Abstract
Chromosome deletions have several applications in the genetic analysis of complex organisms. They can be used as reagents in region-directed mutagenesis, for mapping of simple or complex traits, or to identify biological consequences of segmental haploidy, the latter being relevant to human contiguous gene syndromes and imprinting. We have generated three deletion complexes in ES (Embryonic Stem) cells that collectively span ∼ 40 cM of proximal mouse chromosome 5. The deletion complexes were produced by irradiation of F1 hybrid ES cells containing herpes simplex virus thymidine kinase genes (tk) integrated at the Dpp6, Hdh (Huntington disease locus), or Gabrb1 loci, followed by selection for tk-deficient clones. Deletions centered at the adjacent Hdh and Dpp6 loci ranged up to ∼ 20 cM or more in length and overlapped in an interdigitated fashion. However, the interval between Hdh and Gabrb1 appeared to contain a locus haploinsufficient for ES cell viability, thereby preventing deletions of either complex from overlapping. In some cases, the deletions resolved the order of markers that were previously genetically inseparable. A subset of the ES cell–bearing deletions was injected into blastocysts to generate germline chimeras and establish lines of mice segregating the deletion chromosomes. At least 11 of the 26 lines injected were capable of producing germline chimeras. In general, those that failed to undergo germline transmission bore deletions larger than the germline-competent clones, suggesting that certain regions of chromosome 5 contain haploinsufficient developmental genes, and/or that overall embryonic viability is cumulatively decreased as more genes are rendered hemizygous. Mice bearing deletions presumably spanning the semidominant hammertoe locus (Hm) had no phenotype, suggesting that the classic allele is a dominant, gain-of-function mutation. Overlapping deletion complexes generated in the fashion described in this report will be useful as multipurpose genetic tools and in systematic functional mapping of the mouse genome.
The Human Genome Project is transitioning into the so-called “Post-Genomics” era, in which the emphasis in genome research is switching from structure to function. This change is spawning new technologies and concepts for conducting functional analyses on a comprehensive, large scale, including gene expression chips, protein interaction assays, and computational tools to infer function from DNA sequence. However, a true understanding of the role of a gene in the context of a whole organism depends on analysis of mutations or allelic variation. Presently, the infrastructure and technology for understanding the molecular genetic basis of phenotypically defined mammalian genes has outstripped the supply of human disease alleles and mouse mutants.
As with the two general strategies being used for genomic sequencing —whole genome shotgun versus a systematic sequencing of chromosome regions (Green 1997; Weber and Myers 1997)—there are two general approaches for mutagenesis: whole genome versus region directed. The former approach involves three generation screens for recessive mutations located throughout the genome. The latter typically involves the use of deletions to saturate particular chromosome regions for randomly induced point mutations. The relative merits of each approach have been discussed in detail elsewhere (Schimenti and Bucan 1998) but can be summarized as follows. Genome-wide scans have the advantage of yielding more mutants, but their map locations are unknown. Region-specific mutagenesis has the advantage of restricting the location of new mutations to a known region, thereby facilitating genetic mapping. It is also advantageous for the identification and propagation of steriles and lethals. However, this approach requires the availability of special chromosomes (inversions/deletions), and the mutations identified are limited to a small fraction of the genome. Ready availability of deletion stocks would alleviate this drawback and open new avenues for directed functional analyses of particular chromosomal regions.
Here, we report the generation of three deletion complexes—collections of nested deletions—spanning ∼ 40 cM of mouse chromosome 5. These deletions were induced by irradiation of ES cell clones containing targeted insertions of a negatively selectable (tk) reporter. Deletions centered around two of the three DFPs (Deletion Focal Points) overlap in an interdigitated manner, allowing one to systematically characterize functional regions of this chromosome by pairwise combination of the deletions through mating. These deletions also will serve as reagents in a region-directed ENU (Ethylnitrosourea) saturation mutagenesis screen and in the modeling of the Wolf-Hirschhorn contiguous gene syndrome that resides in this region.
RESULTS
Targeted Insertions of a TK-neo Cassette at the Hdh, Dpp6, and Gabrb1 Loci
To create a series of deletions spanning the proximal region of mouse chromosome 5, the technique of irradiating F1 hybrid ES cells was used (You et al. 1997a, 1997b; Thomas et al. 1998). This strategy entails the targeted insertion of a tk-expressing cassette into a locus of choice, treatment of targeted cells with radiation, and selection for loss of tk expression with the drug FIAU (1–2'-deoxy-2′-fluoro-β-d-arabinofuranosyl-5-iodouracil). The loci chosen for targeting the tk cassette, Dpp6, Hdh, and Gabrb1, are distributed across the region spanned by the rump white (Rw) inversion. The proximal breakpoint of this inversion lies within Dpp6, and the distal end lies near the Kit (formerly W) locus (Fig. 2; Hough et al. 1998). This inversion serves as a balancer that is important for downstream saturation mutagenesis screens (Schimenti and Bucan 1998; see Discussion).
Figure 2.
Deletion complexes on chromosome 5. The proximal region of chromosome 5 is depicted as a horizontal line, with the centromere (circle) on the left. Map positions (in centimorgans) as reported in the MGD are indicated, along with the approximate locations of certain landmark loci, such as hammertoe (Hm), Qdpr, and Kit, also known as dominant white spotting (W). Shh is sonic hedgehog homolog, within which a simple sequence repeat was used to type deletions (see Methods). Microsatellite loci, indicated under the map, have been abbreviated by exchanging the prefix “M” for “D5Mit.” The map positions are based on three sources: MGD values, deletion mapping, and RH mapping. As discussed in the text, analysis of deletion breakpoints revealed two disagreements in microsatellite order with MGD; these markers are indicated with asterisks, and in both cases, the MGD order juxtaposes these two markers with the adjacent, proximal microsatellites on this map. A region surrounding Dpp6 is expanded to integrate data obtained by high resolution RH mapping, as described in the text. Deletions are indicated as horizontal rectangles, either solid, striped, or unfilled, and are color coded with the locus at which they were induced (red, Dpp6; blue, Hdh; green, Gabrb1). The amount of DNA known to be absent in each deletion is spanned by the rectangles. The thin lines extending from the ends of the rectangles indicate the regions in which the deletion breakpoints reside. Those deletions that have been converted into stocks of mice are represented by solid rectangles, and associated allele names are given (the bracketed text corresponds to superscripting). Deletions existing only in ES cells are represented by the striped or unfilled rectangles. In this latter case, the number of independent ES cell lines containing a particular class of deletion (currently indistinguishable with the markers used in this study) is indicated on the right. In some cases, red numbers in parentheses next to these indicate the number of ES cell lines in this deletion class that failed to produce germline chimeras. An ES cell clone containing the Hdhdf5J deletion, indicated by a dashed line, generated a germline chimera that only transmitted the nondeleted chromosome to its offspring. The relative positions of D5Mit421, tlt (tilted), and D5Mit128 are taken from Ying et al. (Ying et al. 1999). Microsatellite markers that are polymorphic between 129 and B6 are in black, whereas those that are not are in red. The following markers had a 129 allele size smaller than B6: D5Mit1, 25, 52, 72, 73, 106, 124, 148, 176, 232, 335, 348, 356, 388,and Dpp6Rep3. The remaining SSRs had larger 129 alleles.
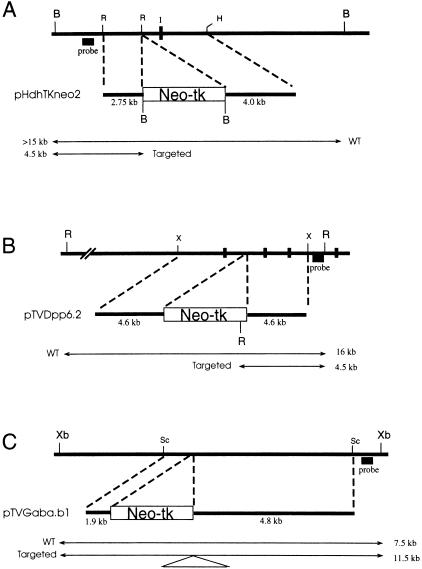
The Hdh locus of v6.4 F1 hybrid ES cells (C57BL/6J × 129/Jae) (You et al. 1998) was targeted (Fig. 1a) with a tk-neomycin resistance (neo) cassette (You et al. 1997a) by modifying a vector previously used to generate a null mutation in this gene (Duyao et al. 1995; Fig. 1a) Eighty-seven of 261 (33.3%) G418-resistant clones examined contained a targeted integration. Vectors (Fig. 1b,c) for the Dpp6 and Gabrb1 loci also resulted in successful targeting by homologous recombination in the v6.4 line, at overall frequencies of 6/347 (1.7%) and 24/93 (26%), respectively.
Figure 1.
Targeting vectors. (A) Hdh targeting vector. Transcriptional orientation of the gene is to the right. Note position of exon 1 (1). (B) Dpp6 targeting vector. (C) Gabrb1 targeting vector. In each panel, the genomic locus is shown at the top as a horizontal thick line. Positions of exons are indicated by vertical broken bars. Probes used to detect targeting events are indicated, and the sizes of endogenous (WT) and targeted restriction fragments are shown next to arrows spanning the endpoints. The names of targeting vectors are shown, with the location of the HSV-tk (tk)/neomycin resistance (neo) cassette indicated in the large rectangular boxes. (B) BamHI; (R ) EcoRI; (H) HindIII; (X or Xb) Xba I; (Sc) SacI.
Before induction of deletions, it is important to select targeted clones that retain germline-colonizing potential. Accordingly, several clones from each targeted locus were first karyotyped, and two or three of those with a euploid chromosome mode were injected into blastocysts to check for germline transmission. Despite carrying tk genes, which are known to disrupt spermatogenesis (Wilkie et al. 1991), at least one clone from each locus yielded germline chimeric males. These clones, or in some cases subclones derived from them, were used for subsequent induction of deletions.
Induction of Deletions
At the Hdh locus, two independently targeted germline competent clones were used to induce deletions by subjecting cultures to 400 Gy of gamma radiation from a 137Cs source (see Methods). Both contained the targeted integration on the C57BL/6J allele (B6). Of the ∼ 800,000 cells surviving the radiation, ∼ 550 survived selection in FIAU, indicating that ∼ 1 in 1455 cells had lost expression of the tk gene. Three hundred eighty-four FIAU-resistant (FIAUR) colonies were picked and tested for G418 sensitivity, and 96 were screened by PCR (Polymerase Chain Reaction) for presence or absence of the tk gene. Only three G418R clones were found, indicating that these clones may not have had true deletions, but underwent inactivation of tk by epigenetic silencing or mutation. Similarly, 1 clone was tk positive by PCR. Overall, the results indicate that the majority of clones were missing the tk and neo genes, presumably by radiation-induced deletion, mitotic recombination, or whole chromosome loss. The frequency of tk loss (∼ 1/1400) was much higher than that (∼ 1/20,000) observed at the D17Aus9 locus (You et al. 1997a).
DNA from these clones then was amplified with primers corresponding to microsatellites polymorphic between the 129 and B6 parental chromosomes. For the initial screen, three markers were used, one just proximal to Hdh (D5Mit148) and two (D5Mit1 and 25) that reside at distances proximal and distal to Hdh. The purpose was to eliminate clones that had either: 1) very small deletions (no B6 alleles deleted); 2) large deletions or whole chromosome loss (two or three markers missing); or possible mitotic recombination (retention of the B6 allele of D5Mit1 but not the distal B6 alleles). A subset of 36 clones appearing to have large interstitial deletions was analyzed with additional markers (Fig. 2 and Table 1; see below).
Table 1.
ES Cell Deletion Lines
(DFP) Deletion focal point. (Class) corresponds to the codes in Figure 2. (Clone) lab codes for all deletion lines in a particular class. (Germ) refers to whether the indicated clones generated germline chimeras. The maximum and minimum deletion sizes in centimorgans were determined using current MGD values.
The cell line with this deletion underwent germline transmission, but the deletion chromosome itself was not transmissable.
Induction of deletions at Dpp6 was conducted in a similar manner, with one exception. Following the demonstration that one targeted clone (B6 allele) retained germline competence, it was found that this culture contained a high background of FIAUR cells. Therefore, several subclones were derived that retained essentially complete FIAU sensitivity, and cultures of six of these subclones were irradiated to induce deletions. FIAUR clones were isolated at a frequency of ∼ 1/16,400. Approximately half were G418S, virtually all of which were shown to have deletions at the molecular level. Thus, the induced deletion rate at Dpp6 was ∼ 1/33,000. As with Hdh, a subset of the deletions that appeared to be extremely large, and might actually represent whole chromosome loss or mitotic recombination, were omitted from subsequent analysis of the remainder with a panel of microsatellite markers.
Recovery of deletions at the Gabrb1 locus was less efficient. Only ∼ 2–5% of FIAU-resistant clones were, in fact, missing the tk gene after irradiation (presumably the rest underwent epigenetic silencing of tk expression), and many of these had undergone either extremely large deletions or mitotic recombination (showing LOH [Loss of Heterozygosity] of D5Mit1, D5Mit244, or both). Nevertheless, 13 interstitial deletions were obtained (of which 12 were analyzed in detail), falling into several classes (see below).
Molecular Mapping of Deletion Breakpoints and Interdigitation of Deletion Complexes
The deficiencies at each of the three DFPs had a range of sizes and breakpoints. As shown in Figure 2, there was a large degree of overlap between the Hdh and Dpp6 deletions, forming an indigitated collection. Some of the deletions were extremely large, extending ∼ 20 cM or more in length (using map positions currently reported in the MGD [Mouse Genome Database]; the minimum and maximum deletion sizes are listed in Table 1). Although the breakpoints of several independent deletions are currently indistinguishable, this is attributable to the limited number of polymorphic microsatellites available for the parental (129 and B6) genotype of the ES cells. Higher resolution analysis of some deletions was attainable by crossing mice derived from deletion-bearing ES cells (see next section) to Mus castaneous and analyzing DNA from the resulting F1 interspecific hybrids.
Interestingly, there was no evidence for overlap of deletions originating from the Hdh and Gabrb1 loci, although several breakpoints from both deletion complexes lie between the Qdpr and Gabrb1 loci, within the D5Mit52 to D5Mit355 interval. Although the apparent lack of overlap may be a consequence of the dearth of polymorphic markers in the region, it is possible that there is a locus or loci in this interval that cannot exist in the haploid state in ES cells. Because the Hdh deletions occurred on the B6 chromosome, and the Gabrb1 deletions occurred on 129, it is unlikely that such a haploinsufficiency is caused by parental chromosome-specific imprinting, although the possibility that two loci are present that are oppositely imprinted remains.
Creation of Mice Bearing Deletions
Several deletion-bearing ES cell clones were injected into B6 blastocysts to generate chimeric animals. Chimeras then were mated to B6 to check for germline transmission. Although all agouti animals from such a mating must be ES-cell derived, some black animals were also ES cell-derived, because the v6.4 cells are heterozygous for a B6 allele at the agouti locus. In most cases, a deletion clone was injected on only one or two occasions, so the failure of a clone to yield germline chimeras may reflect technical issues rather than true ability of an ES cell clone to colonize the germline.
Of 17 injected TK- clones from the Hdh locus, eight were shown capable of making germline chimeras. Two of these were not true deletions; one was a mitotic recombinant, and the other had LOH of several markers apparently caused by a double crossover or event of similar consequence. Another (Hdhdf5J; dashed blue line in Fig. 2) was a large deletion that yielded germline chimeras; however, the agouti progeny produced by these chimeras exclusively inherited the nondeleted (129) chromosome. Five clones bearing deletions of various sizes ultimately were established as lines of mice (solid blue lines in Fig. 2). Cell lines bearing deletions that did not produce germline chimeras are indicated by red numbers in parentheses next to the corresponding deletion class (depicted as patterned lines in Fig. 2). Seven such clones contained deletions extending proximally past D5Mit72 and/or D5Mit348 loci that were not deleted in clones showing germline transmission. This suggests that the primary reason for failure of germline transmission might be related to the deletion size or hemizygosity of particular loci, rather than problems caused by the irradiation and selection.
Similar results were obtained with deletions induced at the Dpp6 locus. Six deletion lines were injected into blastocysts, and four (Dpp6df1J, 2J, 4J and 5J) produced chimeras that transmitted the deletion chromosomes to progeny (Fig. 2). The two cell lines that failed to produce germline chimeras contained extremely large deletions that extended past D5Mit232. Once again, we suspect that the failure of these two lines is related to the large size of the deletions.
A more drastic example of deletion size compromising the ability of ES cells to colonize the germline was experienced with deletions induced at the Gabrb1 locus. Only one of six blastocysts-injected deletion clones yielded germline chimeras, and this particular line contained a small deletion that did not remove any flanking microsatellite markers. Four of the remaining five deletions removed one or more markers, and these either failed to produce chimeric mice or the chimeras derived from them had poor contribution from the injected ES cells, judging from the low proportion of agouti fur in the coat (the C57BL/6J host blastocysts produce a black coat). We conclude that there is a haploinsufficient locus or loci near the Gabrb1 locus that, when present in only a single copy, is incompatible with development of most tissues or cell types. At present, we cannot determine whether this locus (or loci) is located proximal, distal, or on both sides of Gabrb1. Unfortunately, this would appear to preclude the generation of large deletions in this immediate region.
Deletions as Mapping Reagents
An ancillary benefit of the deletions is that they provide an excellent tool for determining the relative orders of polymorphic loci along chromosomes. The map locations of most microsatellites remains exclusively deduced from the relatively small (48 progeny) F2 mapping cross performed by the Whitehead Institute Genome Center. Higher resolution crosses performed by others, including the EUCIB (European Collaborative Interspecific Backcross; www.hgmp.mrc.ac.uk/MBx/MBxHomepage.html) usually include only a subset of these markers and “anchor” loci. In the course of typing the deletions for the array of markers illustrated in Figure 2, we uncovered two inconsistencies in marker order as represented in MGD as of May 2000. D5Mit72 must be proximal rather than distal to D5Mit348, because 15 Hdh deletions and nine Dpp6 deletions removed D5Mit348 but not D5Mit72. Analysis of Gabrb1 deletions indicated that D5Mit113 (42.0 cM; MGD) is distal to D5Mit25 and D5Mit205 (45.0 cM; MGD); one (“E” in Fig. 2) deletion was found that removed the latter two but not the former. However, it is possible that this deletion is complex or not continuous, as has been observed with classically induced deletions (Sharan et al. 1991; Johnson et al. 1995).
The collection of deletion breakpoints also was able to separate markers that were previously nonrecombinant in mapping crosses (and reflected in MGD). One such nonrecombinant group was D5Mit148,176, 388, and 389. Deletion analysis enabled these to be subdivided into three intervals (Fig. 2). It also was possible to separate one microsatellite marker from each of two other clusters that contained three inseparable loci, D5Mit83/304/356 and D5Mit25/153/205 (Fig. 2).
Complementation Analysis Indicates Hammertoe (Hm) Is Not a Null Mutation
Hm is a semidominant mutation located proximal to Hdh and apparently distal to Dpp6. Although it has not been mapped in the same cross as Dpp6, Hm was shown to be extremely close but just proximal to hemimelic extra toes (Hx), recombining once in 3664 offspring (∼ 0.03 cM; Sweet 1982). Hx was shown to be distal to En2 (Robert et al. 1994), which is, in turn, distal to Dpp6 as reported in The Jackson Laboratory's BSS interspecific backcross and two independent crosses (Wada et al. 1993; Montgomery et al. 1994), resulting in MGD consensus locations of 12 and 16 cM for Dpp6 and Hm, respectively. Mice heterozygous for Hm retain skin between their digits, giving the appearance of webbed paws. Homozygotes display a more severe phenotype, in which the digits are severely curled and essentially attached (although not polysyndactylous). The deficiency Hdhdf4J probably removes the Hm locus because it extends proximal to Dpp6 and distal to Hdh. However, Hdhdf4J heterozygotes do not possess webbed paws, indicating that Hm is not a loss-of-function mutation. Furthermore, Hdhdf4J/Hm animals are phenotypically identical to Hm/ +, not having the more severe phenotype characteristic of Hm homozygotes (not shown). These observations indicate that the hammertoe mutation is a gain-of-function allele. We caution, however, that this conclusion relies on the assumption that Hm maps distal to Dpp6.
DISCUSSION
The work described here illustrates the power of the radiation-induced deletion technique for rapidly saturating large chromosome regions with deficiencies of all sizes. Only a single targeted ES cell clone is needed to yield an unlimited number of different deletions in a single experiment, with staggered breakpoints in both directions. Importantly, our data suggest that the exposure of the cells to radiation, which may cause second site mutations and other biological responses, has little deleterious consequence on the germline-colonizing potential of ES cells. Rather, it appears that haploidy of particular genomic regions is the primary determinant of whether a deletion-bearing ES cell can create germline chimeric mice. This is a biological issue that appears to be independent of the technology itself. Similar observations were made in the cases of large deletions created by Cre-loxP recombination (Liu et al. 1998).
There are probably three factors that influence whether a deletion will be lethal or prevent the formation of chimeras. One is the overall size, such that hemizygosity of increasing numbers of genes incrementally decreases fitness. A second factor may have to do with the existence of a strictly haploinsufficient locus, which when deleted, abolishes pluripotentiality of ES cells. In this situation, deletions emanating from a particular DFP would be inviable once they extend past a given point. This might be the case with the Gabrb1 deletions. Notably, we have observed a high rate of peri- and postnatal lethality (60%) of mice heterozygous for the W19H deletion, which covers the D5Mit83-D5Mit112 interval just distal to Gabrb1 (King et al. 1997; G. Leach and M. Bucan, unpubl.). A final possibility is that there exists an imprinted locus, near the DFP, that is active only on the targeted chromosome (in the Gabrb1 case, 129). Because ES cells maintain their genomic imprinting, a deletion of the active allele essentially would create nullizygosity for expression of the imprinted gene; if that gene were required for either germ cell development or early embryogenesis, this would abolish the ability of the ES cell to make germline chimeras. From a technical viewpoint, if one desired to make and propagate mice bearing deletions in a region in which this was the case, then the tk-neo cassette should be targeted to the chromosome bearing the silenced allele, exploiting the preference of an exogenous vector to undergo homologous recombination with an isogenic target (teRiele et al. 1992).
Interestingly, the experiments provided evidence for the existence of a gene or genes that are haploinsufficient for growth of ES cells near the Gabrb1 locus. Evidence for haploinsufficient loci on chromosome 9 in ES cells also was reported by Thomas et al., who created a radiation-induced deletion complex at the Ncam locus (Thomas et al. 1998), and Zheng et al., who induced deletions on chromosome 11 with a targeted Cre-loxP strategy (Zheng et al. 2000).
The utility of deletion complexes, rather than individual deletions, lies in downstream applications for dissecting genome function. In mice, deletion complexes created by whole animal irradiation experiments have been generated around several visibly scorable loci, including albino (Tyrc), brown (Tyrp1b), pink-eyed dilution (p), short ear (Bmp5se), nonagouti (a), dilute (Myo5ad), and piebald (Ednrbs). A systematic characterization of functional units along these chromosomal regions has been performed with the brown and albino deletions in particular to identify genes important in development (Russell et al. 1982; Rinchik and Russell 1990; Holdener-Kenny et al. 1992; Rinchik et al. 1994). In these studies, different deletions from the same deletion complex were crossed to one another or rendered homozygous, and the phenotypes associated with nullizygosity of various regions were delineated and, in one case, ultimately cloned (Schumacher et al. 1996). However, a drawback to this approach is that early acting deletions that are very close to the DFP will mask functions encoded in more distant locations. The availability of adjacent deletion complexes obviates this problem, enabling the creation of complex heterozygotes that are nullizygous for intervals in between the DFPs (You et al. 1997b). The deletion complexes described here enable such an approach for the overlapping Hdh and Dpp6 deletions.
To increase the value of deletion complexes described in this article, we integrated the deletion map with marker-dense RH (Radiation Hybrid) and genetic maps that, for this 40-cM chromosomal region, include > 90 genes and 200 SSLP (Simple Sequence Length Polymorphism) markers (Tarantino et al. 2000). By placing a set of common SSLP markers on all three maps, it is now possible to provisionally determine or predict a set of genes missing in each overlapping deletion. For example, the integrated map allows us to predict that the genes Dpp6, Htr5a, En2, Shh, Il6, and Kcnk3 are deleted in Dpp6df2J (D5Mit73 to D5Mit389 interval). Current efforts to place novel expressed sequence tags on the RH map and to eventually sequence the mouse genome will allow more accurate prediction of genes deleted in the individual lines, and therefore facilitate the identification of candidate genes for abnormal phenotypes associated with these deletions.
A principal motivation for establishing these deletion complexes was to conduct an ENU saturation mutagenesis screen of the Rw inversion region of chromosome 5 (Schimenti and Bucan 1998). The Rw inversion spans a 30-cM region between the Dpp6 and Kit (W) loci (see Fig. 2). The two generation screen for detection of ENU-induced mutations is being performed by crossing animals carrying ENU-exposed chromosomes in trans to the Rw balancer to deletion-bearing mice and phenotyping (for visible anomalies) only those animals that contain the deletion in trans to the mutagenized chromosome. To span the Rw region as efficiently as possible, a set of deletions representing a minimum tiling path is being used. For example, the two deletions Dpp6df1J and Hdhdf2J together span > 22 cM. The value of a nested deletion set is that it serves as a powerful and rapid resource for mapping newly induced recessive mutations. The random nature of the breakpoints on both ends of each deletion confers bidirectional mapping utility.
The ability to create deletion complexes also should prove valuable in modeling human contiguous gene syndromes such as Cri-du-chat, Williams-Beuren, and Jacobson. This generation of a mouse strain bearing a deletion corresponding to the DiGeorge syndrome region demonstrated this point (Lindsay et al. 1999). The deletions described here will be relevant for modeling and mapping loci involved in WHS (Wolf-Hirshhorn syndrome), the critical region of which is located within a megabase of Hdh (Wright et al. 1997). The large collection of Dpp6 and Hdh deletions already existing as mouse strains, plus those remaining cryopreserved in ES cells, potentially can be used to dissect regions corresponding subphenotypes of this syndrome that may be manifested in mice. Indeed, several of the Hdh deletions have presented defects reminiscent of WHS (D. Naf and J.C. Schimenti, unpubl.).
METHODS
Targeting Constructs
Hdh
The plasmid pHdh-2, containing the downstream 4-kb arm of the Hdh targeting vector (Fig. 1) as an EcoRI/HindIII insert, was linearized with NotI, Klenow filled, BamHI digested (cleaving at a site within the polylinker adjacent to the EcoRI site), and ligated to the 2.75-kb upstream arm excised from plasmid pHdh-1 (which contained the arm as an EcoRI fragment) by linearizing with ClaI, Klenow filling, and cleaving with BamHI. This intermediate plasmid was linearized with BamHI (cleaving at the junction between the two arms) and ligated to a 4-kb BamHI tk-neo cassette fragment excised from pBSSKΔBam (You et al. 1997a).
Dpp6
A 9.2-kb XbaI genomic fragment containing three exons of Dpp6, isolated from cosmid cAB1 cloned from a Rw/+ library (Hough et al. 1998) was subcloned into the vector pBSSKΔBam (You et al. 1997a) to create pX8–1. A 4.0-kb BamHI fragment containing a neo-tk cassette then was subcloned, in both orientations, into an unique BamHI site near the center of the 9.2-kb XbaI insert of pX8-1. The resulting targeting vectors were called pTVDpp6.1 and pTVDpp6.2. These vectors were linearized with NotI for electroporation into ES cells.
Gabrb1
A 6.7-kb SacI genomic fragment, isolated from bacterial artificial chromosome 388L8 (Lengeling et al. 1999) and derived from the CITB 129/Sv library (Research Genetics, Huntsville, AL), was subcloned into pBSSKΔBam, and the Neo-tk cassette subsequently was inserted into a unique BamHI site, resulting in 1.9 kb of homology on one side of the cassette, and 4.8 kb on the other. The resulting targeting vector was called pTVGaba.b1. The vector was linearized with NotI for electroporation into ES cells.
ES Cell Culture, Irradiation, and Selection
Detailed protocols for inducing and selecting deletions have been published elsewhere (You et al. 1997b). Briefly, targeted ES cell clones were grown in G418-containing medium on a feeder layer until the time they were trypsinized and irradiated, in suspension, with 400 Gy from a 137Cs source. In general, 2 ×106 irradiated cells were plated onto gelatin-coated, feederless, 150-mm culture dishes, in leukemia inhibitory factor–containing medium without selection. FIAU was added after 72 hours, and this selection was continued for 3 days, with daily medium changes. After selection, the cells were grown until colonies were of a sufficient size to pick into 96-well plates as described (Ramirez-Solis et al. 1993). DNA was prepared either according to that method or by crude lysis overnight at 55 °C in PBND (50 mM KCL, 10 mM Tris, pH 8.3, 2.5 mM MgCl2, 0.1 mg/ml gelatin, 0.45% NP40, 0.45% Tween 20, 50 μg/ml proteinase K) if the samples were to be used exclusively for PCR. One microliter or less of such a lysate was used for PCR.
Microsatellite Marker Analysis
Most microsatellite PCR products were analyzed on 3–3.5% Metaphor gels (FMC). Because these gels may not detect size differences of 1 or 2 bp, it is possible that some of the markers listed as nonpolymorphic may indeed be so. The strains tested for SSLPs were 129/SvJae (extracted from J1 ES cells) and C57BL/6J, the parental strains of the v6.4 ES cells. Markers that we discovered to be polymorphic between 129/SvJae and C57BL/6J are indicated in Figure 2, and the relative allele sizes are indicated in the legend. Two new polymorphic microsatellites were identified to screen all deletions at Dpp6: Dpp6Rep3 (primers ctattcctagagttcacatggtgg and acagaaataccagttcctagaacc) and Dpp6Rep4 (agacaggcagactccctcaagagg and cctcaagtcctctttctgctttgg). A polymophic microsatellite at the Shh locus was amplified with the primers CCTggTCCAACCgAgTgAgAC and CCACggAgTTCTCTgCggAg.
Acknowledgments
This work was supported by grants HD35984 to J.C.S. and HD24180 to M.B. We thank Karen Moore for providing data on 129 versus B6 polymorphisms; Marcy MacDonald for Hdh clones; and Tim O'Brien and Wayne Frankel for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jcs@jax.org; FAX (207) 288-6082.
REFERENCES
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, Ge P, Vonsattel JP, Gusella JF, Joyner AL, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Green P. Against a whole-genome shotgun. Genome Res. 1997;7:410–417. doi: 10.1101/gr.7.5.410. [DOI] [PubMed] [Google Scholar]
- Holdener-Kenny B, Sharan SK, Magnuson T. Mouse albino-deletions: From genetics to genes in development. Bioessays. 1992;14:831–839. doi: 10.1002/bies.950141208. [DOI] [PubMed] [Google Scholar]
- Hough RB, Lengeling A, Bedian V, Lo C, Bucan M. Rump white inversion in the mouse disrupts dipeptidyl aminopeptidase-like protein 6 and causes dysregulation of Kit expression. Proc Natl Acad Sci USA. 1998;95:13800–13805. doi: 10.1073/pnas.95.23.13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DK, Stubbs LJ, Culiat CT, Montgomery CS, Russell LB, Rinchik EM. Molecular analysis of 36 mutations at the mouse pink-eyed dilution (p) locus. Genetics. 1995;141:1563–1571. doi: 10.1093/genetics/141.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Vitaterna MH, Chang AM, Dove WF, Pinto LH, Turek FW, Takahashi JS. The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit. Genetics. 1997;146:1049–1060. doi: 10.1093/genetics/146.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeling A, Wiltshire T, Otmani C, Bucan M. A sequence-ready BAC contig of the GABAA receptor gene cluster Gabrg1– Gabra2-Gabrb1 on mouse chromosome 5. Genome Res. 1999;9:732–738. [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Botta A, Jurecic V, Carattini-Rivera S, Cheah YC, Rosenblatt HM, Bradley A, Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- Liu P, Zhang H, McLellan A, Vogel H, Bradley A. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse chromosome 11. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J, Guarnieri M, Tartaglia K, Flaherty L. High-resolution genetic map and YAC contig around the mouse neurological locus reeler. Mamm Genome. 1994;5:756–761. doi: 10.1007/BF00292008. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R, Davis A, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–877. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- Rinchik E, Russell L. Germ-line deletion mutations in the mouse: Tools for intensive functional and physical mapping of regions of the mammalian genome. Genome Anal. 1990;1:121–158. [Google Scholar]
- Rinchik E, Bell J, Hunsicker P, Friedman J, Jackson I, Russell L. Molecular genetics of the brown (b)-locus region of mouse chromosome 4: Origin and molecular mapping of radiation and chemical induced lethal brown deletions. Genetics. 1994;137:845–854. doi: 10.1093/genetics/137.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert B, Montagutelli X, Houzelstein D, Ferland L, Cohen A, Buckingham M, Guenet JL. Msx1 is close but not allelic to either Hm or Hx on mouse chromosome 5. Mamm Genome. 1994;5:446–449. doi: 10.1007/BF00357006. [DOI] [PubMed] [Google Scholar]
- Russell L, Montgomery C, Raymer G. Analysis of the albino-locus region of the mouse. IV. Characterization of 34 deficiencies. Genetics. 1982;100:427–453. doi: 10.1093/genetics/100.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimenti J, Bucan M. Functional genomics in the mouse: Phenotype-based mutagenesis screens. Genome Res. 1998;8:698–710. doi: 10.1101/gr.8.7.698. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Holdener Kenny B, Ruppert S, Schedl A, Kelsey G, Rinchik EM, Magnuson T. The albino-deletion complex of the mouse: Molecular mapping of deletion breakpoints that define regions necessary for development of the embryonic and extraembryonic ectoderm. Genetics. 1991;129:825–832. doi: 10.1093/genetics/129.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet H. Hm and Hx are not alleles. Mouse News Lett. 1982;66:66. [Google Scholar]
- Tarantino L, Feiner L, Alavizadeh A, Wiltshire T, Hurle B, Ornitz D, Webber A, Raper J, Rowe L, Lengeling A, et al. A high-resolution radiation hybrid map of the proximal portion of mouse chromosome 5. Genomics. 2000;66:65–75. doi: 10.1006/geno.2000.6183. [DOI] [PubMed] [Google Scholar]
- teRiele H, Maandag E, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Nat Acad Sci USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, LaMantia C, Magnuson T. X-ray-induced mutations in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1998;95:1114–1119. doi: 10.1073/pnas.95.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Zimmerman KL, Adamson MC, Yokotani N, Wenthold RJ, Kozak CA. Genetic mapping of the mouse gene encoding dipeptidyl aminopeptidase-like proteins. Mamm Genome. 1993;4:234–237. doi: 10.1007/BF00417570. [DOI] [PubMed] [Google Scholar]
- Weber JL, Myers EW. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- Wilkie TE, Braun RE, Ehrman W, Palmiter R, Hammer R. Germ-line intrachromosomal recombination restores fertility in transgenic MyK-103 male mice. Genes Dev. 1991;5:38–48. doi: 10.1101/gad.5.1.38. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Ricke DO, Denison K, Abmayr S, Cotter PD, Hirschhorn K, Keinanen M, McDonald-McGinn D, Somer M, Spinner N, et al. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6:317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
- Ying HC, Hurle B, Wang Y, Bohne BA, Wuerffel MK, Ornitz DM. High-resolution mapping of tlt, a mouse mutant lacking otoconia. Mamm Genome. 1999;10:544–548. doi: 10.1007/s003359901041. [DOI] [PubMed] [Google Scholar]
- You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, Ticknor C, Jaenisch R, Schimenti J. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nat Genet. 1997a;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]
- You Y, Browning VL, Schimenti JC. Generation of radiation-induced deletion complexes in the mouse genome using embryonic stem cells. Methods. 1997b;13:409–421. doi: 10.1006/meth.1997.0547. [DOI] [PubMed] [Google Scholar]
- You Y, Bersgtram R, Klemm M, Nelson H, Jaenisch R, Schimenti J. Utility of C57BL/6J × 129/SvJae embryonic stem cells for generating chromosomal deletions: Tolerance to gamma radiation and microsatellite polymorphism. Mamm Genome. 1998;9:232–234. doi: 10.1007/s003359900731. [DOI] [PubMed] [Google Scholar]
- Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A. Engineering mouse chromosomes with Cre-loxP: Range, efficiency, and somatic applications. Mol Cell Biol. 2000;20:648–655. doi: 10.1128/mcb.20.2.648-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]