Abstract
We have taken a computational approach to the problem of discovering and deciphering the grammar and syntax of gene regulation in eukaryotes. A logical first step is to produce an annotated catalog of all regulatory sites in a given genome. Likely candidates for such sites are direct and indirect repeats, including three subcategories of indirect repeats: inverted (palindromic), everted, and mirror-image repeats. To that end we have produced a searchable database of inverted repeats of chromosomes III and X of Caenorhabditis elegans, the first completely sequenced multicellular eukaryote. Initial results from the use of this catalog are observations concerning odd/even biases in perfect IRs. The potential usefulness of the catalog as a discovery tool for promoters was shown for some of the genes involved with G-protein functions and for heat shock protein 104 (hsp104).
The completion of the genome sequence of Caenorhabditis elegans marks the beginning of what is likely to be years of database mining of this genome for the purpose of cataloging, organizing, and interpreting actual or putative regulatory motifs by which this multicellular eukaryote coordinates the development and maintenance of differentiation (Clarke and Berg 1998; Goffeau 1998; Brown 1999). The choice of which motifs or types of motifs to search for can be guided in part by the wealth of laboratory research on gene regulation in Drosophila, Caenorhabditis, Strongylocentrotus, and various mammals, as well as the unicellular eukaryote Saccharomyces. That is, a starting place for any search of likely regulatory motifs is to seek ones similar to those already identified in these model organisms.
Transcription profiling has helped to focus some genome-wide studies such as Roth et al. (1998) and van Helden et al. (1998), both of which looked for conserved motifs ∼600–800 bp upstream of well-analyzed assemblages of co-regulated genes in Saccharomyces. Both study groups were optimistic that their search algorithms had found some (but not necessarily all) previously known regulatory motifs and had discovered likely new motifs. Numerous other studies of model multicellular eukaryotes, such as Drosophila and Caenorhabditis, have revealed many conserved regulatory motifs and these have been widely used in chromosome and genome-wide searches (e.g., Wong et al. 1985).
Upon publication of the complete Caenorhabditis genome, an analysis of IRs, as well as other repetitive sequences, was carried out (C. elegans Sequencing Consortium 1998). In that study, inverted repeats (IRs) up to 2000 bp in length with loops of any size within the 2000-bp range were searched for. IRs of that description were found in 3.6% of the Caenorhabditis genome but were most densely clustered on the autosomal arms. Furthermore, a disproportionate number of IRs were found within genes, presumably as parts of introns. Frontali and Pizzi (1999) analyzed recurring patterns of short oligonucleotides of introns and intergenic regions of Caenorhabditis chromosome III and found those areas to be repetitive.
Our initial focus on IRs was based on their known functions in three different types of regulatory mechanism: (1) Some IRs are symmetrical sites for the binding of regulatory factors, especially dimeric factors such as leucine zippers. (2) Some IRs provide opportunities for distinctive internal basepairing and regulation in RNAs. (3) Some IRs use transposon-like mechanisms to rearrange and regulate genes, and transposons themselves are likely to have been the origin for some regulatory sequences (Britten 1997).
We have produced a web-based, searchable, annotated catalog for Caenorhabditis chromosomes III and X comprising all possible IRs of sizes ranging from 20–200 bp with 0–10% mismatches in the stems and non-base-paired loops of up to one-third the length of the IR. The database may be accessed through queries based on size, location, or sequence. Each IR is identified in respect to location, nearest downstream gene, frequency, similar sequences, and links to the Caenorhabditis genome project. Furthermore, there is a selectable option to show a graphical representation of any IR, internally base paired with the minimum number of mismatches.
The constraints of the search were such that it is unlikely that the results show functional transposons, which tend to be hundreds to thousands of base-pairs long with terminal IRs representing a small fraction of sequence, if present at all. Our limit on loop size would exclude most typical transposons. Even MITES (miniature inverted repeat transposable elements) which are abundant in Caenorhabditis and are in the low hundreds of base pairs long are probably not represented in most of the data reported here (Surzycki and Belknap 2000). An exception might be IRs at the high end of the range, about 200 bp long with loops of 60 bp or more, placing them within boundaries of some MITES. (Inverted repeats of that length are available for viewing at the web site but were not the focus of this analysis.)
Moderately repetitive microsatellites (2–5 bp) and minisatellites (∼15 bp), as well as highly repetitive satellites (5–100 bp), are tandemly repeated sequences, some of which appear to propagate themselves by uneven crossovers or replication slippage (as defined in Charlesworth et al. 1994). Some satellite repeats are found associated with or as parts of IRs, as well as in typical tandem arrays. In general the functions of satellite repeats are not well defined or understood. However, in some cases, such as in promoters for heat shock genes, repetitive satellite-like sequences of heat shock elements (HSEs) seem to be important for transcription (e.g., Wiederrecht et al. 1987). Thus, although many smaller satellite repeats are below the limits of detection for this study, the ones that were revealed may be of potential interest as regulators. A case in point is the promoter region of the gene for hsp104 described in this paper.
RESULTS
Short Inverted Repeats Show Specific Biases for Odd or Even Lengths
Tables 1 and 2 show the totals of all non-AT-rich, perfect IRs from length 4–60 bp on chromosomes III and X, along with a list of randomly generated IRs of the same lengths. “AT-rich” and “CG-rich” are defined here as any sequences composed exclusively of As and Ts or Cs and Gs, respectively. All IRs have been unnested, which means that only counts of unique sequences are shown. Only IRs without loops in the middle were counted. In both chromosomes, for lengths 5–19 Bp there is a strong bias toward odd lengths, which is strikingly different from the randomly generated sequences of the same length.
Table 1.
Perfect Loopless Inverted Repeats from Size 4 bp to 60 bp in Chromosome III or Random Sequence
| Length | Sum of non-AT-rich chrom. III | Sum of non-AT-rich using random sequence | Sum of AT-rich chrom. III | Sum of AT-rich using random sequence | Sum of CG-rich chrom. III |
|---|---|---|---|---|---|
| 4 | 209440 | 242685 | 298769 | 410408 | 54611 |
| 5 | 349726 | 389475 | 270417 | 262959 | 23093 |
| 6 | 72480 | 91991 | 83467 | 84158 | 4700 |
| 7 | 119561 | 122669 | 71285 | 53441 | 2627 |
| 8 | 25272 | 30044 | 21354 | 17433 | 640 |
| 9 | 39526 | 36172 | 17258 | 11198 | 246 |
| 10 | 9367 | 9332 | 6426 | 3402 | 132 |
| 11 | 13435 | 10526 | 5356 | 2292 | 35 |
| 12 | 3477 | 2760 | 2151 | 703 | 39 |
| 13 | 5163 | 2947 | 1990 | 450 | 7 |
| 14 | 1509 | 795 | 939 | 146 | 7 |
| 15 | 1844 | 902 | 755 | 96 | 4 |
| 16 | 597 | 186 | 402 | 29 | 1 |
| 17 | 630 | 238 | 299 | 21 | 0 |
| 18 | 217 | 76 | 199 | 2 | 0 |
| 19 | 256 | 60 | 102 | 3 | 0 |
| 20 | 108 | 19 | 116 | 3 | 0 |
| 21 | 106 | 16 | 40 | 0 | 0 |
| 22 | 61 | 2 | 64 | 0 | 0 |
| 23 | 43 | 2 | 4 | 1 | 0 |
| 24 | 43 | 1 | 44 | 0 | 0 |
| 25 | 16 | 2 | 4 | 0 | 0 |
| 26 | 9 | 0 | 41 | 0 | 0 |
| 27 | 6 | 0 | 0 | 0 | 0 |
| 28 | 9 | 0 | 32 | 0 | 0 |
| 29 | 4 | 0 | 3 | 0 | 0 |
| 30 | 14 | 0 | 31 | 0 | 0 |
| 31 | 5 | 0 | 0 | 0 | 0 |
| 32 | 5 | 0 | 29 | 0 | 0 |
| 33 | 3 | 0 | 0 | 0 | 0 |
| 34 | 2 | 0 | 27 | 0 | 0 |
| 35 | 1 | 0 | 0 | 0 | 0 |
| 36 | 2 | 0 | 22 | 0 | 0 |
| 37 | 0 | 0 | 0 | 0 | 0 |
| 38 | 4 | 0 | 19 | 0 | 0 |
| 39 | 2 | 0 | 0 | 0 | 0 |
| 40 | 0 | 0 | 18 | 0 | 0 |
| 41 | 0 | 0 | 0 | 0 | 0 |
| 42 | 1 | 0 | 15 | 0 | 0 |
| 43 | 0 | 0 | 0 | 0 | 0 |
| 44 | 0 | 0 | 11 | 0 | 0 |
| 45 | 0 | 0 | 0 | 0 | 0 |
| 46 | 9 | 0 | 6 | 0 | 0 |
| 47 | 0 | 0 | 0 | 0 | 0 |
| 48 | 0 | 0 | 6 | 0 | 0 |
| 49 | 0 | 0 | 0 | 0 | 0 |
| 50 | 0 | 0 | 5 | 0 | 0 |
| 51 | 0 | 0 | 0 | 0 | 0 |
| 52 | 0 | 0 | 4 | 0 | 0 |
| 53 | 0 | 0 | 0 | 0 | 0 |
| 54 | 1 | 0 | 4 | 0 | 0 |
| 55 | 0 | 0 | 0 | 0 | 0 |
| 56 | 0 | 0 | 4 | 0 | 0 |
| 57 | 0 | 0 | 0 | 0 | 0 |
| 58 | 0 | 0 | 4 | 0 | 0 |
| 59 | 1 | 0 | 0 | 0 | 0 |
| 60 | 0 | 0 | 4 | 0 | 0 |
“AT-rich” and “CG-rich” are defined as being comprised of those bases exclusively. “Non-AT-rich” sequences are heterogeneous in respect to base content. “Perfect” repeats of odd length are defined as including a single base at the central position but otherwise show complete base pairing in the stem. Perfect repeats of even length have no mismatches. Random files for analysis were generated with 36% GC content.
Table 2.
Perfect Loopless Inverted Repeats from Size 4 bp to 60 bp in Chromosome X or Random Sequence
| Length | Sum of non-AT-rich chrom. X | Sum of non-AT-rich using random sequence | Sum of AT-rich chrom. X | Sum of AT-rich using random sequence | Sum of CG-rich chrom. X |
|---|---|---|---|---|---|
| 4 | 293619 | 354399 | 444230 | 597333 | 67087 |
| 5 | 539236 | 567437 | 372913 | 381917 | 30238 |
| 6 | 107428 | 133637 | 113369 | 122313 | 6441 |
| 7 | 178237 | 177227 | 95042 | 78315 | 2727 |
| 8 | 36950 | 43738 | 29496 | 25068 | 682 |
| 9 | 56311 | 53252 | 23833 | 15861 | 378 |
| 10 | 12571 | 13410 | 8013 | 5263 | 180 |
| 11 | 17887 | 15149 | 6173 | 3206 | 44 |
| 12 | 4210 | 4006 | 2502 | 993 | 32 |
| 13 | 6082 | 4282 | 1823 | 601 | 9 |
| 14 | 1608 | 1120 | 905 | 213 | 7 |
| 15 | 2003 | 1214 | 547 | 136 | 2 |
| 16 | 530 | 339 | 448 | 50 | 3 |
| 17 | 657 | 334 | 181 | 28 | 1 |
| 18 | 190 | 130 | 260 | 5 | 0 |
| 19 | 287 | 96 | 62 | 6 | 0 |
| 20 | 97 | 21 | 167 | 0 | 0 |
| 21 | 99 | 31 | 18 | 1 | 0 |
| 22 | 51 | 12 | 128 | 1 | 0 |
| 23 | 35 | 8 | 3 | 0 | 0 |
| 24 | 46 | 2 | 91 | 0 | 0 |
| 25 | 7 | 1 | 2 | 0 | 0 |
| 26 | 8 | 0 | 79 | 0 | 0 |
| 27 | 10 | 0 | 0 | 0 | 0 |
| 28 | 3 | 0 | 75 | 0 | 0 |
| 29 | 2 | 0 | 0 | 0 | 0 |
| 30 | 4 | 0 | 61 | 0 | 0 |
| 31 | 3 | 0 | 0 | 0 | 0 |
| 32 | 5 | 0 | 56 | 0 | 0 |
| 33 | 2 | 0 | 0 | 0 | 0 |
| 34 | 4 | 0 | 52 | 0 | 0 |
| 35 | 0 | 0 | 0 | 0 | 0 |
| 36 | 0 | 0 | 47 | 0 | 0 |
| 37 | 0 | 0 | 0 | 0 | 0 |
| 38 | 1 | 0 | 46 | 0 | 0 |
| 39 | 1 | 0 | 0 | 0 | 0 |
| 40 | 0 | 0 | 38 | 0 | 0 |
| 41 | 0 | 0 | 0 | 0 | 0 |
| 42 | 0 | 0 | 39 | 0 | 0 |
| 43 | 0 | 0 | 1 | 0 | 0 |
| 44 | 0 | 0 | 36 | 0 | 0 |
| 45 | 0 | 0 | 0 | 0 | 0 |
| 46 | 3 | 0 | 34 | 0 | 0 |
| 47 | 0 | 0 | 0 | 0 | 0 |
| 48 | 0 | 0 | 32 | 0 | 0 |
| 49 | 0 | 0 | 0 | 0 | 0 |
| 50 | 0 | 0 | 30 | 0 | 0 |
| 51 | 0 | 0 | 0 | 0 | 0 |
| 52 | 0 | 0 | 30 | 0 | 0 |
| 53 | 0 | 0 | 0 | 0 | 0 |
| 54 | 0 | 0 | 26 | 0 | 0 |
| 55 | 0 | 0 | 0 | 0 | 0 |
| 56 | 0 | 0 | 23 | 0 | 0 |
| 57 | 0 | 0 | 0 | 0 | 0 |
| 58 | 0 | 0 | 20 | 0 | 0 |
| 59 | 0 | 0 | 0 | 0 | 0 |
| 60 | 0 | 0 | 17 | 0 | 0 |
See Table 1 footnote for relevant definitions.
The odd number biases described may well continue for lengths greater than 25 bp; however this would not be reflected in the greatly reduced sample size for longer sequences. Note that a certain amount of odd-number bias is also seen in the random data due to our definition of odds as being perfect, although, in fact, they were allowed a wild card of any base at the middle position. The experimental data show a greater bias, especially for lengths over 9 bp, for which there are many more perfect repeats than what would be expected from random combinations.
Tables 1 and 2 also show the totals of all AT-rich IRs. This is contrasted with the sums of all CG-rich inverted repeats. Here there is a pronounced even bias for AT-rich IRs of 20–60 bp on chromosome III (18–60 on X), Although there is no particular bias for CG-rich repeats and, indeed, there are relatively few of those. For lengths greater than 30 bp on chromosome III, there are no odd lengths of AT-rich IRs and only one on chromosome X after length 25 bp.
Table 3 compares genic and intergenic sequences of 4–25 bp. In general, and for both chromosomes, the biases described above are still evident after this rearrangement of the data. However, decreasing sample size at greater lengths prevents simple extrapolation of these trends. Intergenic non-AT-rich repeats represent 40–70% of chromosome III and 58–74% of X. For AT-rich repeats, 50–78% are intergenic on chromosome III and 54–69% on X. However, the two chromosomes are not directly comparable in that a smaller proportion of chromosome III is composed of intergenic regions.
Table 3.
Counts and Percentages of Genic vs. Intergenic Sequences for Non-AT-Rich and AT-Rich Perfect Loopless Inverted Repeats from Size 4 bp to 25 bp in Chromosomes III and X
| Length | Non-AT-Rich | AT-Rich | ||||||
|---|---|---|---|---|---|---|---|---|
| genic | % genic | intergenic | % intergenic | genic | % genic | intergenic | % intergenic | |
| a. Chromosome III | ||||||||
| 4 | 101986 | 49 | 107454 | 51 | 132454 | 44 | 166315 | 56 |
| 5 | 170671 | 49 | 179055 | 51 | 116919 | 43 | 153498 | 57 |
| 6 | 35749 | 49 | 36731 | 51 | 35258 | 42 | 48209 | 58 |
| 7 | 56782 | 47 | 62779 | 53 | 30219 | 42 | 41066 | 58 |
| 8 | 11985 | 47 | 13287 | 53 | 9148 | 43 | 12206 | 57 |
| 9 | 17769 | 45 | 21757 | 55 | 7517 | 44 | 9741 | 56 |
| 10 | 4157 | 44 | 5210 | 56 | 2692 | 42 | 3734 | 58 |
| 11 | 5941 | 44 | 7494 | 56 | 2279 | 43 | 3077 | 57 |
| 12 | 1457 | 42 | 2020 | 58 | 898 | 42 | 1253 | 58 |
| 13 | 2203 | 43 | 2960 | 57 | 846 | 43 | 1144 | 57 |
| 14 | 610 | 40 | 899 | 60 | 408 | 43 | 531 | 57 |
| 15 | 781 | 42 | 1063 | 58 | 328 | 43 | 427 | 57 |
| 16 | 270 | 45 | 327 | 55 | 156 | 39 | 246 | 61 |
| 17 | 244 | 39 | 386 | 61 | 125 | 42 | 174 | 58 |
| 18 | 79 | 36 | 138 | 64 | 71 | 36 | 128 | 64 |
| 19 | 96 | 38 | 160 | 63 | 46 | 45 | 56 | 55 |
| 20 | 44 | 41 | 64 | 59 | 36 | 31 | 80 | 69 |
| 21 | 34 | 32 | 72 | 68 | 13 | 33 | 27 | 68 |
| 22 | 13 | 21 | 48 | 79 | 14 | 22 | 50 | 78 |
| 23 | 13 | 30 | 30 | 70 | 2 | 50 | 2 | 50 |
| 24 | 26 | 60 | 17 | 40 | 10 | 23 | 34 | 77 |
| 25 | 7 | 44 | 9 | 56 | 1 | 25 | 3 | 75 |
| b. Chromosome X | ||||||||
| 4 | 191546 | 42 | 265782 | 58 | 205155 | 34 | 395292 | 66 |
| 5 | 327528 | 41 | 473331 | 59 | 167322 | 33 | 333276 | 67 |
| 6 | 66200 | 40 | 97509 | 60 | 50884 | 33 | 105333 | 67 |
| 7 | 103472 | 40 | 158151 | 60 | 41155 | 32 | 86530 | 68 |
| 8 | 21744 | 39 | 34537 | 61 | 13774 | 32 | 29074 | 68 |
| 9 | 31800 | 38 | 51586 | 62 | 10329 | 32 | 22314 | 68 |
| 10 | 7222 | 37 | 12109 | 63 | 4312 | 32 | 9040 | 68 |
| 11 | 9845 | 36 | 17230 | 64 | 2828 | 32 | 5982 | 68 |
| 12 | 2450 | 36 | 4310 | 64 | 1711 | 32 | 3628 | 68 |
| 13 | 3104 | 34 | 6084 | 66 | 854 | 32 | 1783 | 68 |
| 14 | 870 | 34 | 1680 | 66 | 924 | 33 | 1913 | 67 |
| 15 | 1006 | 32 | 2100 | 68 | 253 | 31 | 561 | 69 |
| 16 | 291 | 31 | 651 | 69 | 659 | 34 | 1273 | 66 |
| 17 | 337 | 31 | 766 | 69 | 94 | 35 | 173 | 65 |
| 18 | 120 | 29 | 292 | 71 | 519 | 35 | 965 | 65 |
| 19 | 139 | 31 | 307 | 69 | 28 | 33 | 58 | 67 |
| 20 | 61 | 27 | 161 | 73 | 438 | 36 | 786 | 64 |
| 21 | 52 | 33 | 107 | 67 | 11 | 46 | 13 | 54 |
| 22 | 32 | 26 | 93 | 74 | 386 | 37 | 671 | 63 |
| 23 | 21 | 35 | 39 | 65 | 2 | 33 | 4 | 67 |
| 24 | 19 | 26 | 55 | 74 | 342 | 37 | 587 | 63 |
| 25 | 8 | 32 | 17 | 68 | 1 | 33 | 2 | 67 |
For lengths greater than 25 bp, low sample size prevents simple extrapolation of trends. Note that proportionally less of chromosome III is comprised of intergenic sequences and therefore the two chromosomes are not strictly comparable.
Repeats Are Either Evenly Distributed Across Chromosomes or Are More Prevalent on the Chromosome Arms
Perfect repeats of sizes 4–30 bp were examined in respect to their distribution on chromosomes III and X. Also repeats composed exclusively of A and T were analyzed separately. For both chromosomes, all repeats of 4–9 bp were distributed evenly. At increasing lengths the repeats of chromosome III were found to be more abundant on the arms forming a bowl-shaped distribution. The distribution for chromosome X was more even and only slightly bowl-shaped. The AT-rich sequences of chromosome III were more abundant on the arms with a bowl-shaped distribution, becoming more marked at greater lengths (15–30 bp). The AT-rich sequences of chromosome X were fairly evenly distributed, with only slight bias toward the arms. However, spikes of AT-rich repeats interrupted the even distribution on the X, indicating clusters of very AT-rich sequence.
Several Clusters of Inverted Repeats of 25–60 bp Are Highly Conserved and Contain Known Conserved Motifs
Perfect IRs of >25 bp have a strong possibility of being biologically significant and therefore are worth investigating further. There are no IRs >25 bp generated from random sequence. The presence of IRs of those lengths and greater on chromosomes III and X suggests that they are the result of evolutionary selection and conservation. In particular, Table 1 shows at least two anomalous clusters of non-AT-rich perfect IRs: 30 bp and 46 bp, as well as about 8 other perfect, loopless IRs between 25 and 60 bp.
In order to show the usefulness of this software and graphical display as a discovery tool for promoters, some repeats were selected for additional analysis. For the two largest anomalous clusters in chromosome III (30 bp and 46 bp), conserved motifs were searched by compiling a list of all the nucleotide strings 5 bp, in proximity to the pivot points of the IRs, and then querying the literature as to whether any were published motifs. Three motifs were found, one of which, GTGAC, was chosen for further detailed analysis (Tables 4,5).
Table 4.
Perfect, Loopless, Intergenic Inverted Repeats Size 25–60 bp with GTGAC Motif-Chromosome III
| Starting bp | Size (bp) of inverted repeat | Between these two genes on direct strand (protein ID from GenBank) | Between these two genes on indirect strand (protein ID from GenBank) | ||
|---|---|---|---|---|---|
| 773,376 | 46 | 3844610 (unknown) | 2088857 (unknown) | 2088860 (unknown) | 2088861 (unknown) |
| 7,671,060 | 46 | 388605 (homology with vitronectin receptor α subunit) | 388597 (unknown) | 388598 (unknown) | 3878108 (similar sugar transporter) |
| 9,333,805 | 46 | 3879514 (unknown) | 3874820 (probable rab gap domain) | 3874822 (similar UMP synthetase) | 3878636 (similar cAMP dependent protein kinase) |
| 12,071,592 | 46 | 3880555 (similar serine threonine kinase) | 3878440 (similar central domain tyrosinase) | 3880554 (unknown) | 3878436 (unknown) |
| 12,071,593 | 44 | 3880555 (similar serine threonine kinase) | 3878440 (similar central domain tyrosinase) | 3880554 (unknown) | 3878436 (unknown) |
| 4,281,164 | 38 | 485148 (unknown) | 485147 (unknown) | 485149 (unknown) | 488150 (unknown) |
| 4,237,391 | 30 | 485157 (similar tyrosine kinase receptor) | 746490 (similar yeast SSDI protein) | 485160 (unknown) | 485159 (unknown) |
| 10,201,272 | 30 | 3925251 (unknown) | 3880324 (unknown) | 3925249 (similar zinc finger C2H2 type) | 3880322 (unknown) |
| 2,479,727 | 28 | 3874994 (similar pyridine nucleotide di-sulphide oxide reductase class I) | 3874288 (unknown) | 3880235 (similar family I of G-protein-coupled receptors) | 3880236 (similar cyclophilin-like NK tumor recognition) |
| 8,317,436 | 26 | 3873800 (similar TPR domain) | 3877610 (similar domain to disheveled, Egl-10, Pleckstrin) | 3873792 (putative aspartyl tRNA synthetase) | 3877607 (similar super oxide dismutase) |
| 10,461,309 | 25 | 3880323 (unknown) | 3880319 (similar ankrin repeat) | 3880324 (unknown) | 3880320 (similar α tubulin subunit) |
| 10,325,368 | 25 | 3925251 (unknown) | 3880324 (unknown) | 3925249 (similar zinc finger C2H2 type) | 3880322 (unknown) |
Perfect loopless inverted repeats size 25–60 bp with GTGAC motif found in the intergenic regions of Caenorhabditis chromosome III.
Table 5.
Perfect, Loopless Inverted Repeats Size 25–60 bp with GTGAC Motif Found within Genes of Chromosome III
| Starting bp | Size of inverted repeat (bp) | Protein ID number from GenBank (within this gene) |
|---|---|---|
| 644,673 | 46 | 485123 (like lipase in short region) |
| 3,174,166 | 46 | 3873684 (unknown) |
| 5,326,052 | 46 | 485139 (acetylcholine receptor) |
| 5,792,108 | 46 | 1072163 (protein kinase) |
| 11,958,991 | 46 | 3879796 (unknown) |
| 4,427,747 | 38 | 2873385 (dauer development regulatory protein) |
| 5,778,038 | 30 | 532821 (unknown) |
| 6,021,399 | 28 | 459001 (tsr repeats as in thrombospondin and prosperdin) |
| 3,664,678 | 28 | 687891 (unknown) |
| 2,531,460 | 28 | 3876659 (proline rich domain partly rd protein) |
Perfect loopless inverted repeats size 25–60 bp with GTGAC motif found within genes of Caenorhabditis chromosome III.
Table 4 lists all 12 intergenic repeats containing GTGAC, a sequence that binds estrogen receptor and AP-1 factor in mammals, sometimes as a palindrome (Weisz and Rosales 1990). The motif has not been reported previously in Caenorhabditis. Note that the only perfect lengths that contain this motif are 46, 44, 38, 30, 28, 26, and 25 bp and that those sequences are distributed fairly evenly across the 13 million bp length of chromosome III. It is remarkable that all non-AT-rich, 46-bp perfect IRs on chromosome III are identical. The following is half of the sequence of the repeat. (See also Fig. 1.)
Figure 1.
The structures of all twenty-one 46 bp perfect or imperfect inverted repeats that contain the GTGAC motif on Caenorhabditis elegans chromosome III. Note that the drawings of the repeats are for the purpose of visualizing the symmetries and are not meant to imply that these occur in vivo or in vitro or that they have been analyzed thermodynamically for base pairing potential.
ATGTATTTAAATACATTTGTGAC
Furthermore, subsets of this sequence are conserved in the shorter IRs containing GTGAC.
In Table 5, the ten IRs with a GTGAC motif, which are found within genes, are listed. These also have a conserved full-length sequence, but unlike their intergenic counterparts, most are clustered within genes located between about 2.5–6.5 million bp on chromosome III.
Because all the perfect IRs of length 46 were identical and contained the GTGAC motif, we looked at irregular IRs (with GTGAC) of that length for up to 10% mismatches on the stem, including loops of up to one-third of the length of the IR. There were 12 such repeats. Figure 1 shows the configurations for all 46-bp IRs on chromosome III with up to 10% mismatches. There is a noticeable tendency for those repeats to exhibit conserved symmetry even if mismatched.
The search for perfect IRs with GTGAC was repeated for the X chromosome. Only six were found, two of 26 bp, one of 30 bp, and three of 46 bp. The nearest downstream genes were of unknown identity except for a collagen gene (GenBank PID 3874534) and a glycogenin (PID 3879754). All of the 46-bp repeats on the X chromosome were identical in sequence to those of the same length on III.
Two other motifs from IRs of 30 bp were examined: TAGGTCA and CTAAAT. Neither of these has been reported in Caenorhabditis. TAGGTCA is a motif of RZR (retinoid-related orphan receptors)(Carlberg et al., 1994). Furthermore, a sequence for estrogen response element (ERE) GGTCA and its inverted complement TGACC are found within TAGGTCA and its inverted complement TGACCTA (Krawczyk et al. 1993). The TAGGTCA motif was searched for all perfect, loopless IRs of lengths 25–60 bp on chromosome III and found exclusively in those of length 30 bp. We found TAGGTCA present in two copies of a 30-bp IR upstream of a gene for 5 estradiol 17 beta hydrogenase 3.
CTAAAT is a motif involved in regulation of iron metabolism in Pseudomonas (Rombel et al. 1995). The motif CTAAAT, initially found on chromosome III in three 30-bp IRs, was also searched within perfect, loopless repeats of 25–60 bp and was found only at lengths 25 (one copy), 28 (three copies), and 38 (two copies). Genes associated with this motif (with their GenBank PID numbers) were P59 protein (861,392), cox-17 (3,790,743), protein phosphatase (849,241) and nucleolus-cytoplasm shuttle phosphoprotein (669,020), as well as four genes of unknown function.
These searches for IRs with TAGGTCA and CTAAAT were repeated for the X chromosome. Remarkably, there were no IRs either perfect or imperfect with those motifs, with one exception: two imperfects of size 25 contained TAGGTCA, both associated with genes of unknown functions.
GTGAC and TAGGTCA Are in Several Repeats Associated with hsp104
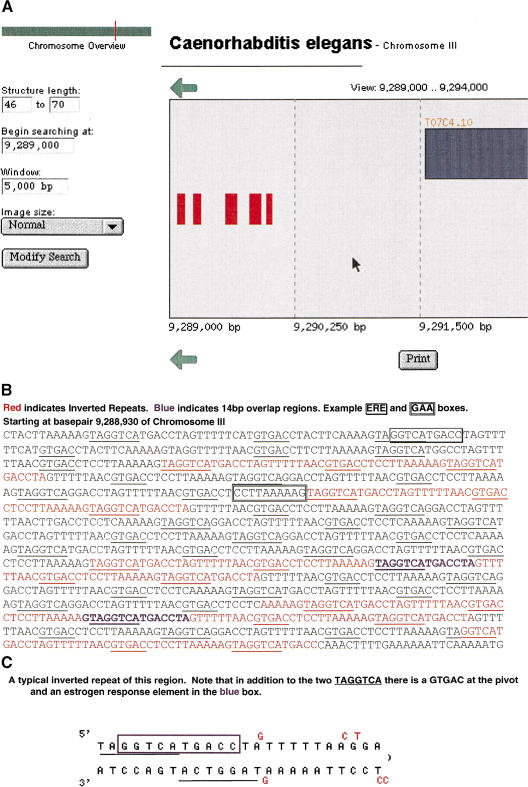
Interest in the GTGAC and TAGGTCA motifs led us to the discovery of eight IRs in the promoter region of a gene for hsp104 (PID # 3979898)(Fig. 2). The eight repeats are clustered in five regions, the first two of which are IRs of 54 bp each. The third region contains two overlapping 54-bp IRs. The overlap area is a 14-bp IR. The fourth region has three overlapping IRs, two of 54 bp and one of 66 bp, with another 14-bp repeat in an overlap region. The fifth region is a 50-bp IR (Fig. 2a,b). All eight repeats are conserved in respect to sequence and shape. All but the 50-bp repeat include GTGAC and two TAGGTCAs as direct repeats (Fig. 2); the 50-bp repeat contains just one each of the two motifs. Furthermore, the background sequence in this area is full of other GTGACs and TAGGTCAs, as well as irregular IRs with >10% mismatches and therefore below the resolution limit for this search.
Figure 2.
Inverted repeats in promoter region of a gene for heat shock protein hsp104. Note that this locus is rich with many additional inverted repeats of the type highlighted except that they fall below the limits of resolution of the search that stipulated no more than 10% mismatches on the stem. Motifs in this region include numerous GAA boxes and estrogen response elements (ERE) both identified in other heat shock promoters as well as GTGAC and TAGGTCA motifs. Two examples of GAA and ERE are boxed in this figure. The locus resembles the satellite-like configuration reported in other heat shock promoters.
DISCUSSION
The current wealth of sequence data has been amassed at a much faster rate than the rate at which we can convert it into useful biological information. Multiple independent approaches are needed to organize these data into usable results. The approach we took for this study was to construct a new tool to analyze potential promoters by focusing on certain types of structural symmetries reflected in DNA sequence. A searchable annotated catalog of IRs of Caenorhabditis chromosomes III and X was generated for this study. It continues to be useful as a discovery tool in searches for and analyses of promoters. Both interesting numerical trends and conserved motifs within IRs were found and analyzed.
Odd/Even Bias in Short Perfect Inverted Repeats
Athough there is no simple numerology to gene regulation, and mere counting will not reveal any complex patterns, there are some intriguing numerical trends that are worth noting. Just as Chargaff (1950) observed that C = G and T = A, simple patterns, in some cases, have important but as yet unknown biological significance.
This analysis of chromosomes III and X of Caenorhabditis has revealed, so far, two numerical trends: a bias for odd-length non-AT-rich IRs and a bias for even-length AT-rich IRs. Our discovery tool is set up so that researchers may visit the web site and perhaps discover other interesting patterns. A bias for odd-length non-AT-rich IRs between 5 bp and 19 bp has not been reported in the literature. Nor has the pronounced bias for even-length AT-rich IRs between 20 bp and 60 bp. However, Cox and Mirkin (1997), comparing the genomes of several prokaryotes and eukaryotes (although not Caenorhabditis) noted that most long IRs were <10% GC content. We also found a paucity of GC-rich IRs.
In prokaryotes, it has been observed that IRs of 4, 5, and 6 bp are avoided because they would often comprise restriction sites (Gelfand and Koonin 1997). Also Robinson et al. (1995) found a particular octameric IR was especially abundant in cyanobacteria. The biases we are reporting for Caenorhabditis chromosomes III and X, although they cannot yet be explained, suggest a strong evolutionary selection that might also exist in other eukaryotic genomes.
Distribution of Inverted Repeats on Chromosomes III and X
Results from this analysis of distribution did not differ markedly from that of the C. elegans Sequencing Consortium (1998), even though its distribution study included irregular repeats up to 2000 bp, while our analysis was of all perfect repeats of size 4–30 bp, with an additional analysis of repeats composed exclusively of A and T. We did not include longer IRs in the analysis because of their scarcity. The C. elegans Sequencing Consortium found that IRs were fairly uniform across the X chromosome, with only a slight bias to the chromosome arms. The consortium's analysis of chromosome III showed a more bowl-shaped distribution, with a bias to the arms. We confirmed these results. In addition, by separating out the data for AT-rich IRs, we found some pronounced spikes or clusters on the X that are not as obvious when all of the data are viewed together. This suggests that the X has denser clusters of AT-rich sequence than chromosome III. Further analysis of the AT-rich regions was not pursued in this study; the data are available at our web site.
Genes Putatively Associated with the Functions of G-Proteins and GTGAC
Cascades involving G-proteins are universal throughout the eukaryotes and are used in a wide diversity of situations in which a signal is transferred from a cell surface receptor to the inside of the cell. It is likely that eukaryotic multicellularity was a selection pressure for the fine-tuning of this and other means of cell–cell communication. A typical G-protein cascade begins with the binding of a receptor by a hormone, neurotransmitter, or other factor or by analogous drugs. Binding stimulates the assembly of G-proteins, which in turn initiates the activity of effector molecules, followed by the release of second messengers such as cAMP. The second messengers activate protein kinases specific to threonine and serine, which mediate cell responses, including transcription of genes.
The well-studied ras family of oncoproteins is a family of modified G-proteins involved in signal transduction cascades sensitive to mitogens. One of the results of such a cascade may be the activation of transcription factor AP-1 (activator protein-1, composed of products of fos and jun). The promoter region to which AP-1 binds often has a GTGAC motif sometimes within or next to an IR. Weisz and Rosales (1990) were first to report GTGAC as part of an imperfect palindrome capable of binding both estrogen receptor and AP-1 within a promoter region of c-fos in mammalian cells. Wasylyk et al. (1991) showed the same motif in an inverted repeat of the AP-1 binding region upstream of a gene for matrix degradation in mammals.
The GTGAC motif can also appear upstream of ras genes themselves. For example, in Drosophila as well as in mammals, the ras promoter region is bidirectional. The ras promoter of Drosophila is situated between ras and rop (ras opposite) and regulates both by means of a site that includes an AP-1 binding region with the GTGAC motif (Lightfoot et al. 1994).
There has been no previous description of GTGAC motifs in Caenorhabditis. It makes sense that the many components of a specific G-protein-related cascade would be coordinately regulated. Thus, our observations, described below, concerning common motifs in promoters of genes of some G-related proteins may well have regulatory significance.
Table 4 shows all of the perfect, intergenic IRs of 25 bp to 60 bp containing a GTGAC motif on chromosome III. Such repeats were found only at lengths 46, 44, 38, 30, 28, 26, and 25 bp. Furthermore, in all cases of these perfect IRs, GTGAC was positioned to be half of the pivot point of the palindrome. For each repeat, the immediate upstream and downstream genes for both direct and indirect strands were noted. Of the 48 genes (or predicted genes) listed, 21 have no identified homologies or similarity with known genes. Of the 27 genes that are meaningfully annotated, seven are tentatively associated with the functioning of G-proteins. One of these is similar to a family I G-protein-coupled receptor, three are similar to protein kinases, one has a tyrosinase domain, one has a probable rab-gap domain, and one has a domain similar to that of disheveled and egl-1 associated with G-protein regulation (Elmore et al. 1998).
Ten IRs with the GTGAC motif are found within genes of chromosome III, presumably as parts of introns (Table 5). Of these, two may have an association with G proteins: a protein kinase and a gene with repeats similar to those of thrombospondin (Frazier et al. 1999). Therefore, the total of genes tentatively associated with both G proteins and GTGAC is at least ten, counting only those genes either immediately up- or downstream or containing GTGAC.
Irregular IRs of 46 bp (with mismatches up to 10% and loops up to one-third the length) and containing GTGAC were searched. In many cases, the symmetry of the IR is preserved in spite of mismatches (Fig. 1). Furthermore, in many cases, GTGAC preserves its position as half of the pivot point of the palindrome or is maintained as an asymmetrical part of the pivot. Most of the genes associated with these mismatch repeats were of unknown function.
These results suggest the potential usefulness of search algorithms for identifying potential promoter motifs. In particular, the DNA motif GTGAC, especially as part of an IR may be part of a cis-regulatory system for genes associated with the functions of G-proteins. It should be noted, however, that there are at least 14 genes on chromosome III identified at NCBI as possibly coding for G-protein receptors per se and only two of these were found to be associated with GTGAC IRs either up- or downstream, with that motif as half of a perfect 10-bp palindrome. One is upstream of PID 3880235 (Table 4) and the other is upstream of PID 3881443. (Both are GenBank protein ID numbers.) Furthermore, there are 28 genes on the X chromosome identified at as G-protein receptors, none of which were associated with GTGAC motif repeats of the type described in this study.
Other Motifs: TAGGTCA and CTAAAT Associated with Inverted Repeats
Two other published motifs, TAGGTCA and CTAAAT, were present on chromosome III as parts of perfect IRs. Both examples of TAGGTCA were within 30-bp repeats, upstream of a gene for 5 estradiol 17 beta hydrogenase 3. Carlberg et al. (1994) reported this motif as part of a retinoid-related orphan receptor that may function as part of a palindrome or direct repeat and Krawczyk et al. (1993) describe an ERE-motif GGTCA, which is reported here as part of TAGGTCA. The location of the two motifs upstream of 5 estradiol 17 beta hydrogenase 3 may be significant in that 17 beta-estradiol can cause up-regulation of some genes via ERE (Lee and Mouradian 1999).
The genes associated with IRs containing CTAAAT do not have obvious connections; they are the genes for P 59 protein, cox 17, protein phosphatase, and nucleolus-cytoplasm shuttle phosphoprotein. Rombel et al. (1995) reported that motif as part of a consensus G/CCTAAAT-CCC, a putative transcription-factor-binding site for bacterial iron regulation.
Genes Similar to Yeast hsp104 and yK10 with GTGAC and TAGGTCA Motifs
A search of chromosome III for irregular, 46-bp IRs with both GTGAC and TAGGTCA motifs turned up eight IRs, all clustered within a single promoter region flanked by a gene with similarity to yeast gene yK10 on the 5′ side and two yeast hsp104 genes in tandem on the 3′ side. The entire locus looks like a satellite repeat region, possibly of the type containing heat shock response elements (HSEs), reported to be significant in the regulation of some hsp genes of yeast, fruit fly and mammal (Fernandes et al. 1994; Krawczyk et al. 1993). Identifying characteristics of HSEs include EREs TGACC, present here as part of the IR of TACGGTCA and GAA boxes, and their IRs present in large numbers throughout the background (LaVolpe et al. 1988; La Volpe 1994). Although it is beyond the purview of this paper to speculate on the cis regulation that might be occurring at this site, it should be noted that the relationships between these repeats seems to be quite complex. Some of the repeats overlap, and there are many copies of the various HSEs in and around the repeats. Furthermore, many similar IRs containing >10% mismatches can be found in this area (Fig. 2).
However, there are five other hsp genes identified on chromosome III and six on the X chromosome. None of these have promoter regions with IRs containing either of the two motifs (within our range of resolution). Nevertheless, the striking enrichment of IRs with two motifs in the promoter area of one hsp104 gene is likely to be significant. HSEs are known to act over long distances (Lewin 2000), leaving open a possible influence on other heat shock genes at other loci.
The potentials for the use of IRs are likely to extend much further than the most obvious palindromic configurations presented here. It should be considered that a given IR might appear nested within other repeats, oriented in other ways with respect to other sequences, or with the two halves separated by thousands of base pairs of other sequences. It is premature to draw final conclusions about the roles of IRs or to limit searches to only some types or orientations. The grammar and syntax of gene regulatory mechanisms in multicellular eukaryotes are likely to be not only complex but full of redundancies and irregularities, as in any evolving language. Particular sequences of IRs should be searched to determine the extent to which they are also direct repeats, everted repeats, and mirror-image repeats. That is, although the various categories of repeats have distinct designations, they are in fact not mutually exclusive. Any sequence by virtue of its position and orientation may be any one or all of the types of repeats. Furthermore, the looping of DNA regulatory sites may alter the effective orientation of a repeat within a transcription complex. Indeed structure, position, and orientation are more important than exact sequence in regulatory sites. We see our annotated catalog as a tool for discovery in silico (Clarke and Berg 1998) and an appropriate complement to the genetic analysis of regulation.
METHODS
A dynamic programming algorithm was used to search for IRs of 40–200 bp (both perfect and imperfect) and 4–60 bp (perfect only) on chromosomes III and X. The algorithm, a longest-common subsequence variant of edit distance, maximizes the number of exact match base pairs. For this study, perfect IRs of even length are symmetrical and contain no mismatches. Perfects of odd length are defined to include a single base pair only at the center of the repeat. IRs (40–200 bp) were reported if at least 90% of the base pairs on the stem matched exactly and if hairpin loops were no larger than one-third the length of the IR. Loops were defined as being comprised of sequences with no possibility of internal base pairing. However, “blebs”, mismatched bases in the stem, may include sequences with possible base pairings. In no case is it assumed that any base pairing occurs in vivo. Figures depicting internal base pairing are for the purposes of visualizing the repeats.
The web site http://genomics.wheatoncollege.edu provides tools to search and view the results, including a search engine (Fig. 3) to locate IRs, a utility to draw optimal structures including loops, and a browser (Fig. 4) to allow the user to walk up and down a chromosome and view the relative positions of IRs and genes. In a search, IRs that are AT-rich (defined by us to be comprised exclusively of As and Ts) may be filtered out. Furthermore, all IRs have been unnested such that only unique sequences are reported, not those that are embedded symmetrically within larger sequences. The drawings of genes are linked to GenBank, specifically to the protein descriptions for each of the genes on chromosomes III and X.
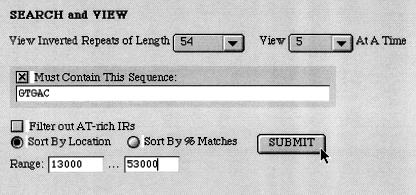
Figure 3.
The search tool locates (IRs) and reports on other IRs of like size and shape. Searches may be limited to particular motifs and may be restricted to omit AT-rich sequences. It is available at http://genomics.wheatoncollege.edu.
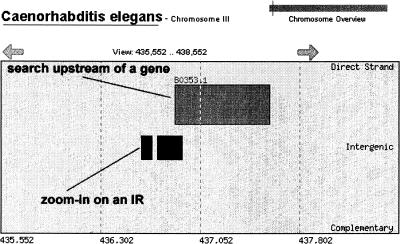
Figure 4.
The browse tool highlights direct (5′-3′) and complementary (3′-5′) genes and the relative locations of IRs. Links to (NCBI) are made by clicking on particular genes. The tool is available at http://genomics.wheatoncollege.edu.
The sequence files for Caenorhabditis chromosome III (worm_III.fna) and chromosome X (worm_X.fna) were downloaded from the GenBank (NCBI) ftp site (ncbi.nlm.nih.gov) in the directory /genbank/genomes/C_elegans/ (CHR_III, May 24, 1999 and CHR_X, November 7, 1999). Control searches were run for perfect IRs of lengths 4–60 bp on random input files of the same number of base pairs as worm_III.fna and worm_X.fna (∼11million bp and 16 million bp, respectively) and with a GC content of 36% as reported by the C. elegans Sequencing Consortium (1998). As the other chromosomal sequences of Caenorhabditis become more finalized, especially in the repetitive regions, those also will be searched and added to the web site.
Acknowledgments
The equipment was purchased in part by funds from the National Science Foundation. Additional funding was provided by a Wheaton Foundation grant and a Shouse Fellowship. We thank Richard Durban, Sanger Institute, for e-mail discussions early in this project and Robert Obar, Cereon Genomics, for critical guidance throughout the project.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL BDyer@Wheatoncollege.edu; FAX (508) 285-8278.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.122700.
REFERENCES
- Britten R. Mobile elements inserted in the distant past have taken on important functions. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- Brown SM. Dealing with genome project data. Biotechniques. 1999;26:266–268. [Google Scholar]
- Carlberg C, van Huijsduijen HR, Staple JK, DeLamenter JF, Becker-Andre M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Endocrin. 1994;8:757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the Nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chargaff E. Chemical specificity of nucleic acids and the mechanism of their enzymatic degradation. Experientia. 1950;6:201–209. doi: 10.1007/BF02173653. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Clarke N, Berg J. Zinc fingers in Caenorhabditis elegans: Finding families and probing pathways. Science. 1998;282:2081–2022. doi: 10.1126/science.282.5396.2018. [DOI] [PubMed] [Google Scholar]
- Cox R, Mirkin SM. Characteristic enrichment of DNA repeats in different genomes. PNAS. 1997;13:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore T, Rodriguez A, Smith DP. dRGS7 encodes a Drosophila homolog of EGL-10 and vertebrate RGS7. DNA Cell Biol. 1998;17:983–989. doi: 10.1089/dna.1998.17.983. [DOI] [PubMed] [Google Scholar]
- Fernandes M, Xiao H, Lis JT. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor–heat shock element interactions. Nucleic Acids Res. 1994;22:167–173. doi: 10.1093/nar/22.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier WA, Gao AG, Dimitry J, Chung J, Brown EJ, Lindberg FP, Linder ME. The thrombospondin receptor integrin-associated protein (CD47) functionally couples to heterotrimeric Gi. J Biol Chemistry. 1999;274:2554–2560. doi: 10.1074/jbc.274.13.8554. [DOI] [PubMed] [Google Scholar]
- Frontali C, Pizzi E. Similarity in oligonucleotide usage in introns and intergenic regions contributes to long-range correlation in the Caenorhabditis genome. Gene. 1999;232:87–95. doi: 10.1016/s0378-1119(99)00111-0. [DOI] [PubMed] [Google Scholar]
- Gelfand MS, Koonin EV. Avoidance of palindromic words in bacterial and archaeal genomes: A close connection with restriction enzymes. Nucleic Acids Res. 1997;25:2430–2439. doi: 10.1093/nar/25.12.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A. Genomic-scale analysis goes upstream? Nature Biotech. 1998;16:907–908. doi: 10.1038/nbt1098-907. [DOI] [PubMed] [Google Scholar]
- Krawczyk Z, Schmid W, Harkonen P, Wolniczek P. The ERE-like sequence from the promoter region of the testis specific hsp70-related gene is not estrogen responsive. Cell Biol Int. 1993;17:245–253. doi: 10.1006/cbir.1993.1061. [DOI] [PubMed] [Google Scholar]
- La Volpe A. A repetitive DNA family, conserved throughout the evolution of free-living nematodes. J Mol Evol. 1994;39:473–477. doi: 10.1007/BF00173416. [DOI] [PubMed] [Google Scholar]
- La Volpe A, Ciaramella M, Bazzicalupo P. Structure, evolution and properties of a novel repetitive DNA family in Caenorhabditis elegans. Nucleic Acids Res. 1988;16:8213–8231. doi: 10.1093/nar/16.17.8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Mouradian MM. Up-regulation of D1A dopamine receptor gene transcription by estrogen. Mol Cell Endocrin. 1999;156:151–157. doi: 10.1016/s0303-7207(99)00133-1. [DOI] [PubMed] [Google Scholar]
- Lewin B. Genes VII. New York: Oxford University Press; 2000. [Google Scholar]
- Lightfoot K, Maltby L, Duarte R, Veale R, Segev O. Conserved cis-elements bind a protein complex that regulates Drosophila ras2/rop bidirectional expression. Brit J of Cancer. 1994;69:264–273. doi: 10.1038/bjc.1994.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Robinson PJ, Gupta A, Bleasby AJ, Whitton BA, Morby AP. Singular over-representation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res. 1995;23:729–735. doi: 10.1093/nar/23.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombel IT, McMorren BJ, Lamont IL. Identification of a DNA sequence motif required for expression of iron-regulated genes in pseudomonads. Mol Gen Genet. 1995;246:519–528. doi: 10.1007/BF00290456. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole genome mRNA quantification. Nature Biotech. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Surzycki SA, Belknap WR. Repetitive-DNA elements are similarly distributed on Caenorhabditis elegans autosomes. PNAS. 2000;97:245–249. doi: 10.1073/pnas.97.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Helden J, Andre B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- Wasylyk C, Gutman A, Nicholson R, Wasylyk B. The c-Ets oncoprotein activates the stromelysin promoter through the same elements as several non-nuclear oncoproteins. EMBO J. 1991;10:1127–1134. doi: 10.1002/j.1460-2075.1991.tb08053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz A, Rosales R. Identification of an estrogen response element upstream of the human c-fos gene that binds the estrogen receptor and the AP-1 transcription factor. Nucleic Acid Res. 1990;18:5097–5106. doi: 10.1093/nar/18.17.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G, Shuey DJ, Kibbe WA, Parker CS. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987;48:507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Wong YC, Pustell J, Spoerel N, Kafatos FC. Coding and potential regulatory sequences of a cluster of chorion genes in Drosophila melanogaster. Chromosoma. 1985;92:124–135. doi: 10.1007/BF00328464. [DOI] [PubMed] [Google Scholar]