwaki H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;351:820–4. doi: 10.1016/j.bbrc.2006.10.128. [DOI] [PubMed] [Google Scholar]
- 12.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florek M, Haase M, Marzesco AM, Freund D, Ehninger G, Huttner WB, Corbeil D. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 2005;319:15–26. doi: 10.1007/s00441-004-1018-z. [DOI] [PubMed] [Google Scholar]
- 14.Hilbe W, Dirnhofer S, Oberwasserlechner F, Schmid T, Gunsilius E, Hilbe G, Woll E, Kahler CM. CD133 positive endothelial progenitor cells contribute to the tumour vasculature in non-small cell lung cancer. J Clin Pathol. 2004;57:965–9. doi: 10.1136/jcp.2004.016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepherd CJ, Rizzo S, Ledaki I, Davies M, Brewer D, Attard G, de Bono J, Hudson DL. Expression profiling of CD133+ and CD133- epithelial cells from human prostate. Prostate. 2008;68:1007–24. doi: 10.1002/pros.20765. [DOI] [PubMed] [Google Scholar]
- 16.Marzesco AM, Janich P, Wilsch-Brauninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB. Release of extracellular membrane particles carrying the stem cell marker Three X. laevis homologues of prominin 25 prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci. 2005;118:2849–58. doi: 10.1242/jcs.02439. [DOI] [PubMed] [Google Scholar]
- 17.Corbeil D, Roper K, Hellwig A, Tavian M, Miraglia S, Watt SM, Simmons PJ, Peault B, Buck DW, Huttner WB. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J Biol Chem. 2000;275:5512–20. doi: 10.1074/jbc.275.8.5512. [DOI] [PubMed] [Google Scholar]
- 18.Corbeil D, Joester A, Fargeas CA, Jaszai J, Garwood J, Hellwig A, Werner HB, Huttner WB. Expression of distinct splice variants of the stem cell marker prominin-1 (CD133) in glial cells. Glia. 2009;57:860–74. doi: 10.1002/glia.20812. [DOI] [PubMed] [Google Scholar]
- 19.Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, Kumaramanickavel G, John S, Nancarrow D, Roper K, Weigmann A, Huttner WB, Denton MJ. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet. 2000;9:27–34. doi: 10.1093/hmg/9.1.27. [DOI] [PubMed] [Google Scholar]
- 20.Zacchigna S, Oh H, Wilsch-Brauninger M, Missol-Kolka E, Jaszai J, Jansen S, Tanimoto N, Tonagel F, Seeliger M, Huttner WB, Corbeil D, Dewerchin M, Vinckier S, Moons L, Carmeliet P. Loss of the cholesterol-binding protein prominin-1/CD133 causes disk dysmorphogenesis and photoreceptor degeneration. J Neurosci. 2009;29:2297–308. doi: 10.1523/JNEUROSCI.2034-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbeil D, Roper K, Hannah MJ, Hellwig A, Huttner WB. Selective localization of the polytopic membrane protein prominin in microvilli of epithelial cells - a combination of apical sorting and retention in plasma membrane protrusions. J Cell Sci. 1999;112:1023–33. doi: 10.1242/jcs.112.7.1023. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Chen Y, Lillo C, Chien J, Yu Z, Michaelides M, Klein M, Howes KA, Li Y, Kaminoh Y, Chen H, Zhao C, Al-Sheikh YT, Karan G, Corbeil D, Escher P, Kamaya S, Li C, Johnson S, Frederick JM, Zhao Y, Wang C, Cameron DJ, Huttner WB, Schorderet DF, Munier FL, Moore AT, Birch DG, Baehr W, Hunt DM, Williams DS, Zhang K. Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J Clin Invest. 2008;118:2908–16. doi: 10.1172/JCI35891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaelides M, Gaillard MC, Escher P, Tiab L, Bedell M, Borruat FX, Barthelmes D, Carmona R, Zhang K, White E, McClements M, Robson AG, Holder GE, Bradshaw K, Hunt DM, Webster AR, Moore AT, Schorderet DF, Munier FL. The PROM1 mutation p.R373C causes an autosomal dominant bull's eye maculopathy associated with rod, rod-cone, and macular dystrophy. Invest Ophthalmol Vis Sci. 2010;51:4771–80. doi: 10.1167/iovs.09-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Permanyer J, Navarro R, Friedman J, Pomares E, Castro-Navarro J, Marfany G, Swaroop A, Gonzalez-Duarte R. Autosomal recessive retinitis pigmentosa with early macular affectation caused by premature truncation in PROM1. Invest Ophthalmol Vis Sci. 2010;51:2656–63. doi: 10.1167/iovs.09-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florek M, Bauer N, Janich P, Wilsch-Braeuninger M, Fargeas CA, Marzesco AM, Ehninger G, Thiele C, Huttner WB, Corbeil D. Prominin-2 is a cholesterol-binding protein associated with apical and basolateral plasmalemmal protrusions in polarized epithelial cells and released into urine. Cell Tissue Res. 2007;328:31–47. doi: 10.1007/s00441-006-0324-z. [DOI] [PubMed] [Google Scholar]
- 26.Huttner HB, Janich P, Kohrmann M, Jaszai J, Siebzehnrubl F, Blumcke I, Suttorp M, Gahr M, Kuhnt D, Nimsky C, Krex D, Schackert G, Lowenbruck K, Reichmann H, Juttler E, Hacke W, Schellinger PD, Schwab S, Wilsch-Brauninger M, Marzesco AM, Corbeil D. The stem cell marker prominin-1/CD133 on membrane particles in human cerebrospinal fluid offers novel approaches for studying central nervous system disease. Stem Cells. 2008;26:698–705. doi: 10.1634/stemcells.2007-0639. [DOI] [PubMed] [Google Scholar]
- 27.Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Bräuninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–95. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jászai J, Fargeas CA, Haase M, Farkas LM, Huttner WB, Corbeil D. Robust expression of Prominin-2 all along the adult male reproductive system and urinary bladder. Histochem Cell Biol. 2008;130:749–59. doi: 10.1007/s00418-008-0445-4. [DOI] [PubMed] [Google Scholar]
- 29.Jászai J, Janich P, Farkas LM, Fargeas CA, Huttner WB, Corbeil D. Differential expression of Prominin-1 (CD133) and Prominin-2 in major cephalic exocrine glands of adult mice. Histochem Cell Biol. 2007;128:409–19. doi: 10.1007/s00418-007-0334-2. [DOI] [PubMed] [Google Scholar]
- 30.Kemper K, Tol MJ, Medema JP. Mouse tissues express multiple splice variants of prominin-1. PLoS ONE. 2010;5:e12325. doi: 10.1371/journal.pone.0012325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fargeas CA, Huttner WB, Corbeil D. Nomenclature of prominin-1 (CD133) splice variants - an update. Tissue Antigens. 2007;69:602–6. doi: 10.1111/j.1399-0039.2007.00825.x. [DOI] [PubMed] [Google Scholar]
- 32.Fargeas CA, Joester A, Missol-Kolka E, Hellwig A, Huttner WB, Corbeil D. Identification of novel Prominin-1/CD133 splice variants with alternative C-termini and their expression in epididymis and testis. J Cell Sci. 2004;117:4301–11. doi: 10.1242/jcs.01315. [DOI] [PubMed] [Google Scholar]
- 33.Jászai J, Fargeas CA, Florek M, Huttner WB, Corbeil D. Focus on molecules: prominin-1 (CD133). Exp Eye Res. 2007;85:585–6. doi: 10.1016/j.exer.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 34.Besharse JC, Hollyfield JG, Rayborn ME. Turnover of rod photoreceptor outer segments. II. Membrane addition and loss in relationship to light. J Cell Biol. 1977;75:507–27. doi: 10.1083/jcb.75.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollyfield JG, Besharse JC, Rayborn ME. Turnover of rod photoreceptor outer segments. I. Membrane addition and loss in relationship to temperature. J Cell Biol. 1977;75:490–506. doi: 10.1083/jcb.75.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–83. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 37.Moritz OL, Tam BM, Knox BE, Papermaster DS. Fluorescent photoreceptors of transgenic Xenopus laevis imaged in vivo by two microscopy techniques. Invest Ophthalmol Vis Sci. 1999;40:3276–80. [PubMed] [Google Scholar]
- 38.Green CB, Cahill GM, Besharse JC. Tryptophan hydroxylase is expressed by photoreceptors in Xenopus laevis retina. Vis Neurosci. 1995;12:663–70. doi: 10.1017/s0952523800008956. [DOI] [PubMed] [Google Scholar]
- 39.Bertioli D. Rapid amplification of cDNA ends. Methods Mol Biol. 1997;67:233–8. doi: 10.1385/0-89603-483-6:233. [DOI] [PubMed] [Google Scholar]
- 40.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–9. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Wong S, Laflèche J, Crowe S, Mesana TG, Suuronen EJ, Ruel M. In vitro functional comparison of therapeutically relevant human vasculogenic progenitor cells used for cardiac cell therapy. J Thorac Cardiovasc Surg. 2010;140:216–24. doi: 10.1016/j.jtcvs.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 43.Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–72. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohshima Y, Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987;195:247–59. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skowasch D, Jabs A, Andrie R, Dinkelbach S, Luderitz B, Bauriedel G. Presence of bone-marrow- and neural-crest-derived cells in intimal hyperplasia at the time of clinical in-stent restenosis. Cardiovasc Res. 2003;60:684–91. doi: 10.1016/j.cardiores.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 48.Ye J, Song YS, Kang SH, Yao K, Kim JC. Involvement of bone marrow-derived stem and progenitor cells in the pathogenesis of pterygium. Eye (Lond) 2004;18:839–43. doi: 10.1038/sj.eye.6701346. [DOI] [PubMed] [Google Scholar]
- 49.Hao SG, Sun GL, Wu WL, Wu YL. Studies on the dynamics of biological characteristics of CD133+ cells from human umbilical cord blood during short-term culture. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2003;11:569–75. [PubMed] [Google Scholar]
- 50.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–5. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girish V, Vijayalakshmi A. Affordable image analysis using NIH Image/ImageJ. Indian J Cancer. 2004;41:47. [PubMed] [Google Scholar]
- 52.McGrail M, Batz L, Noack K, Pandey S, Huang Y, Gu X, Essner JJ. Expression of the zebrafish CD133/prominin1 genes in cellular proliferation zones in the embryonic central nervous system and sensory organs. Dev Dyn. 2010;239:1849–57. doi: 10.1002/dvdy.22307. [DOI] [PubMed] [Google Scholar]
- 53.Corbeil D, Fargeas CA, Huttner WB. Rat prominin, like its mouse and human orthologues, is a pentaspan membrane glycoprotein. Biochem Biophys Res Commun. 2001;285:939–44. doi: 10.1006/bbrc.2001.5271. [DOI] [PubMed] [Google Scholar]
- 54.Miraglia S, Godfrey W, Buck D. A response to AC133 hematopoietic stem cell antigen: human homologue of mouse kidney prominin or distinct member of a novel protein family? Blood. 1998;91:4390–1. [PubMed] [Google Scholar]
- 55.Shmelkov SV, St Clair R, Lyden D, Rafii S. AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 2005;37:715–9. doi: 10.1016/j.biocel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Corbeil D, Roper K, Weigmann A, Huttner WB. AC133 hematopoietic stem cell antigen: human homologue of mouse kidney prominin or distinct member of a novel protein family? Blood. 1998;91:2625–6. [PubMed] [Google Scholar]
- 57.Zelhof AC, Hardy RW, Becker A, Zuker CS. Transforming the architecture of compound eyes. Nature. 2006;443:696–9. doi: 10.1038/nature05128. [DOI] [PubMed] [Google Scholar]
- 58.Steiner PA. Bardet-Biedl syndrome. J Am Optom Assoc. 1990;61:852–5. [PubMed] [Google Scholar]
- 59.Stark DR. Hedgehog signalling: pulling apart patched and smoothened. Curr Biol. 2002;12:R437–9. doi: 10.1016/s0960-9822(02)00920-x. [DOI] [PubMed] [Google Scholar]
- 60.Mombaerts P. The human repertoire of odorant receptor genes and pseudogenes. Annu Rev Genomics Hum Genet. 2001;2:493–510. doi: 10.1146/annurev.genom.2.1.493. [DOI] [PubMed] [Google Scholar]
- 61.Mitrovich QM, Anderson P. mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol. 2005;15:963–7. doi: 10.1016/j.cub.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 62.Yano Y, Saito R, Yoshida N, Yoshiki A, Wynshaw-Boris A, Tomita M, Hirotsune S. A new role for expressed pseudogenes as ncRNA: regulation of mRNA stability of its homologous coding gene. J Mol Med. 2004;82:414–22. doi: 10.1007/s00109-004-0550-3. [DOI] [PubMed] [Google Scholar]
- 63.Yoo M, Steggles AW. The characterization of three types of partially processed mRNA and two pseudogenes for human liver cytochrome b5. Biochem Biophys Res Commun. 1989;163:18–24. doi: 10.1016/0006-291x(89)92092-5. [DOI] [PubMed] [Google Scholar]
- 64.MacLeod AR, Houlker C, Reinach FC, Talbot K. The mRNA and RNA-copy pseudogenes encoding TM30nm, a human cytoskeletal tropomyosin. Nucleic Acids Res. 1986;14:8413–26. doi: 10.1093/nar/14.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tam BM, Qazalbash A, Lee HC, Moritz OL. The dependence of retinal degeneration caused by the rhodopsin P23H mutation on light exposure and vitamin a deprivation. Invest Ophthalmol Vis Sci. 2010;51:1327–34. doi: 10.1167/iovs.09-4123. [DOI] [PubMed] [Google Scholar]
- 66.Tam BM, Moritz OL. Dark rearing rescues P23H rhodopsin-induced retinal degeneration in a transgenic Xenopus laevis model of retinitis pigmentosa: a chromophore-dependent mechanism characterized by production of N-terminally truncated mutant rhodopsin. J Neurosci. 2007;27:9043–53. doi: 10.1523/JNEUROSCI.2245-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tam BM, Moritz OL. Characterization of rhodopsin P23H-induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47:3234–41. doi: 10.1167/iovs.06-0213. [DOI] [PubMed] [Google Scholar]
- 68.Moritz OL, Tam BM, Hurd LL, Peranen J, Deretic D, Papermaster DS. Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol Biol Cell. 2001;12:2341–51. doi: 10.1091/mbc.12.8.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tabara H, Grishok A, Mello CC. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–1. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 70.Timmons L, Tabara H, Mello CC, Fire AZ. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol Biol Cell. 2003;14:2972–83. doi: 10.1091/mbc.E03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li M, Rohrer B. Gene silencing in Xenopus laevis by DNA vector-based RNA interference and transgenesis. Cell Res. 2006;16:99–105. doi: 10.1038/sj.cr.7310013. [DOI] [PubMed] [Google Scholar]
- 72.Nakano H, Amemiya S, Shiokawa K, Taira M. RNA interference for the organizerspecific gene Xlim-1 in Xenopus embryos. Biochem Biophys Res Commun. 2000;274:434–9. doi: 10.1006/bbrc.2000.3178. [DOI] [PubMed] [Google Scholar]
- 73.Chen CM, Chiu SL, Shen W, Cline HT. Co-expression of Argonaute2 Enhances Short Hairpin RNA-induced RNA Interference in Xenopus CNS Neurons In Vivo. Front Neurosci. 2009;3:63. doi: 10.3389/neuro.17.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]