Abstract
Inflammatory bowel disease appears to result from an abnormal host immune response to the intestinal microbiota. Experimental models have allowed the dissection of the complex dialogue between the host and its microbiota. Through genetic manipulation of the host genome the immune compartments, cells, molecules, and genes that are critical for maintenance of intestinal homeostasis are being identified. Genetic association studies in humans have identified over 100 susceptibility loci. Although there is remarkable coherence between the experimental model and the human genetic data, a full understanding of the mechanisms involved in genetic susceptibility to IBD and of gene-gene and gene-environmental interactions, will require a “next generation” of experimental models.
Introduction
Inflammatory bowel diseases (IBD) constitute a significant health burden in developed countries impacting the quality of life of some 1.4 million individuals in North America and 2.2 million individuals in Europe (Loftus, 2004). The two main forms of IBD are ulcerative colitis and Crohn's disease, which occur at a comparable incidence of three to twenty new cases per 100,000 persons per year in the USA and Canada and have an equivalent prevalence in males and females. While the nature and anatomical location of the inflammatory pathology differ between the two disorders, it is thought that both arise as a result of an abnormal immune response to the intestinal microbiota in genetically predisposed individuals. In Crohn's disease inflammation is transmural, can be granulomatous, and occurs in any part of the gastro-intestinal tract although the ileum is mainly affected, whereas in ulcerative colitis, the pathology impacts primarily the colonic mucosa (Podolsky, 2002). Since the 1940s, the incidence of IBD has dramatically increased in countries with a more ‘westernized’ lifestyle, suggesting the influence of environmental factors, including lifestyle, hygiene, diet and use of antibiotics, all of which may alter the microbiota in favor of disease onset and/or progression (Shanahan and Bernstein, 2009). Similar to other multi-factorial diseases, the rate of incidence of Crohn's disease among monozygotic twins is higher than for dizygotic twins. In addition, familial and ethnic aggregations and segregation studies also strongly suggest the influence of genetic factors in the etiology of IBD, and more precisely the presence of recessive genetic determinants with incomplete penetrance (Cho, 2008). Detailed linkage analyses in IBD-affected families using genome-wide scanning, and more recently genome-wide association studies (GWAS), have revealed more than 100 genetic loci that show significant association with IBD (Franke et al., 2010a; McGovern et al., 2010). Although these loci account for only 20% of the estimated genetic risk for IBD, they point to specific pathways with implications in intestinal homeostasis. Namely, NOD2-dependent innate immunity, the inflammasome pathway, autophagy and Interleukin-23 (IL-23)-IL-17 circuitry have surfaced as potentially necessary for epithelial barrier integrity, bacterial handling and immune tolerance.
IBD patients are generally treated with anti-inflammatory and immunosuppressive drugs, antibiotics, biologicals such as anti-TNF therapies and/or surgery (Van Assche et al., 2009). While managing some symptoms of the disease, these strategies do not cure IBD patients from their periodic and life-long illness. Alternative approaches aim at modifying the microbiota, through probiotics or bacteriotherapy (Gareau et al., 2010), or at modulating the polarity of the immune response as with helminth therapy (Weinstock and Elliott, 2009). A better understanding of the etiopathology and mechanistic basis of IBD is thus needed for the development of novel targeted therapeutics. Experimental animal models of the disease, although not fully representative of human IBD, recapitulate aspects of the problem and have provided important insights into the role of the pathways, cells and molecules required for intestinal homeostasis. Notably, the results of human genetic studies are remarkably coherent with the data derived from experimental models. In this review, we examine some of the most commonly used animals models of IBD -genetic, chemical or induced by immune cell transfer - and explore their relevance in dissecting mucosal immunity and mechanisms of intestinal pathologies.
The host-microbiota ‘dialogue’
The human body is colonized at birth by a wide spectrum of microorganisms that numerically exceed host cells by approximately ten fold. The intestinal tract is the densest microbial ecosystem found in nature harboring 500–1000 different species of bacteria, protozoa and fungi. Collectively, these microorganisms contain around 100 times as many genes as there are in the human genome (Gill et al., 2006), and it is argued that this vast microbiome is a strong determinant of health and disease. Our co-existence with the microbiota is a dynamic and mutually beneficial one. In addition to a profound influence on host metabolism (Backhed et al., 2004; Backhed et al., 2005) and intestinal development, the commensal flora shapes the immune system both at mucosal surfaces (Hand and Belkaid, 2010; Rescigno, 2009) and systemically (Clarke et al., 2010), and actively protects from enteric pathogenic infections by colonization resistance and by synthesizing factors promoting mutualism. For instance, polysaccharide A produced by Bacteroides fragilis suppresses IL-17 production in the intestine and promotes the function of IL-10-producing CD4+ T cells, which confers protection in an experimental model of colitis induced by Helicobacter hepaticus (Mazmanian et al., 2008). Similarly, induction of a transforming growth factor-β (TGF-β) rich environment by indigenous Clostridium species promotes T regulatory (Treg) cell accumulation in the colon and resistance to colitis (Atarashi et al., 2011). Sensing of the commensal flora by the innate immune system is also critical for intestinal homeostasis and tissue repair following injury. Consistently, depletion of commensal microorganisms by antibiotics treatment, or deficiency in central innate immunity effectors such as the NLRP3 inflammasome and its central enzyme caspase-1, or the Toll-like receptors (TLRs)-IL-1 receptor family (IL-1R, IL-18R) common adaptor MyD88, impairs mucosal regeneration following injury by oral administration of dextran sulfate sodium (DSS), which leads to mucosal injury and colitis (Saleh and Trinchieri, 2011).
Whereas the microbiota is protective and necessary for host physiology, it becomes a liability when coupled with a physical, chemical or immunological insult in genetically predisposed individuals. For instance, segmented filamentous bacteria (SFB), found in mice from certain production facilities, promote Th17 cell development in the intestine, which is linked to local as well as systemic pathologic effects impacting autoimmunity and cancer (Ivanov et al., 2009; Westbrook et al., 2009; Wu et al., 2010). Accumulating evidence indicates that host genotypes reciprocally affect microbiota composition, which in turn alters host responses. The most striking example is that of Tbx21-/- (Tbx21 encodes the transcription factor T-bet) Rag2-/- (TRUC) mice that develop a spontaneous and transferrable form of ulcerative colitis (Garrett et al., 2007; Garrett et al., 2009). Deficiency of T-bet in the innate immune system leads to exaggerated tumor necrosis factor (TNF) production by dendritic cells, which together with the absence of Treg cells creates a chronic inflammatory state that modulates the composition of the microflora. Specifically, two bacterial species, Proteus mirabilis and Klebsiella pneumoniae, are associated with the transfer of colitis to wild-type mice (Garrett et al., 2010). Thus, the environment-microbiota-genetics interaction might explain in part why disease develops in only a small fraction of individuals carrying common risk alleles of disease susceptibility genes. It is clear that the microbiota is the buffer between the environment and the host, and much effort is now invested at deciphering its functionality in health and disease. Various human microbiome projects including the NIH Human Microbiome Project (http://commonfund.nih.gov/hmp/) and the International Human Microbiome Consortium (http://www.human-microbiome.org/) have been established with the aim of defining microbes and microbial genes (metagenome) that are present at different anatomical sites.
The role of the intestinal epithelial barrier in gut homeostasis and pathologies
Intestinal epithelial cells and the barrier they form are critical for host-microbiota mutualism in the gut. Various animal models derived primarily from gene targeting approaches and/or chemical models of colitis (e.g. DSS) have investigated the role of the epithelial barrier in intestinal homeostasis and have revealed critical epithelial mechanisms deregulated in IBD (Table 1). The intestinal epithelium is composed of a single layer of intestinal epithelial cells (IECs), including absorptive enterocytes, goblet cells, Paneth cells and enteroendocrine cells, which collectively cover a surface area of 400 m2, tightly packed into numerous villi and crypts. The integrity of this barrier is crucial to shield the host from commensal microorganisms and foodstuffs in the intestinal lumen and to prevent pathologic antigen-specific immune responses, as evident in IBD and celiac disease.
Table 1. Experimental models of colitis.
| Gene involved | Mechanism | Microbiota or Environment | |
|---|---|---|---|
| Spontaneous | |||
| TRUC | Tbx21-/-Rag2-/- | ↑TNF (innate) ↓Treg (adaptive) | Klebsiella pneumoniae Proteus mirabilis |
| MUC2 | Muc2-/- | ↓mucin (epithelial) | – |
| NEMO | IkbkgIEC | ↑apoptosis (epithelial) | – |
| IKKa and IKKb | Ikbka-IkbkbIEC | ↑apoptosis (epithelial) | – |
| MDR1 | Abcb1b-/- | ↑xenobiotic substances (epithelial) | – |
| TGFb | Tgfb1-/- | ↓Treg (adaptive) | – |
| TGFbRII | Tgfbr2-/- | ↓Treg (adaptive) | – |
| IL-10 | Il10-/- | ↓Treg (adaptive) | – |
| IL-10R1 | Il10r1-/- | ↓Treg (adaptive) | – |
| IL-2 | Il2-/- | ↓Treg (adaptive) | – |
| FOXP3 | Foxp3-/- | ↓Treg (adaptive) | – |
| Microbial | |||
| H. hepaticus | – | ↑Th17 (adaptive) | Bacteroides fragilis (protective-↑IL-10-↓IL-17) |
| Segmented filamentous bacteria (SFB) | – | ↑Th17 (adaptive) | – |
| C. jejuni | Muc1 | ↓mucin (epithelial) | – |
| H. hepaticus | Nod2 | ↓Defensin (epithelial) HD5IEC transgenic (protective) | – |
| Chemical | |||
| DSS | |||
| Tnfaip3IEC | ↑apoptosis (epithelial) | – | |
| Tlr2-/-, Tlr4-/- | ↓Tissue repair (innate) | – | |
| Myd88-/- | ↓Tissue repair (innate) | – | |
| Nlrp3-/- | ↓Tissue repair (innate) | – | |
| Casp1-/- | ↓Tissue repair (innate) | – | |
| Il18-/- | ↓Tissue repair (innate) | – | |
| Il18r1-/- | ↓Tissue repair (innate) | – | |
| Nod2+/+ | MDP protective ↑Tissue repair (innate) | – | |
| – | Clostridium species (protective-↑TGF-β-↑Treg) | ||
| Atg16l1-/- | ↑inflammasome (innate) | ||
| Atg16l1HM | ↓Paneth cell function | Norovirus+microbiota | |
| Immune | |||
| CD4+CD45+Rb(hi) transfer | |||
| Mylk transgenic | Disrupted tight junctions (epithelial) | – | |
| Stat4 transgenic | ↑Th1 (adaptive) | ||
| – | IL-23 neutralization protective ↓Th17 (adaptive) | – | |
| – | CD4+CD25+ cotransfer (↑Treg) protective (adaptive) | – | |
| Microbiota-reactive memory CD4+ Th1 cells transfer | – | ↑Th1 (adaptive) | |
| microbiota-reactive memory CD4+ Th17 cells transfer | – | ↑Th17 (adaptive) |
Each IEC subtype contributes to barrier integrity through specialized mechanisms (Fig. 1). The enterocytes migrate out of the crypts to form the surface epithelium and ensure barrier function through an apical brush border and intercellular tight junctions. Targeted disruption of tight junctions in genetically modified mice results in increased susceptibility to chemically induced colitis (Su et al., 2009), which is consistent with observations of altered tight junction expression and/or localization in IBD patients (Mankertz and Schulzke, 2007). Goblet cells reinforce the enterocyte fence by producing mucus that forms a protective polysaccharide glycocalyx bilayer, in which the firm inner layer is devoid of bacteria (Johansson et al., 2008). Gene targeting approaches of Muc1 or Muc2, which encode key constituents of mucus, have similarly highlighted the critical role of this physical barrier in preventing disease onset. It has been shown that Muc1 ablation enhances small intestinal damage following infection with C. jejuni (McAuley et al., 2007) and, more strikingly, mucin-2-deficiency results in spontaneous chronic colitis and colitis-associated colorectal cancer (Heazlewood et al., 2008; Van der Sluis et al., 2006; Velcich et al., 2002). Notably, altered expression and glycosylation of mucin-1 have been reported in IBD (Campbell et al., 2001) and MUC1 is one of the newly implicated loci in Crohn's disease pathogenecity (Franke et al., 2010a).
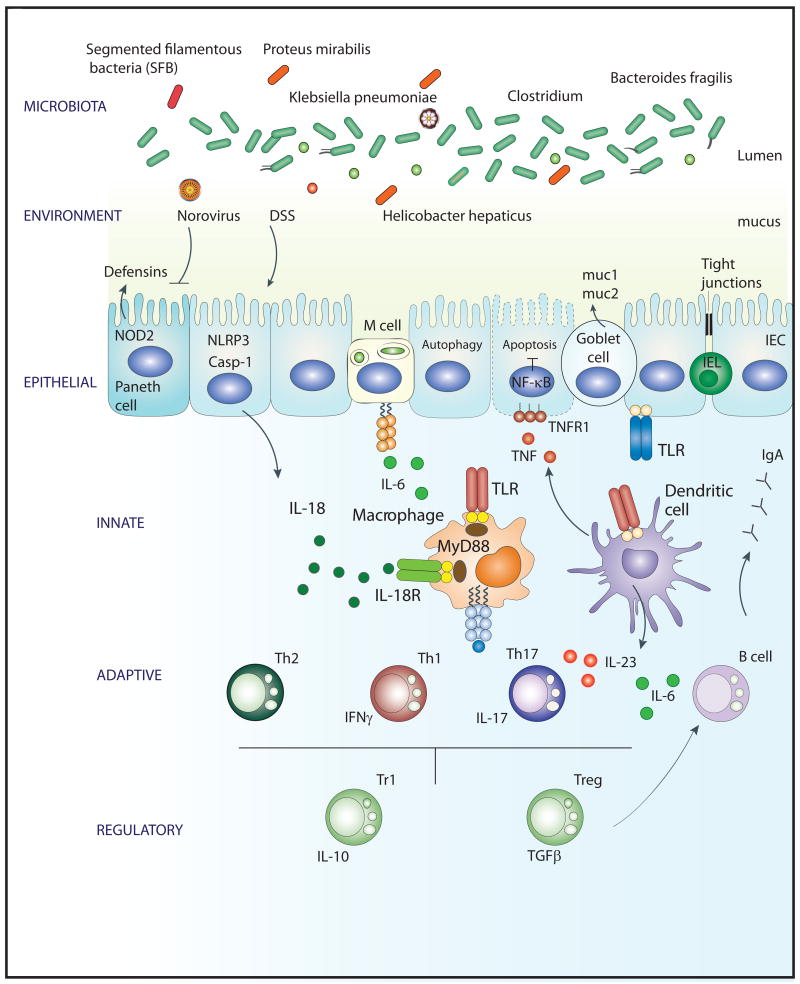
Figure 1.
Intestinal homeostasis is maintained through a dialogue between the host immune system (innate, adaptive and regulatory) and the microbiota, a crosstalk modified by host genetics (e.g. susceptibility loci identified by GWAS) and environmental triggers (e.g. DSS, norovirus, H. hepaticus). Various commensal bacterial species enhance mutualism with the host (e.g. Bacteroides fragilis and clostridium species) and others induce intestinal pathology (e.g. SFB, Klebsiella pneumonia and Proteus mirabilis). Epithelial defenses include mucus production by goblet cells, defensin production by paneth cells and colonocytes, intact tight junctions between intestinal epithelial cells (IECs), IEC anti-apoptotic mechanisms and autophagy. Pattern recognition receptors of the innate immune system, including TLRs, NLRP3 and NOD2, sense microbial and danger motifs and are required for mucosal regeneration and tissue repair. In addition, they are necessary for activation and sculpting of the adaptive and regulatory immune responses.
Paneth cells produce anti-microbial peptides including α-defensins (cryptidins), β-defensins and cathelicidins, and modulation of α-defensin expression or their maturation by matrix metalloproteinase (MMP)-7 in genetically modified mice have revealed an important homeostatic role of defensins on intestinal microbial ecology (Salzman et al., 2010). Indeed, transgenic expression of human α-defensin 5 (HD5) in mice has led to significant reduction in SFB and IL-17-producing T cells in the lamina propria, which is in support of earlier findings that Crohn's disease patients exhibit defective defensin production (Wehkamp et al., 2005).
To ensure barrier integrity, the epithelial layer is constantly renewed through daily cycles of IEC proliferation, migration and apoptosis along the crypt-villus axis, and disruption of these homeostatic processes is linked to disease. For instance, targeted mouse strains with defects in IEC survival, through ablation of the NF-κB pro-survival pathway, develop spontaneous IBD-like intestinal pathologies. Notably, enterocyte-specific deletion of NF-κB essential modulator (NEMO) or both IκB kinase (IKK) α and IKKβ leads to spontaneous IEC apoptosis and massive intestinal inflammation (Nenci et al., 2007). Inhibition of TNFR1 signaling in these mice prevents IEC apoptosis and the consequent development of intestinal inflammation, which is consistent with the positive effects of biologicals targeting the TNF pathway on restoring barrier function in IBD patients (Zeissig et al., 2004). Consistently, IEC-specific deletion of TNFAIP3 (encoding A20), a gene mutated in Crohn's disease (Welcome Trust Case Control Consortium, 2007) that encodes an E3 ligase-editing enzyme involved in dampening NF-κB signaling and TNF-induced apoptosis, leads to massive IEC apoptosis and increased susceptibility to experimental colitis (Vereecke et al., 2010). Another animal model that further illustrates the link between impaired IEC integrity and IBD is that of defective multidrug resistance (MDR) 1 function. Deficiency in MDR1, a trans-membrane P-glycoprotein involved in xenobiotic substances transport and removal, results in spontaneous development of severe IBD, which is dependent on the microflora and the non-hematopoietic compartment of the gut, as revealed by bone-marrow chimera experiments (Panwala et al., 1998).
Crosstalk of ‘first responders’ innate immune cells with the microbiota
It is increasingly appreciated that intestinal homeostasis and IEC barrier integrity are a function of the crosstalk between the microbiota and the innate immune system. The latter is equipped with a variety of pattern-recognition receptors (PRRs) expressed in myeloid cells as well as IECs including among others TLRs and Nod-like receptors (NLRs) that sense microbial-associated motifs and/or host-derived damage signals and trigger a physiological level of inflammation that favors stem cell proliferation, IEC survival and restitution (Saleh and Trinchieri, 2011). The acute DSS model is useful for the study of innate immune mechanisms of colitis and tissue repair. DSS, a sulfated polysaccharide administered to mice in drinking water, is directly cytotoxic to enterocytes of the basal crypts and leads to barrier damage, microbial translocation and induction of a colitis phenotype that is independent of the adaptive immune system (Dieleman et al., 1994). In contrast to DSS, colitis induced with the haptens TNBS, DNBS or oxazolone instilled into the colon depends both on an acute oxidative injury with innate activation and on CD4+ T cell stimulation by autologous or microbial antigens (reviewed in (Wirtz and Neurath, 2007)). Using the DSS model, it has been recently demonstrated that similarly to TLRs (Rakoff-Nahoum et al., 2004), the Nlrp3 inflammasome-caspase-1-IL-18-IL-18R-MyD88 axis of innate immunity is required for tissue repair and protection from colitis (Allen et al., 2009; Dupaul-Chicoine et al., 2010; Salcedo et al., 2010; Zaki et al., 2010). MyD88-dependent signaling regulates the positioning of prostaglandin-endoperoxide synthase 2 (Ptgs2)-expressing mesenchymal cells to the base of the crypt surrounding colonic epithelial progenitors, which is needed for their compensatory proliferation following damage (Brown et al., 2007). Consistent with the protective role of the Nlrp3 inflammasome pathway in experimental models of colitis, a single nucleotide polymorphism (SNP) in the 3′ region of the human NLRP3 gene, which confers decreased NLRP3 expression, is associated with Crohn's disease in individuals of European descent (Villani et al., 2009). However, the healing role of innate immunity in the gut is not confined to the TLR and inflammasome pathways (Fig. 1). Indeed, CARD15 that encodes the cytosolic peptidoglycan receptor NOD2 was the first susceptibility gene identified for Crohn's disease (Hugot et al., 2001; Ogura et al., 2001). Using the DSS model, it has been shown that stimulation of this pathway through administration of the NOD2 cognate agonist muramyl dipeptide (MDP) protected experimental mice from colitis (Watanabe et al., 2008), arguing for a role of NOD2 in mucosal regeneration. Through the generation of two NOD2-deficient mice (Kobayashi et al., 2005; Watanabe et al., 2004) and one human CARD15 gene knock-in mouse (Maeda et al., 2005), NOD2 has been implicated in a number of additional homeostatic processes in the intestine including the production of defensins (Biswas et al., 2010; Wehkamp et al., 2004), the polarization of the adaptive immune response towards a Th2 cell-type cellular response (Magalhaes et al., 2008), and the control of autophagy (Cooney et al., 2010; Travassos et al., 2010), however the exact mechanisms of NOD2 actions remain unclear (see below). PRR sensing of the microbiota in the gut thus leads to multiple mechanisms that collectively contribute to intestinal homeostasis: Through activation of central inflammatory pathways, including NF-κB, MAPK and inflammasome pathways, PRR signaling drives the synthesis of mitogenic factors, including IL-6, IL-11, IL-18, COX-2, and anti-apoptotic factors, such as BCL-XL, which stimulate IEC proliferation and survival, respectively (Saleh and Trinchieri, 2011). In addition, PRRs control microbial ecology in the gut through the regulation of defensin production (Biswas et al., 2010; Wehkamp et al., 2004), and modulate the adaptive immune response by stimulating the expression of factors, such TSLP and IL-33 that direct Th2 differentiation (see below).
CD4+ effector T cells reactive to the microbiota mediate intestinal inflammation in most models
Early studies demonstrated that the critical effector cell in most models of experimental colitis that involve the adaptive immune system is the CD4+ T cell (Elson and Weaver, 2009). Although CD4 Th1, Th2, and Th17 effector cell subsets can all cause colitis, there are only a few instances in which CD4+ Th2 cells mediate colitis; the most studied being the TCRα-deficient mouse (Mizoguchi et al., 2000). In this model a CD4+ TCRβ homodimer-positive cell produces IL-4 and mediates disease (Takahashi et al., 1997).
Other models in which Th2 cells or cytokines have been implicated include oxazalone colitis in SJL mice (Boirivant, 1998), Wiskott Aldrich syndrome protein deficient mouse colitis (Nguyen, 2007), and the chronic phase of ileitis in SAMP1/Yit ileitis (Bamias, 2005). These are quite different models and it is unknown whether they share any common pathway that activates pathogenic Th2 effector cells. Candidate molecules for such a role might include NOD2 and thymic stromal lymphopoeitin (TSLP), which have been shown in humans to modulate dendritic cells to promote Th2 responses (Butler, 2007; Rimoldi, 2005).
Among the models useful for studying effector T cell mechanisms in IBD, the most fertile has been the adoptive transfer of CD45RBhi CD4+ naïve or CD4+ bacterial-reactive memory T cells into immunodeficient recipients (Cong et al., 1998; Powrie et al., 1994). Following such transfer the colitis that ensues is marked by increased production of IL-12p70 and IFN-γ in the lesions. Transfer of T-bet-deficient CD4+CD45RBhi does not result in colitis, supporting an important role for CD4+ Th1 cells in disease pathogenesis (Neurath et al., 2002). IL-12p70 signals Th1 cells via the transcription factor STAT4, and STAT4 transgenic mice have been generated (Wirtz et al., 1999). These mice develop Th1cell-mediated colitis only after an immunization that activates innate antigen presenting cells and adaptive CD4+ T cells, indicating a role for both cell types in the pathogenesis. An important role of innate cells in T cell transfer colitis has been shown by a recent study demonstrating that expansion in the numbers of the transferred CD4+ T cells by homeostatic proliferation in the recipients is absolutely required for colitis, and that this expansion in numbers is dependent on microbiota stimulation of IL-6 secretion via an innate MyD88-dependent mechanism (Feng et al., 2010). Interestingly, a second critical requirement for transfer colitis is the activation and differentiation of the transferred T cells by microbiota antigens (Feng et al., 2010). When both requirements are met, proliferation and differentiation of Th1 and Th17 effector cells occurs and results in colitis.
The role of the CD4+ Th17 cell subset in experimental colitis has been recognized more recently (Hue et al., 2006; Yen et al., 2006). T cell transfer into IL-23-deficient Rag1-/- mice does not result in colitis (Hue et al., 2006), and neutralization of IL-23 reverses active colitis (Elson et al., 2007). IL-23 is produced by innate immune cells and has effects on both innate and adaptive cells. Among CD4+ cells, Th17 cells have receptors for IL-23 and are dependent on IL-23 for maintenance and survival. Transfer of microbiota-reactive memory CD4+ Th17 cells induced colitis at 1/10 the dose than did transfer of CD4+ Th1 cells, indicating that the Th17 cell subset has potent colitogenic activity (Elson et al., 2007). Interestingly, during active colitis, the lamina propria contains CD4+ T cells expressing both IL-17 and IFNγ. Recent data indicate that CD4+ Th17 cells can convert to this double positive subset and then to IL-17-IFNγ+ Th1 cells, both in vitro and in vivo (Lee et al., 2008). This Th17 cell plasticity is dependent on the cytokine milieu. In contrast, conversion of Th1 cells to Th17 cells has not yet been observed. IL-17 and IL-23 are increased in the lesions of patients with Crohn's disease, but so are IFNγ and IL-12 (Fujino et al., 2003; Fuss et al., 2006). Although the data are clear that both Th1 and Th17 cells can mediate colitis, the role that each subset plays, when and how each contributes, and the consequences of Th17 cell plasticity all remain to be elucidated.
CD4+ Th effector cells have specific receptors that respond to antigen-MHC, raising the question of which microbiota antigens drive IBD. It is known that there is an increased adaptive immune response, in the form of serum IgG or IgA antibodies to a small set of microbial antigens in humans with IBD (Mow et al., 2004). Serologic expression cloning was thus used to identify the microbiota antigens involved in a mouse model of spontaneous colitis (Lodes et al., 2004). A limited number of protein antigens were found, among which one quarter were previously unknown bacterial flagellins. Interestingly, half of patients with Crohn's disease, but not those with ulcerative colitis or controls, had reactivity to the prototypic flagellin of this cluster, named CBir1. Both colitic mice and humans with Crohn's disease had increased CD4+ T cell reactivity to CBir1 flagellin (Lodes et al., 2004; Shen et al., 2008). Seroreactivity to CBir1 in patients with Crohn's disease correlates with a more complicated course of disease and a need for surgery (Targan, et al., 2005). The sharing of CD4+ T cell and B cell reactivity to flagellin in both mouse and humans indicates that the abnormal adaptive T cell response in both species may be remarkably similar.
Treg cell response to the microbiota during homeostasis and inflammation
As discussed, the innate compartment of the intestine provides a microenvironment that promotes the induction of Treg cells in the intestine (Barnes and Powrie, 2009; Rescigno and Di Sabatino, 2009). These Treg cells are largely directed at microbiota and food antigens. TGFβ and retinoic acid are two factors that are enriched in the intestine that promote the induction of Treg cells. The importance of Treg cells in maintaining intestinal homeostasis has been demonstrated by the targeted deletion of certain genes expressed by Treg cells, which results in colitis in conventionally reared but not germ-free mice, including those that encode TGFβ, TGFβRII, IL-10, IL-10R1, IL-2, and Foxp3 (Elson and Weaver, 2009). Under homeostatic conditions, the gut lamina propria has the largest proportion of CD4+ Treg cells of any tissue, including Foxp3+IL-10-, Foxp3+IL-10+, and Foxp3-IL-10+ (Tr1) cells (Maynard et al., 2007). Other cell types have also been implicated in intestinal regulation, including γδ T cells, CD8+ T cells, NK cells, and B cells, and these cells can cooperate, for example Foxp3+ Treg cells interact with IgA+ B cells in the intestine to reduce the uptake of microbiota antigens and prevent systemic T cell activation (Cong et al., 1998) (Fig. 1). Among CD4+ Treg cells both thymus-derived natural Foxp3+ Treg cells and peripherally induced Foxp3+ Treg cells are required to maintain intestinal homeostasis (Haribhai et al., 2009).
The adoptive T cell transfer model has demonstrated that co-transfer of CD4+CD25+ Treg cells with CD4+CD45RBhi T cells prevents colitis (Asseman et al., 1999). The exact mechanism by which Treg cells do so remains undefined, but Treg cells deficient in the mucosal α4β7 integrin homing receptor are effective (Denning et al., 2005), indicating that Treg cells can exert their inhibitory effects outside the intestinal lamina propria, possibly by inhibiting spontaneous proliferation of the adoptively transferred CD4+CD45RBhi T cells (Feng et al., 2010). Although transfer of large numbers of CD4+CD25+ Treg cells to mice with active colitis can quell inflammation and restore homeostasis, this takes many months to occur (Mottet et al., 2003). Thus, Treg cells are much more effective at preventing colitis than at reversing it once established. In the absence of critical molecules such as IL-10 or TGFβ, Treg cells are not able to prevent colitis, which develops spontaneously, is progressive, and ultimately fatal.
The relative ineffectiveness of Treg cells during active inflammation is due to a number of mechanisms. First, TLR ligand-induced inflammatory cytokines such as IL-6 make T effector cells more resistant to inhibition (Pasare and Medzhitov, 2003). More recently it has been recognized that CD4+ Treg cells can convert to effector CD4+ phenotypes such as Th1- or Th17-type cells in inflamed tissues (Oldenhove et al., 2009; Wohlfert and Belkaid, 2010). The inflammatory milieu may determine which CD4 subset Treg cells convert into, e.g., a Th1- dominated inflammation appears to induce Foxp3+ IFN-γ+ T cells, and it has been proposed that such converted Foxp3+ IFN-γ+ T cells may be more effective in dampening Th1 cell-mediated inflammation (Belkaid and Rouse, 2005). In other systems using Rosa26 lineage tracing, Treg cells have been shown to convert completely into CD4+ effector T cell subsets that then contribute to inflammation (Zhou et al., 2009). Similar conversion likely also occurs in IBD. Such plasticity may explain why Treg cells are more effective at preventing IBD than in reversing it.
Why have mechanisms evolved to overrule Treg cell function during active inflammation, or put another way, what are the roles of Treg cells in the intestine? The answer to this question is best viewed in the context of intestinal infections, which have likely exerted evolutionary selection on the development of Treg cells. The microbiota are an effective barrier to pathogens. Interestingly, enteric pathogens such as Salmonella and Citrobacter actively induce intestinal inflammation, which, in turn, alters microbiota composition, reducing strict anaerobes such as the Firmicutes, and allowing proteobacteria, including these pathogens, to expand (Lupp et al., 2007; Stecher et al., 2007). Treg cell inhibition of such inflammation would oppose such changes in the microbiota, thus maintaining colonization resistance. However, once the pathogen is established and the immune response is engaged, the role of Treg cells is thought to be mainly in the limitation of tissue damage (Belkaid and Rouse, 2005). The ability of Treg cells to convert to effector phenotypes indicates that they serve a second role, that of a “ready reserve” that be called upon to bolster the effector response to the pathogen and protect the host.
How do these models apply to human IBD? Treg cells are more difficult to study in humans because there are no truly specific markers to identify them. Given that limitation, the proportion and ex vivo function of Treg cells in human IBD are normal (Holmén et al., 2006; Maul et al., 2005; Saruta et al., 2007). It is unknown whether human Treg cells lose function or convert to effector T cell subsets in vivo. Yet a SNP in the IL10 gene confers susceptibility to both Crohn's disease and ulcerative colitis (Franke et al., 2010b; McGovern et al., 2010) and mutation in the IL10R gene resulted in early onset in IBD in two kindreds (Glocker et al., 2009). Whether defects in Treg cells contribute to human IBD remains an open question, but if so, it is likely to be a relative deficiency manifest mainly in the intestine.
The “next generation” of experimental models of IBD
Over the past two decades gene targeting in mice has identified many genes that are required for maintenance of intestinal inflammation and prevention of inflammation. Many of these gene-targeted mouse lines have added greatly to our understanding of the pathogenesis of IBD. The IBD phenotype of mice with the same gene deficiency but on different genetic backgrounds varies markedly (Bristol et al., 2000) due to gene modifiers (Beckwith et al., 2005). The C57Bl6 strain, which is now commonly used, is relatively resistant to colitis. Similar genetic variation exists in humans, as has been demonstrated by GWAS of IBD (Franke et al., 2010b; McGovern et al., 2010). Understanding the mechanisms by which the human gene loci uncovered by GWAS confer susceptibility to IBD will require a “next generation” of experimental models. This will be challenging because human genes are not likely to be globally deficient, but rather relatively impaired in function and this will need to be factored into the models.
Mouse models have been used to study several of the human susceptibility genes. For instance, two NOD2-deficient mice and a NOD2 (CARD15) knock-in mouse have been generated but no consensus on the exact mechanism of the CARD15 gene abnormality has been achieved. Studies on human cells with the NOD2 mutation revealed a function of NOD2 in inducing IL10 gene transcription, a role not conserved for the mouse orthologues (Noguchi et al., 2009). Furthermore chronic stimulation of human cells revealed abnormalities not observed with acute stimulation (Hedl et al., 2007). Because IBD is a chronic disease, design of experiments and experimental models will need to allow for these potential confounders.
The gene that encodes IL-23 receptor is strongly associated with Crohn's disease and ulcerative colitis in GWAS studies, as are multiple genes in the IL-23 pathway (Cho and Weaver, 2007). IL-23 has effects on both innate and adaptive immune cells and in mice its effects are most prominent in the intestine (Uhlig et al., 2006). IL-23p19 was first cloned in the mouse and has been shown to be required for colitis to develop in multiple models (Yen et al., 2006). Neutralization of IL-23 completely reversed active colitis in one model (Elson et al., 2007), which is consistent with data showing that the dominant IL23R SNP, which is protective in IBD, encodes for an alternative splicing of the gene resulting in a soluble receptor antagonist of IL-23 (Yu and Gallagher, 2010). Thus, mouse and human studies on the role of IL-23-Th17 cell pathway in IBD are remarkably convergent and are likely to lead to new therapeutic approaches. Indeed, therapeutic trials on IL-23p19 monoclonal antibodies are underway in Crohn's disease.
Discovery that ATG16L1, an autophagy gene, conferred susceptibility for Crohn's disease was unexpected (Hampe 2007; Rioux et al., 2007). The ATG16L1 SNP is a common genetic variation present in 40-50% of the population, thus the vast majority of persons with this SNP do not develop IBD. The Atg16l1 gene has been deleted in mice, but is embryonic lethal. Cells obtained from embryos of Atg16L1-deficient mice exhibited heightened inflammasome activation and IL-1β production when stimulated with LPS (Saitoh et al., 2008). Studies in mice that are hypomorphic, but not completely deficient in Atg16L1 (Atg16l1HM; approximately 1% of normal expression), have identified abnormalities in ileal Paneth cells consisting of abnormal inclusions and escape of antimicrobial peptides, such as lysozyme, into the cytoplasm. Interestingly, similar abnormalities of Paneth cells have been found in ileal tissue of Crohn's disease patients but only in those who have the ATG16L1 SNP (Cadwell et al., 2008). Subsequent studies in Atg16l1HM have shown that murine norovirus infection is required for the Paneth cell phenotype to express, even though the Paneth cells themselves are not infected by this virus (Cadwell et al., 2010). Although there was no spontaneous inflammation in norovirus-infected Atg16l1HM mice, these mice were more susceptible to DSS, leading to a TNF-dependent phenotype resembling aspects of IBD (Cadwell et al., 2010). These studies are among the first to reveal an important interaction among environmental components, namely norovirus infection and the microbiota, with the ATG16L1 gene variant, with both components being required for the IBD susceptibility phenotype. This is notable because environmental factors are known to be major contributors to IBD susceptibility, but the identity and mechanisms of these environmental factors is largely unknown. A human intestinal virome has been described recently, which was unique to each individual studied (Reyes, 2010). The contribution of the gut virome to IBD and to the variability in disease course amongst individuals has not yet been explored.
At present only a few IBD susceptibility genes have resulted in gene targeted mice and mechanistic studies. Although gene-deficient mice can be useful for such studies, to understand the role of these genes in humans, conditional deletion and inducible gene knock-in mouse models will be needed. These are more complex to generate but the technology exists and is fairly widely available. Such models will be necessary to understand the complex gene-gene and gene-environment interactions that contribute to IBD. Stressors such as DSS may be needed for these next generation models, but will not be a substitute for them. The recent discovery that the NOD2 gene regulates autophagy is an example of the potential importance of gene-gene interactions in IBD susceptibility (Cooney et al., 2010; Travassos et al., 2010). Adding to this complexity, it is clear that the composition of the microbiota can impact the immune system in major ways, and thus the microbiota composition of various models will need to be defined and controlled in these next generation models.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med. 1999;190:995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, NY. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bamias G, Martin C, Mishina M, Ross WG, Rivera-Nieves J, Marini M, Cominelli F. Gastroenterology. 2005;128:654–666. doi: 10.1053/j.gastro.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31:401–411. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Beckwith J, Cong Y, Sundberg JP, Elson CO, Leiter EH. Cdcs1, a major colitogenic locus in mice, regulates innate and adaptive immune response to enteric bacterial antigens. Gastroenterology. 2005;129:1473–1484. doi: 10.1053/j.gastro.2005.07.057. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Rouse BT. Natural regulatory T cells in infectious disease. Nat Immunol. 2005;6:353–360. doi: 10.1038/ni1181. [DOI] [PubMed] [Google Scholar]
- Biswas A, Liu YJ, Hao L, Mizoguchi A, Salzman NH, Bevins CL, Kobayashi KS. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc Natl Acad Sci U S A. 2010;107:14739–14744. doi: 10.1073/pnas.1003363107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boirivant M, Fuss IJ, Chu A, S W. J Exp Med. 1998;188:1929–1939. doi: 10.1084/jem.188.10.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristol IJ, Farmer MA, Cong Y, Zheng XX, Strom TB, Elson CO, Sundberg JP, Leiter EH. Heritable susceptibility for colitis in mice induced by IL-10 deficiency. Inflamm Bowel Dis. 2000;6:290–302. doi: 10.1002/ibd.3780060407. [DOI] [PubMed] [Google Scholar]
- Brown SL, Riehl TE, Walker MR, Geske MJ, Doherty JM, Stenson WF, Stappenbeck TS. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Invest. 2007;117:258–269. doi: 10.1172/JCI29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M, Chaudhary R, van Heel DA, Playford RJ, Ghosh S. Journal of Crohn's and Colitis. 2007;1:106–115. doi: 10.1016/j.crohns.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Cadwell K, Liu J, Brown S, Miyoshi H, Loh J, Lennerz J, Kishi C, Kc W, Carrero J, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng ACY, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-Plus-Susceptibility Gene Interaction Determines Crohn's Disease Gene Atg16L1 Phenotypes in Intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu LG, Rhodes JM. Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconj J. 2001;18:851–858. doi: 10.1023/a:1022240107040. [DOI] [PubMed] [Google Scholar]
- Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- Cho JH, Weaver CT. The genetics of inflammatory bowel disease. Gastroenterology. 2007;133:1327–1339. doi: 10.1053/j.gastro.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Brandwein SL, McCabe RP, Lazenby A, Birkenmeier EH, Sundberg JP, Elson CO. CD4+ T cells reactive to enteric bacterial antigens in spontaneously colitic C3H/HeJBir mice: increased T helper cell type 1 response and ability to transfer disease. J Exp Med. 1998;187:855–864. doi: 10.1084/jem.187.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJP, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Denning TL, Kim G, Kronenberg M. Cutting edge: CD4+CD25+ regulatory T cells impaired for intestinal homing can prevent colitis. J Immunol. 2005;174:7487–7491. doi: 10.4049/jimmunol.174.12.7487. [DOI] [PubMed] [Google Scholar]
- Dieleman LA, Ridwan BU, Tennyson GS, Beagley KW, Bucy RP, Elson CO. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology. 1994;107:1643–1652. doi: 10.1016/0016-5085(94)90803-6. [DOI] [PubMed] [Google Scholar]
- Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Elson CO, Cong Y, Weaver CT, Schoeb TR, McClanahan TK, Fick RB, Kastelein RA. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–2370. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- Elson CO, Weaver CT. In: In Vivo Models of IBD. In Inflammatory Bowel Disease: Translating Basic Science into Clinical Practice. Targan SR, Shanahan F, Karp LC, editors. Oxford: Wiley-Blackwell; 2009. pp. 25–51. [Google Scholar]
- Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. 2010;207:1321–1332. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nature Genet. 2010a;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nature Genet. 2010b;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss IJ, Becker C, Yang Z, Groden C, Hornung RL, Heller F, Neurath MF, Strober W, Mannon PJ. Both IL-12p70 and IL-23 are synthesized during active Crohn's disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm Bowel Dis. 2006;12:9–15. doi: 10.1097/01.mib.0000194183.92671.b6. [DOI] [PubMed] [Google Scholar]
- Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7:503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host & Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Punit S, Gallini CA, Michaud M, Zhang D, Sigrist KS, Lord GM, Glickman JN, Glimcher LH. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16:208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Hand T, Belkaid Y. Microbial control of regulatory and effector T cell responses in the gut. Curr Opin Immunol. 2010;22:63–72. doi: 10.1016/j.coi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, Li SH, Simpson PM, Chatila TA, Williams CB. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci USA. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmén N, Lundgren A, Lundin S, Bergin AM, Rudin A, Sjövall H, Ohman L. Functional CD4+CD25high regulatory T cells are enriched in the colonic mucosa of patients with active ulcerative colitis and increase with disease activity. Inflamm Bowel Dis. 2006;12:447–456. doi: 10.1097/00054725-200606000-00003. [DOI] [PubMed] [Google Scholar]
- Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, Powrie F, Maloy KJ. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Lee Y, Turner H, Maynard C, Oliver J, Chen D, Elson C, Weaver C. Late Developmental Plasticity in the T Helper 17 Lineage. Immunity. 2008;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host & Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M. Nod2 mutation in Crohn's disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- Magalhaes JG, Fritz JH, Le Bourhis L, Sellge G, Travassos LH, Selvanantham T, Girardin SE, Gommerman JL, Philpott DJ. Nod2-dependent Th2 polarization of antigen-specific immunity. J Immunol. 2008;181:7925–7935. doi: 10.4049/jimmunol.181.11.7925. [DOI] [PubMed] [Google Scholar]
- Mankertz J, Schulzke JD. Altered permeability in inflammatory bowel disease: pathophysiology and clinical implications. Curr Opin Gastroenterol. 2007;23:379–383. doi: 10.1097/MOG.0b013e32816aa392. [DOI] [PubMed] [Google Scholar]
- Maul J, Loddenkemper C, Mundt P, Berg E, Giese T, Stallmach A, Zeitz M, Duchmann R. Peripheral and intestinal regulatory CD4+ CD25(high) T cells in inflammatory bowel disease. Gastroenterology. 2005;128:1868–1878. doi: 10.1053/j.gastro.2005.03.043. [DOI] [PubMed] [Google Scholar]
- Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, Florin TH, Hill GR, Korolik V, McGuckin MA. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313–2324. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genetics. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi A, Mizoguchi E, Saubermann LJ, Higaki K, Blumberg RS, Bhan AK. Limited CD4 T-cell diversity associated with colitis in T-cell receptor alpha mutant mice requires a T helper 2 environment. Gastroenterology. 2000;119:983–995. doi: 10.1053/gast.2000.18153. [DOI] [PubMed] [Google Scholar]
- Mottet C, Uhlig H, Powrie F. Cutting Edge: Cure of colitis by CD4+CD25+ regulatory T cells. J Immunol. 2003;170:3939–3943. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- Mow WS, Vasiliauskas EA, Lin YC, Fleshner PR, Papadakis KA, Taylor KD, Landers CJ, Abreu-Martin MT, Rotter JI, Yang H, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn's disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, Mizoguchi A, Mizoguchi E, Mudter J, Galle PR, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J Exp Med. 2002;195:1129–1143. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DD, Maillard MH, Cotta-de-Almeida V, Mizoguchi E, Klein C, Fuss I, Nagler C, Mizoguchi A, Bhan AK, Snapper SB. Gastroenterology. 2007;133:1188–1197. doi: 10.1053/j.gastro.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Homma Y, Kang X, Netea MG, Ma X. A Crohn's disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1. Nat Immunol. 2009;10:471–479. doi: 10.1038/ni.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Oldenhove G, Bouladoux N, Wohlfert E, Hall J, Chou D, Dos Santos L, O'Brien S, Blank R, Lamb E, Natarajan S, et al. Decrease of Foxp3(+) Treg Cell Number and Acquisition of Effector Cell Phenotype during Lethal Infection. Immunity. 2009;5:772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwala CM, Jones JC, Viney JL. A novel model of inflammatory bowel disease: mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J Immunol. 1998;161:5733–5744. [PubMed] [Google Scholar]
- Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science. 2003;299:1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- Powrie F, Correa-Oliveira R, Mauze S, Coffman RL. Regulatory interactions between CD45RBhigh and CD45RBlow CD4+ T cells are important for the balance between protective and pathogenic cell-mediated immunity. J Exp Med. 1994;179:589–600. doi: 10.1084/jem.179.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rescigno M. Before they were gut dendritic cells. Immunity. 2009;31:454–456. doi: 10.1016/j.immuni.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Rescigno M, Di Sabatino A. Dendritic cells in intestinal homeostasis and disease. J Clin Invest. 2009;119:2441–2450. doi: 10.1172/JCI39134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi M, Chieppa M, Salucci V, Avogadri F, Sonzogni A, Sampietro GM, Nespoli A, Viale G, Allavena P, Rescigno M. Nat Immunol. 2005;6:507–514. doi: 10.1038/ni1192. [DOI] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, Wang E, Ma W, Haines D, O'hUigin C, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207:1625–1636. doi: 10.1084/jem.20100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev. 2011;11:9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saruta M, Yu QT, Fleshner PR, Mantel PY, Schmidt-Weber CB, Banham AH, Papadakis KA. Characterization of FOXP3+CD4+ regulatory T cells in Crohn's disease. Clin Immunol. 2007;125:281–290. doi: 10.1016/j.clim.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Shanahan F, Bernstein CN. The evolving epidemiology of inflammatory bowel disease. Curr Opin Gastroenterol. 2009;25:301–305. doi: 10.1097/MOG.0b013e32832b12ef. [DOI] [PubMed] [Google Scholar]
- Shen C, Landers C, Derkowski C, Elson C, Targan S. Enhanced CBir1-specific innate and adaptive immune responses in Crohn's disease. Inflamm Bowel Dis. 2008;14:1–11. doi: 10.1002/ibd.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella entérica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS biology. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, Abraham C, Turner JR. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I, Kiyono H, Hamada S. CD4+ T-cell population mediates development of inflammatory bowel disease in T-cell receptor alpha chain-deficient mice. Gastroenterology. 1997;112:1876–1886. doi: 10.1053/gast.1997.v112.pm9178680. [DOI] [PubMed] [Google Scholar]
- Targan SR, Landers CJ, Yang H, Lodes MJ, Cong Y, Papadakis KA, Vasiliauskas E, Elson CO, Hershberg RM. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn's disease. Gastroenterology. 2005;128:2020–2028. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, Stepankova R, Robinson N, Buonocore S, Tlaskalova-Hogenova H, Cua DJ, et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Van Assche G, Vermeire S, Rutgeerts P. Immunosuppression in inflammatory bowel disease: traditional, biological or both? Curr Opin Gastroenterol. 2009;25:323–328. doi: 10.1097/MOG.0b013e32832c073a. [DOI] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- Vereecke L, Sze M, Guire CM, Rogiers B, Chu Y, Schmidt-Supprian M, Pasparakis M, Beyaert R, van Loo G. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J Exp Med. 2010;207:1513–1523. doi: 10.1084/jem.20092474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock JV, Elliott DE. Helminths and the IBD hygiene hypothesis. Inflamm Bowel Dis. 2009;15:128–133. doi: 10.1002/ibd.20633. [DOI] [PubMed] [Google Scholar]
- Welcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook AM, Wei B, Braun J, Schiestl RH. More damaging than we think: systemic effects of intestinal inflammation. Cell Cycle. 2009;8:2482–2483. doi: 10.4161/cc.8.16.9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz S, Finotto S, Kanzler S, Lohse AW, Blessing M, Lehr HA, Galle PR, Neurath MF. Cutting edge: Chronic intestinal inflammation in STAT-4 transgenic mice: Characterization of disease and adoptive transfer by TNF- plus IFN-gamma-producing CD4(+) T cells that respond to bacterial antigens. J Immunol. 1999;162:1884–1888. [PubMed] [Google Scholar]
- Wirtz S, Neurath MF. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev. 2007;59:1073–1083. doi: 10.1016/j.addr.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Wohlfert E, Belkaid Y. Plasticity of T reg at infected sites. Muc Immunol. 2010;3:213–215. doi: 10.1038/mi.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RY, Gallagher G. A naturally occurring, soluble antagonist of human IL-23 inhibits the development and in vitro function of human Th17 cells. J Immunol. 2010;185:7302–7308. doi: 10.4049/jimmunol.1002410. [DOI] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeissig S, Bojarski C, Buergel N, Mankertz J, Zeitz M, Fromm M, Schulzke JD. Downregulation of epithelial apoptosis and barrier repair in active Crohn's disease by tumour necrosis factor alpha antibody treatment. Gut. 2004;53:1295–1302. doi: 10.1136/gut.2003.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martínez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]