Abstract
Background
Cryptosporidium spp. is prevalent globally, pigs are an important Cryptosporidium reservoir. In China, little data regarding rates of Cryptosporidium infections in pigs are available. The present study was therefore aimed at characterizing the distribution of Cryptosporidium species in pigs from two different cities, Shaoxing and Shanghai, from the Yangtze River delta.
Methodology/Principal Findings
Nested PCR to amplify the 18S rRNA locus on DNA extracted from fecal samples (n = 94) revealed the positive rate of Cryptosporidium in pigs from two cities was approximately 17.0%. The positive rates in Shanghai and Shaoxing were 14.3% and 25.0% respectively. Amplified sequences were verified by sequencing. The identified strain belonged to the C. pig genotype II using BLAST analysis in the NCBI database.
Conclusion/Significance
Our finding of Cryptosporidium pig genotype II in pigs in the Yangtze River delta area suggests that pig farms in this region must be considered a public health threat and proper control measures be introduced.
Introduction
Cryptosporidium is a protozoan parasite that can cause severe diarrhea, anorexia and weight loss, especially in neonatal and immunocompromised animals. To date, more than 20 Cryptosporidium species have been identified [1], [2], however, more than 60 genotypes remain undefined [1], [2], [3]. Cryptosporidium species can parasitize a wide spectrum of animals and are the cause of zoonotic infections affecting humans and infections are typically caused by several species (and genotypes) including C. parvum, C. hominis, C. canis, C. felis, C. cuniculus and C. meleagridis. Less frequently associated with human infections are strains including C. suis, C. muris, C. ubiquitum and the rare Cryptosporidium pig genotype II [4], [5], [6], [7]. Pigs are an important Cryptosporidium reservoir making it important to understand the prevalence of Cryptosporidium species in swine as a means of controlling and preventing cryptosporidiosis in both animals and humans. In China, however, little data regarding rates of Cryptosporidium infections in pigs are available. The present study was therefore aimed at characterizing the distribution of Cryptosporidium species in pigs from two different cities, Shaoxing and Shanghai, from the Yangtze River delta.
Materials and Methods
Sampling
In total, 94 fecal samples (50 g) were collected randomly from different animals immediately after defecation into individually labelled plastic bags with a gloved hand from 6 pig farms in Shanghai and 1 from Shaoxing between April 2009 and October 2009.
Stool Sample Processing
Approximately 20 g of each sample was suspended in 50 ml of deionized water, vortexted and filtered through the sieve. Filtrates were poured back into the same centrifuge tube and centrifuged at 3000 rpm for 10 min. Pellets were then resuspended in 15 ml fresh deionized water and 1.5 ml were transferred into a 2 ml microcentrifuge tube and centrifuged at 3000 rpm for 10 min. Supernatants were discarded and pellets stored at −70°C until use.
DNA Extraction and PCR Amplification
Genomic DNA was extracted using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA). Supernatants containing DNA were stored at −20°C until use. Nested PCR was used to amplify an approximately 840 base pair (bp) long fragment corresponding to the Cryptosporidium 18S rRNA gene using two sets of oligonucleotide primers: TTCTAGAGCTAATACATGCG and CCCATTTCCTTCGAAACAGGA for primary PCR and GGAAGGGTTGTATTTATTAGATAAAG and CTCATAAGGTGCTGAAGGAGTA for secondary PCR [8], [9], [10]. The PCR reactions were carried out in 25 µl volumes containing 12.5 µl 2× Taq Green Master Mix (Promega, Madison, WI), 1 µl of each primer (10 µM), 9.5 µl nuclease-free water (Promega) and 1 µl DNA template. For amplification, templates were subjected to a hot start at 94°C for 1 min followed by 35 cycles of 94°C for 10 s, 55°C for 30 s and 72°C for 1 min followed by a final extension at 72°C for 10 min. Secondary PCR products were analyzed by 2% agarose gel electrophoresis and ethidium bromide staining.
DNA Sequencing and Analysis
Positive secondary PCR products were subjected to two directional sequencing with secondary primers by the Shanghai Biotechnology Co. Ltd. (Shanghai, China). Amplified sequences were blasted against sequences in the NCBI database and then deposited in GenBank. Cryptosporidium isolates identified were compared phylogenetically using the MEGA 4.1 software.
Results
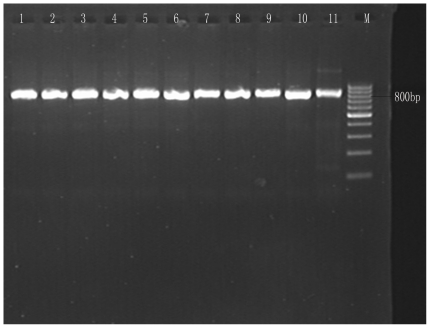
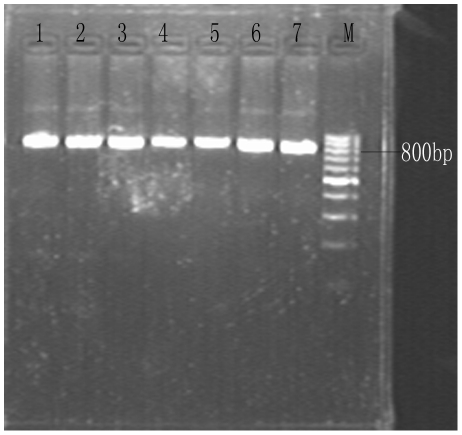
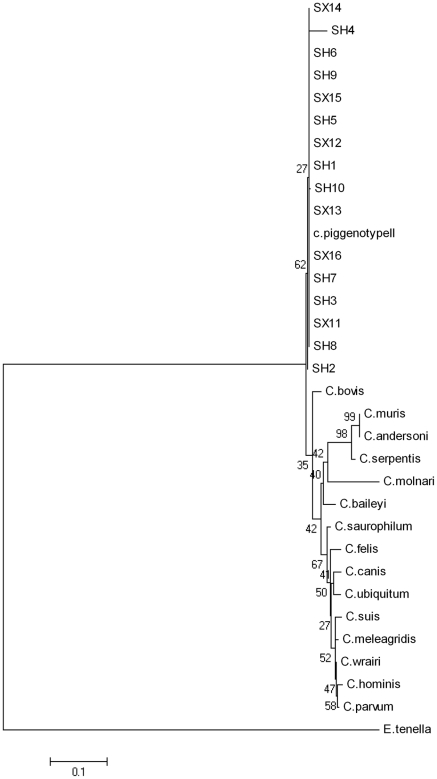
Fecal samples (n = 94) were collected from swine farms (24 samples from Shaoxing and 70 from Shanghai). PCR amplification of the 18S rRNA gene locus identified 16/94 (17.0%) Cryptosporidium species with 14.3% (10/70) in Shanghai (Figure 1) and 25.0% (6/24) in Shaoxing (Figure 2). BLAST analysis of positive secondary PCR products revealed that all sequences belonged to the Cryptosporidium pig genotype II. Phylogenetic relationships between Cryptosporidium sequences identified from samples collected from Shanghai and Shaoxing (SH1–10 and SX11–16) and the Cryptosporidium pig genotype II (GenBank: GU254170.1) demonstrated in Figure 3.
Figure 1. Agarose gel electrophoresis of Shanghai sequences.
PCR amplification products of respective Cryptosporidium isolates were subjected to 2% agarose gel electrophoresis. Lane 1, positive control, lanes 2–11 18S rRNA amplification products and M, 100 bp DNA ladder. PCR products are approximately 840 bp.
Figure 2. Agarose gel electrophoresis of Shaoxing sequences.
PCR amplification products of respective Cryptosporidium isolates were subjected to 2% agarose gel electrophoresis. Lanes 1–6 represent secondary 18S rRNA amplification products, lane 7 positive control and M, 100 bp DNA ladder. PCR products are approximately 840 bp.
Figure 3. Phylogenetic analysis.
18S rRNA sequences from the respective Cryptosporidium isolates identified were compared phylogenetically using the MEGA 4.1 software. The phylogenetic tree obtained demonstrates the genetic diversity of the Cryptosporidium genotype II isolates identified from stool samples from Shanghai (SH1–10) and Shaoxing (SX11–16). Analysis was based on the nested-PCR of the18S rRNA locus sequence using the tree method of Neighbor Joining.
The nucleotide sequences of Cryptosporidium pig genotype II from pigs in this study deposited in GenBank under accession numbers HQ844719–HQ844728 (SH1–SH10) and HQ844729–HQ844734 (SX11–SX16).
Discussion
Cryptosporidium is shed in feces as oocysts that can be transmitted to humans or animals by contaminated water or food. Pigs have been recognized as important reservoirs for several Cryptosporidium species/genotypes, some of which are zoonotic pathogens. In the present study, all of the positive samples belonged to the Crytosporidium pig genotype II following BLAST analysis against NCBI sequences separately and the phylogenetic analysis..
In Denmark, Langkjaer et al. (2007) genotyped Cryptosporidium species at the 18S rDNA and/or the hsp 70 gene. This analysis identified Cryptosporidium suis and/or the Cryptosporidium pig genotype II in 183 pigs, in addition to demonstrating an age-specific distribution of species/genotypes. In contrast to the present study the Danish study identified infections with more than 1 species/genotype [11]. In Western Australia, Johnson et al. (2008) identified in pre- and post-weaned pigs bred indoors and outdoors a 21.1% prevalence of Cryptosporidium suis and Cryptosporidium genotype II infections using a nested PCR based 18S rRNA amplification approach, however, the Australian study did not identify age-dependent associations with respect to infection status [12]. In the Czech Republic, Kvac et al. (2009) reported that the Cryptosporidium infection prevalence in slaughtered finisher pigs and sows was 29% and 10%, respectively, using microscopic analysis. In finishers, mixed C. suis and the Cryptosporidium pig genotype II infections were detected and C. parvum infections in sows were identified [13]. Another study by the same group also showed the age-related prevalence of Cryptosporidium suis, C. muris and Cryptosporidium pig genotype II [14]. These three species and genotypes were also identified in pig slurry samples from an Irish study [15].
In China, studies carried out in different provinces demonstrated using morphologic analysis that 1) C. parvum infections were detected in 58/577 pigs in Anhui province with differences in infection rates dependent on age but not sex [16], 2) C. suis infections in Chongqing province were identified in 14 pigsties independent of age [17], 3) in Eastern China, including Jiangsu province and Shanghai, C. parvum-like genotype infections were identified in pigs using reverse transcription PCR amplification of SSU rRNA [18] and 4) in the present study, only the Cryptosporidium pig genotype II was identified in the two districts examined. Additional studies will be needed to identify additional geographic areas where these infections may be present to fully define the nature of Cryptosporidium species infections in pigs.
Several Cryptosporidium species/genotypes isolated from pigs in China, including C. suis, C. muris, C. parvum and the Cryptosporidum pig genotype II have been associated with zoonotic infections. In this study, we identified for the first time the presence of the Cryptosporidium pig genotype II in the Yangtez River delta suggesting that prevalence of Cryptosporidium infections in swine populations must be carefully monitored since pigs are an important Cryptosporidium reservoir that could facilitate zoonotic infections affecting human populations.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Y.S. was supported by grants from the Chinese Special Program for Scientific Research of Public Health No. 200802012 and the Chinese National Key Program for Infectious Diseases No. 2008ZX10004-002. J.C. was supported by grant from the Chinese National Key Program for Infectious Diseases No. 2009ZX10004-201. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Plutzer J, Karanis P. Genetic polymorphism in Cryptosporidium species: an update. Vet Parasitol. 2009;165:187–199. doi: 10.1016/j.vetpar.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Fayer R. Taxonomy and species delimitation in Cryptosporidium. Exp Parasitol. 2010;124:90–97. doi: 10.1016/j.exppara.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao L, Fayer R. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol. 2008;38:1239–1255. doi: 10.1016/j.ijpara.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Xiao L, Ryan U. Fayer R, Xiao L, editors. Molecular epidemiology. Cryptosporidium and Cryptosporidiosis, second ed. 2008. pp. 119–163. second ed: CRC Press and IWA Publishing, Boca Raton, FL.
- 6.Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Xiao L, et al. Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerg Infect Dis. 2009;15:829–830. doi: 10.3201/eid1505.081419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kvac M, Kvetonova D, Sak B, Ditrich O. Cryptosporidium pig genotype II in immunocompetent man. Emerg Infect Dis. 2009;15:982–983. doi: 10.3201/eid1506.07621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang J, Alderisio KA, Xiao L. Distribution of cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71:4446–4454. doi: 10.1128/AEM.71.8.4446-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao L, Alderisio K, Limor J, Royer M, Lal AA. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol. 2000;66:5492–5498. doi: 10.1128/aem.66.12.5492-5498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langkjaer RB, Vigre H, Enemark HL, Maddox-Hyttel C. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology. 2007;134:339–350. doi: 10.1017/S0031182006001533. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J, Buddle R, Reid S, Armson A, Ryan UM. Prevalence of Cryptosporidium genotypes in pre and post-weaned pigs in Australia. Exp Parasitol. 2008;119:418–421. doi: 10.1016/j.exppara.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Kvac M, Sak B, Hanzlikova D, Kotilova J, Kvetonova D. Molecular characterization of Cryptosporidium isolates from pigs at slaughterhouses in South Bohemia, Czech Republic. Parasitol Res. 2009;104:425–428. doi: 10.1007/s00436-008-1215-x. [DOI] [PubMed] [Google Scholar]
- 14.Kvac M, Hanzlikova D, Sak B, Kvetonova D. Prevalence and age-related infection of Cryptosporidium suis, C. muris and Cryptosporidium pig genotype II in pigs on a farm complex in the Czech Republic. Vet Parasitol. 2009;160:319–322. doi: 10.1016/j.vetpar.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Xiao L, Moore JE, Ukoh U, Gatei W, Lowery CJ, et al. Prevalence and identity of Cryptosporidium spp. in pig slurry. Appl Environ Microbiol. 2006;72:4461–4463. doi: 10.1128/AEM.00370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Chang-cheng, Li Pei-ying, Wu Lin, Chen Ling-zhi, Wei-yi Y. Epidemiological survey on swine cryptosporidiosis in Anhui province. Animal Husbandry & Veterinary Medicine. 2005;37:18–20. [Google Scholar]
- 17.Zhou Rong-qiong, Huang Han-cheng, Wei Guang-he, Liu Ping, Li Qiang, et al. Epidemiology Survey of Pig Cryptosporidium in Chongqing and Identification of Cryptosporidium Species. Journal of Southwest China Normal University (Natural Science) 2007;32:82–85. [Google Scholar]
- 18.Chen F, Huang K. Prevalence and phylogenetic analysis of Cryptosporidium in pigs in eastern China. Zoonoses Public Health. 2007;54:393–400. doi: 10.1111/j.1863-2378.2007.01078.x. [DOI] [PubMed] [Google Scholar]