Abstract
RACK1 proteins belong to the eukaryote WD40-repeat protein family and function as spatial regulators of multiple cellular events, including signaling pathways, the cell cycle and translation. For this latter role, structural and genetic studies indicate that RACK1 associates with the ribosome through two conserved positively charged amino acids in its first WD40 domain. Unlike RACK1s, including Trypanosoma brucei RACK1 (TbRACK1), only one of these two positively-charged residues is conserved in the first WD40 domain of the Leishmania major RACK1 ortholog, LACK. We compared virulence-attenuated LACK single copy (LACK/-) L. major, with L. major expressing either two LACK copies (LACK/LACK), or one copy each of LACK and TbRACK1 (LACK/TbRACK1), to evaluate the function of these structurally distinct RACK1 orthologs with respect to translation, viability at host temperatures and pathogenesis. Our results indicate that although the ribosome-binding residues are not fully conserved in LACK, both LACK and TbRACK1 co-sedimented with monosomes and polysomes in LACK/LACK and LACK/TbRACK1 L. major, respectively. LACK/LACK and LACK/TbRACK1 strains differed in their sensitivity to translation inhibitors implying that minor sequence differences between the RACK1 proteins can alter their functional properties. While biochemically distinguishable, both LACK/LACK and LACK/TbRACK1 lines were more tolerant of elevated temperatures, resistant to translation inhibitors, and displayed robust pathogenesis in vivo, contrasting to LACK/- parasites.
Introduction
Eukaryote RACK1 proteins are highly conserved members of the WD40-repeat protein family adopting a modular seven-bladed ß-sheet propeller structure [1], [2] that regulate a variety of cellular pathways [3], [4], [5]. Studies in mammals, yeasts, plants and the trypanosomatid protozoan, T. brucei, confirm important functions for RACK1 proteins in cell signaling, division, differentiation and translation [6], [7]. Studies of the RACK1 orthologs CPC2 and ASC1 in Schizosaccharomyces pombe and Saccharomyces cerevisiae, respectively, indicate these proteins associate with the translation machinery and regulate protein expression [7], [8].
The physiological importance of RACK1 proteins is well documented. CPC2-deficient S. pombe display a delayed progression through the cell cycle, and an aberrant response to environmental stimuli that induce G1 arrest [9]. These findings, together with data suggesting that ASC1 is important for adhesin-dependent growth and temperature tolerance in S. cerevisiae, demonstrate the importance of RACK1 proteins in cellular responses to environmental changes [10], [11]. Recent structural studies have identified a ribosome-binding motif consisting of a tripeptide arginine, aspartate, and lysine (RDK) in the WD40 domain of the S. cerevisiae RACK1 protein [12], [13]. This motif is highly conserved in RACK1 orthologs of other eukaryotes, including T. brucei RACK1 (TbRACK1). Paradoxically, although TbRACK1 is evolutionarily closely-related to the Leishmania RACK1 ortholog, LACK, the RDK tripeptide is not conserved.
Mutation of the conserved RDK in yeast RACK1 decreases tolerance to translation inhibitors, confirming the functional importance of ribosomal association with RACK1 via this motif [12]. Studies in mammalian cells demonstrate RACK1 associates with both ribosomes and protein kinase C (PKC) thus linking cell signaling cascades to translation [14], [15]. Further, RACK1 is also required for PKC-dependent phosphorylation and release of mammalian translation initiation factor 6 (eIF6) from the 60S ribosomal subunit, prior to assembly of the 80S ribosome [16], [17]. Studies in other eukaryotes also link RACK1 functions in translation to other cellular pathways. For example, data suggest that the function of TbRACK1 in cytokinesis is dependent upon its function in translation [18], [19]. The same studies also identified eukaryote elongation factor 1A (eEF1A) as a TbRACK1-binding protein; consistent with TbRACK1 functioning in translation elongation, T. brucei depleted for TbRACK1 display an increased sensitivity to the elongation inhibitor, anisomycin [18].
Our previous studies demonstrate that the Leishmania major RACK1 ortholog, LACK, is essential for parasite viability, survival at host temperatures, and robust infection of host macrophages [20]. The diploid genome of L. major has four copies of the LACK gene, organized as two tandem copies arranged head-to-tail on each homologous chromosome. These gene copies are indistinguishable in stage-specific expression, and are predicted to express proteins of identical sequence [20]. One allele, containing two of these four copies can be deleted without affecting viability or pathogenesis relative to wild-type (WT) L. major [20]. Targeted deletion of a third copy of LACK results in parasites with reduced levels of LACK that show reduced viability and severely attenuated virulence [20]. Parasites with three LACK copies deleted are referred to as LACK/- in this report. In contrast, targeted replacement of a third LACK copy with an Xho I restriction site-tagged LACK gene yields fully viable parasites [20]. These L. major lines, referred to as LACK/LACK in this report, contain one endogenous LACK copy, followed by a second, targeted LACK copy downstream, thus maintaining LACK expression from two LACK gene copies. Multiple attempts to delete all four copies of LACK failed, indicating that at least one copy of LACK is essential for parasite survival. Despite its importance in parasite viability and virulence, molecular mechanisms underlying LACK function in Leishmania have not yet been elucidated.
Although eukaryotic RACK1 orthologs are highly conserved, recent studies have identified subtle species-specific functional motifs [21], [22]. Some of these species-specific functional differences may result from sequence divergence. For example, although an RDK ribosome-binding motif is conserved in yeast, mammalian and T. brucei RACK1 orthologs, it is not conserved in LACK proteins of Leishmania sp.; in the latter, the highly conserved lysine is substituted with a glycine. Therefore, we hypothesized that LACK's ribosome association may differ from that observed for RACK1 in other eukaryotes, with implications for its function in translation.
We reasoned that the altered RDK motif in LACK might be compensated by other sequence alterations in LACK that maintain its structure and function. Therefore, rather than mutate the putative ribosome-binding motif in LACK, we replaced the second copy of LACK in LACK/LACK parasites with TbRACK1, to create a line referred to as LACK/TbRACK1 in this report. By doing so, any differences observed between LACK and TbRACK1 could be attributed to differences in function rather than effects of mutation-induced disruption of protein structure. We evaluated the viability and virulence of LACK/TbRACK1 parasites in comparison to LACK/LACK parasites, and identified differences in LACK's and TbRACK1's protein synthetic functions using translation inhibitors, and polysome association.
Results
LACK-deficient L. major show cell cycle defects at mammalian temperatures
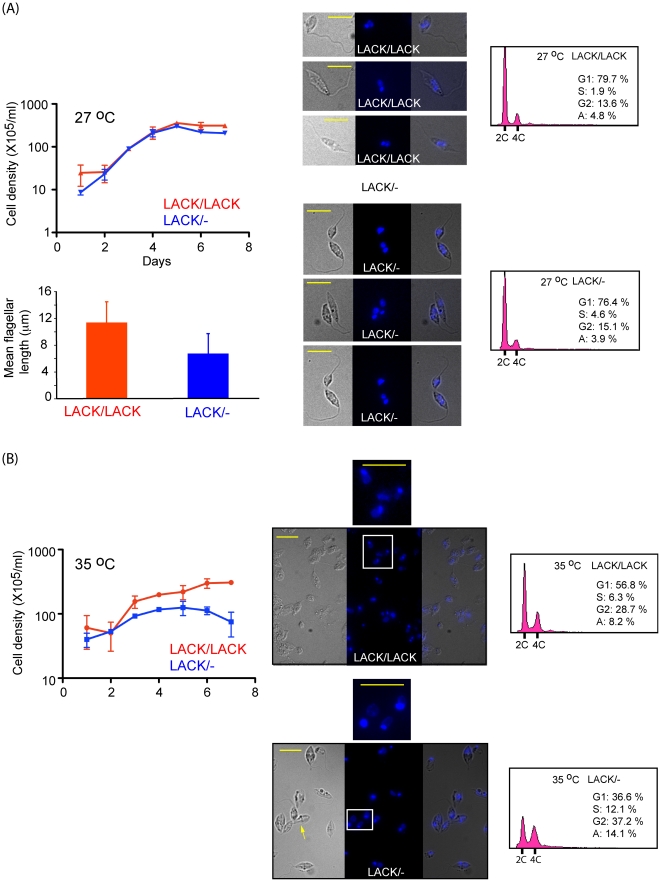
Previously, we demonstrated that LACK/- L. major lines caused dramatically attenuated disease in infected mice, and were significantly less viable at mammalian body temperature in vitro [20]. In contrast, LACK/LACK lines were as virulent as WT L. major in infected mice, and displayed no defect in viability at mammalian body temperature [20]. To further investigate these defects, we compared the morphology and DNA content of LACK/LACK and virulence-attenuated LACK/- L. major isolated at day 4 from promastigote cultures grown at 27°C and 35°C, using immunofluorescence microscopy. These results, shown in Figure 1, indicate that LACK/- parasites display an altered morphology relative to LACK/LACK parasites at both temperatures. At 27°C (Figure 1A), LACK/- parasites appear rounded with shorter flagellae; the latter phenotype is indicated by the bar-graph in Figure 1A. At 35°C (Figure 1B), LACK/- parasites were enlarged relative to LACK/LACK parasites (yellow arrow), with apparently greater nuclear and kinetoplast DNA content as evaluated by DAPI staining. This altered morphology coincided with a growth defect at 27°C, which was greatly accentuated at 35°C.
Figure 1. The effect of host temperature upon LACK-deficient L. major growth, morphology and the cell cycle.
Parasites were seeded in medium 199/10% FBS at 5×105/ml or 1×106/ml and incubated at either 27°C (A) or 35°C (B), respectively. Cell densities were determined daily for seven days by enumeration of diluted parasites using a hemocytometer and plotted as indicated. At day 4 of incubation, culture samples were removed, diluted, and the parasites examined microscopically at 630× magnification for cellular morphology and DNA content by differential interference contrast microscopy (DIC) (left panels) and fluorescence microscopy (middle panel). A merged DIC/fluorescence image is shown in the right panel. Flagella lengths were determined using ImageJ software and indicated by the bar-chart in (A), using sample sizes of 13 and 9 for LACK/LACK and LACK/- promastigotes, respectively. White boxes in (B) indicate areas of images enlarged for clarity, as shown at right of lower panels. Scale bars (10 µm) are denoted by short yellow lines. Yellow arrow indicates rounded, enlarged morphologies of LACK/- parasites as described in the text. The cell cycle of day 4 parasites was also analyzed by flow cytometry, gating out cell doublets, at 27°C and 35°C, as shown in the right panels. % of the cell population corresponding to cell cycle phases are indicated: G1 (Gap1), S (Synthesis), G2 (Gap2). Aneuploid cells (A) with a DNA content greater than 4C are also indicated (A).
To further examine and quantify the difference in DNA content, we compared LACK/LACK and LACK/- lines by flow cytometric analyses of log-phase cells stained with propidium iodide (Figure 1, right panels as indicated). As shown in the figure, the cell-cycle profiles of both LACK/LACK and LACK/- parasite lines were comparable at 27°C, with a 2C∶4C ratio of approximately 6∶1. In contrast, significant differences were observed at 35°C. At this temperature LACK/LACK parasites displayed a 2C∶4C ratio of 2∶1, consistent with an accumulation in G2 and M as observed by others [19]. This accumulation was strikingly pronounced in LACK/- parasites, resulting in a 2C∶4C ratio of 1∶1. We interpret this difference to indicate that LACK/- parasites have an impaired stress response relative to LACK/LACK parasites. Coincidentally, TbRACK1-depleted T. brucei also accumulate post-S phase, and are blocked in cytokinesis [19]. Based on these empirical observations, LACK and TbRACK1 may have equivalent functions in the cell-cycle of these related trypanosomatids. Because the cell-cycle functions of TbRACK1 are dependent on its role in translation, we tested whether LACK co-sedimented with monosomes and polysomes as observed for other RACK1 orthologs.
LACK co-sediments with a ribosomal protein in Leishmania monosomes and polysomes
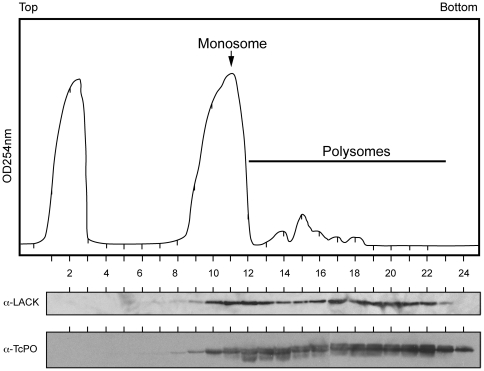
The RDK ribosome-binding motif is conserved in other RACK1 proteins, but is altered in LACK. Therefore, we tested whether LACK associates with Leishmania ribosomes by co-sedimentation analyses on discontinuous sucrose gradients (Figure 2). As shown in the figure, LACK co-sedimented in monosome and polysome fractions with the 60S ribosomal stalk protein PO [18], but was absent from low molecular weight fractions at the top of the sucrose gradient. Although we clearly observe a monosome peak, and polysome-containing fractions, we did not observe strongly distinct 40S and 60S ribosome subunit peaks, similar to previous findings in L. infantum [23]. In control experiments (data not shown), treatment with EDTA disrupted this profile, and resulted in LACK redistributing to the top of the gradient. It is of interest to note that the distribution of LACK in these gradients differs from the distribution of other RACK1 proteins, such as those from S. pombe and T. brucei. For example, TbRACK1, the LACK ortholog from T. brucei, a trypanosomatid closely related to Leishmania, can also be detected in non-ribosomal fractions. LACK's distribution closely resembles that observed for the S. cerevisiae RACK1 ortholog, which only associates with ribosome-containing fractions.
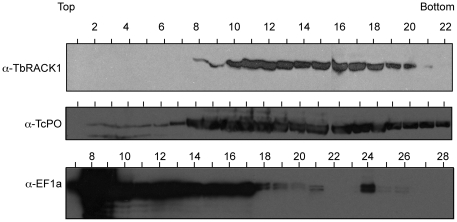
Figure 2. Sucrose gradient polysome analysis of LACK.
L. major lysates were prepared for sucrose gradient fractionation as described in Materials and Methods, then applied to a 10–50% sucrose step gradient and centrifuged for 160 min at 35,000 rpm. Following rRNA monitoring (OD254) and fractionation of the centrifuged gradient into 0.5 ml aliquots, proteins were purified from each fraction by methanol/chloroform precipitation, resolved on SDS/PAGE gels, blotted and probed with either anti-LACK antisera or anti-TcPO antisera. Fractions are numbered relative to the top and bottom of the sucrose gradient, as indicated.
These data indicate that although the RDK motif is not conserved in LACK, it continues to associate with the Leishmania protein synthesis machinery. Three possibilities could account for this association: 1) LACK associates with ribosomes in a manner distinct from the other RACK1 proteins; 2) LACK contains other sequence changes that compensate for the absence of the RDK motif; or 3) there are compensatory changes in other components of the Leishmania translation apparatus that permit association with LACK. One way to dissect these possibilities is by replacing LACK with another RACK1 protein known to be functional in its native context.
If other components of the Leishmania translation apparatus have compensatory changes to permit association with LACK, it is expected that these changes would prevent this heterologous RACK1 protein from functioning in the context of Leishmania. If LACK contains compensatory mutations elsewhere within it to permit association with the same ribosomal site as the other RACK1 proteins, then too it is predicted that the heterologous RACK1 protein will not function in Leishmania. However, if LACK and the heterologous RACK1 protein functionally associate with ribosomes at distinct sites or by distinct mechanisms, it is anticipated that the heterologous RACK1 protein will substitute for LACK in Leishmania. An implication of the latter possibility is that inhibitors of the protein synthesis machinery may have differential effects on LACK and the heterologous RACK1 protein.
Leishmania require at least one copy of LACK to survive, and LACK/LACK parasites cannot be distinguished in viability and virulence from WT L. major. Therefore, we replaced one copy of LACK in LACK/LACK parasites with the RACK1 protein from T. brucei, TbRACK1. We chose TbRACK1 because Leishmania and T. brucei are closely related trypanosomatids; yet, the potential ribosome-binding motif is conserved in TbRACK1, but not LACK. Elegant studies by Ruben and co-workers have characterized the function of TbRACK1 in depth [18], further increasing the value of comparing these two proteins from closely related parasites. We were also curious to determine whether TbRACK1 would distribute to non-ribosomal fractions in Leishmania, as it does in T. brucei. A failure to do so would indicate that the intracellular milieu within these trypanosomatids differs.
Targeted replacement of a LACK gene copy with TbRACK1 in L. major
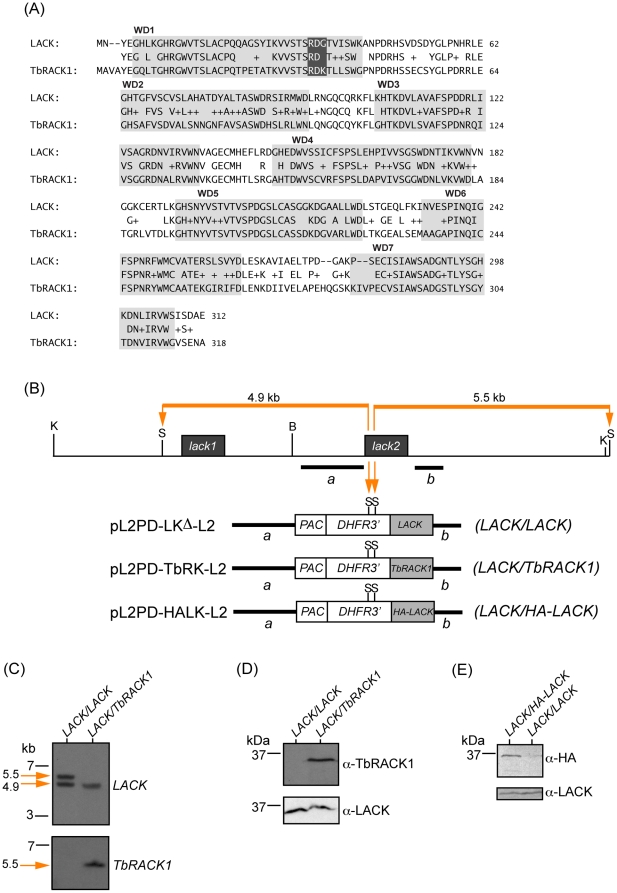
An alignment of the LACK and TbRACK1 sequence is shown in Figure 3A. Overall, the proteins are 62% identical and 78% similar. As might be expected, sequences within the WD40 repeats are generally more conserved than the intervening sequences. The RDK motif in the first WD40 repeat that is conserved in RACK1 orthologs, but not LACK, is highlighted in the figure. An overview of the gene replacement strategy used to create the LACK/TbRACK1 strain is shown in Figure 3B. We have used a similar strategy in the past to replace LACK alleles. Briefly, the second copy of LACK (lack2) in the LACK/LACK strain was replaced with a copy of TbRACK1 or HA-tagged LACK by homologous recombination. Targeting was achieved using lack2 5′ and 3′ flanks, along with a marker conferring resistance to puromycin (PAC) to introduce TbRACK1 or HA-LACK into the lack2 genomic site. Previously, an identical strategy was used with the construct pL2PD-LKΔ-L2 to create the LACK/LACK strain used in these studies [20]. Southern blots were conducted to confirm successful targeting and strain identity (Figure 3C). As shown in the figure, following transfection of lack++/−− with the indicated fragments from pL2PD-LKΔ-L2, pL2PD-TbRK-L2 or pL2PD-HALK-L2 and puromycin selection, lack2 was replaced by LACK, TbRACK1 or HA-tagged LACK respectively. Hybridization with LACK sequences revealed both LACK copies in the LACK/LACK (indicated by LACK-hybridizing Stu I fragments of 5.5 kb and 4.9 kb in Figure 3C, upper panel) and LACK/HA-LACK strains (data not shown), and a single copy of LACK in the LACK/TbRACK1 strain (indicated by a single LACK-hybridizing fragment of 4.9 kb in Figure 3C, lower panel). Hybridization with TbRACK1 sequences revealed a single copy of this gene to be present on a 5.5 kb Stu I fragment exclusively in the LACK/TbRACK1 parasites, as expected. Lastly, we confirmed the expression of TbRACK1 and HA-LACK in the LACK/TbRACK1 and epitope-tagged LACK/HA-LACK strains by immunoblot with either anti-LACK, anti-TbRACK1 or anti-HA (Figures 3D and 3E, respectively). As anticipated, TbRACK1 and HA-LACK were expressed in the LACK/TbRACK1 and LACK/HA-LACK strains, respectively, but not the LACK/LACK strain. After confirming TbRACK1 expression in the LACK/TbRACK1 strain, we examined the biological and biochemical properties of this strain relative to LACK/LACK parasites. The LACK/HA-LACK strain was used for co-immunoprecipitation studies using anti-HA antibodies. We also confirmed that phenotypic differences observed between control and LACK/- strains were indeed attributable to LACK deficiency, by introducing a LACK expression plasmid into the LACK/- line as previously described [24], see Figure S1.
Figure 3. Replacement of the L. major lack2 gene with TbRACK1.
(A) Alignment of the protein sequences for L. major LACK and TbRACK1, as indicated, was performed using BLAST (bl2seq; blast.ncbi.nlm.nih.gov). Gray boxes denote each WD40 ß-propeller blade domain, numbered as indicated. Identical conserved amino acids between LACK and TbRACK1 are indicated by single letter code. Similar amino acids are indicated by a plus sign. White letters indicate the putative ribosome-binding motif. (B) Physical map of the L. major lack genes and targeting constructs used to replace lack2. Restriction enzyme sites for Kpn I, Stu I, and Bam HI are denoted as K, S and B, respectively. Targeting fragments a and b used for the constructs, as described in Materials and Methods, are indicated by heavy black lines. Orange bars and arrows indicate 5.5 kb and 4.9 kb Stu I fragments expected following targeting of lack2 with the indicated constructs. (C) Southern analysis of L. major transfectant DNAs. Genomic DNAs from L. major transfectants were digested with Stu I, size-fractionated and blotted as described in Materials and Methods. The blots were then hybridized with either the LACK or TbRACK1 coding sequences, as indicated at right of panels. (D) Western analysis of L. major transfectants. Extracts from 2×107 transfectant parasites for each of the indicated lines were size-fractionated, blotted and probed with either anti-LACK or anti-TbRACK1 antisera, as indicated and described in Materials and Methods.
Introduction of TbRACK1 rescues the growth defect in LACK/- parasites
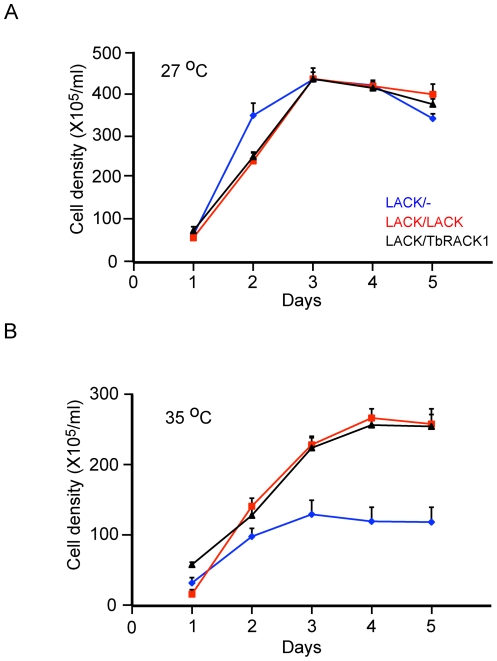
The data in Figure 1 indicate that LACK/- parasites have a severe growth defect at the host temperature of 35°C, although their axenic growth at 27°C is less impaired. Therefore, we examined the growth properties of LACK/TbRACK1 parasites at 27°C and 35°C (Figure 4). As shown in the figure, replacement with TbRACK1 restores normal growth at 35°C, which is indistinguishable from the growth of LACK/LACK parasites.
Figure 4. Growth of LACK/TbRACK1 L. major at host temperatures.
Parasites were inoculated into medium at 5×105/ml and incubated at either 27°C or 35°C as indicated. Cells were counted daily for five days, using a hemocytometer for two independent sets of clones performed at least three times. The data, with standard error bars, are representative of two independent sets of clones repeated three times.
Having observed LACK/TbRACK1 parasites to grow with the same kinetics as LACK/LACK parasites, we performed biochemical analyses to assess differences in protein synthesis, and LACK (or TbRACK1) associating with the translation apparatus. Because both LACK/LACK and LACK/TbRACK1 parasites grow equivalently, any observed differences can be attributed to differences between LACK and TbRACK1, rather than non-specific effects of cell viability or growth rate.
TbRACK1 co-sediments with a ribosomal marker in monosome and polysome fractions
We examined the distribution of TbRACK1 in monosomes and polysomes using extracts of LACK/TbRACK1 parasites fractionated on discontinuous sucrose gradients. The distribution of TbRACK1 (Figure 5) was identical to that observed for LACK (Figure 2). TbRACK1 was associated with both monosomes and polysomes, and was absent in non-ribosomal fractions. These results imply that the presence of TbRACK1 in non-ribosomal fractions of fractionated T. brucei gradients represent differences in the intracellular milieu between Leishmania and T. brucei rather than an intrinsic difference between these RACK1 orthologs. Our findings permit us to conjecture that a similar difference may underlie differences in the distribution pattern of RACK1 in S. cerevisiae and S. pombe.
Figure 5. Sucrose gradient polysome analysis of TbRACK1 expressed in L. major.
LACK/TbRACK1 L. major lysates were prepared, fractionated and blotted as described in Fig. 2. Blots were probed with anti-LACK, anti-TcPO and anti- eEF1A antisera as indicated. Fractions are numbered relative to the top and bottom of the sucrose gradient, as indicated.
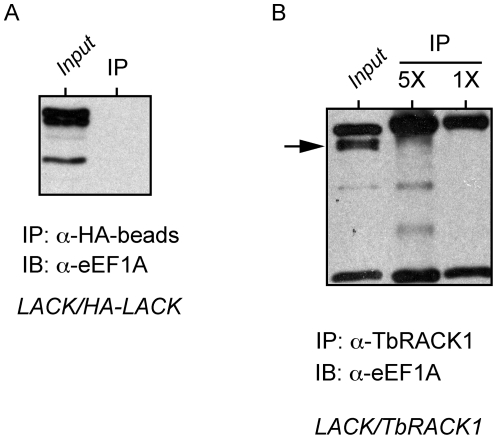
Previous studies in T. brucei indicate that TbRACK1 interacts with translation elongation factor eEF1A, and indeed co-sediments with eEF1A in the non-ribosomal fractions of T. brucei extracts on sucrose gradients [18]. eEF1A from T. brucei and L. major are 92% identical, and therefore we were surprised to find that in LACK/TbRACK1 L. major, TbRACK1 was excluded from non-ribosomal fractions of this gradient. To further elucidate an association between TbRACK1 or LACK1 with eEF1A in Leishmania extracts, we performed co-immunoprecipitations using an antibody against eEF1A. Reciprocal co-immunoprecipitations were also performed using antibodies against LACK, HA-epitope and TbRACK1. In repeated attempts, we did not detect an association between LACK, HA-LACK or TbRACK1 and Leishmania eEF1A (Figure 6 and data not shown). Because Leishmania eEF1A is 92% identical to T. brucei eEF1A, our results imply that in T. brucei, TbRACK1 interacts with eEF1A through other proteins that must differ between Leishmania and T. brucei. This finding correlates with the previously published observation that recombinant TbRACK1 interacts with eEF1A in extracts of T. brucei, or in extracts partially purified using a calmodulin column, but not when E. coli –expressed recombinant forms of both proteins obtained were used [18]. Our results suggest that this interaction may occur through a T. brucei-specific intermediate that is present in T. brucei extracts or co-purifies with eEF1A on a calmodulin column, and therefore is not observed when both proteins were expressed in a recombinant form.
Figure 6. Co-immunoprecipitation analyses of potential interactions between LACK and TbRACK1, and eEF1A.
L. major lines expressing either an HA-epitope-tagged form of LACK or TbRACK1 were created and analysed as depicted for LACK/TbRACK1 in Figure 3. Lysates from these two lines were analysed by co-immunoprecipitation under conditions previously described [18], using either anti HA-antibody coupled to agarose beads or anti-TbRACK1 antisera incubated with protein A/G agarose in panels A and B, respectively. Total lysate inputs are indicated. Note that for the total lysate input in Figure 6B, the immunoprecipitating antibody has also been added to the lysate. Arrow indicates L. major eEF1A.
Irrespective of a functional interaction with eEF1A, we were curious whether LACK's co-sedimentation with monosomes and polysomes correlated with a function in translation, similar to TbRACK1 and other RACK1 orthologs. Because LACK/- parasites are attenuated in virulence and growth at 35°C, we hypothesized this difference to arise from defects in translation that can be overcome by additional LACK protein. A corollary suggested by this hypothesis is that exposure to translation inhibitors may further discriminate between LACK/- and LACK/LACK parasites. This was tested by exposing parasites to inhibitors of translation initiation and elongation.
LACK/- parasites are more susceptible to translation inhibitors than LACK/LACK and LACK/TbRACK1 parasites
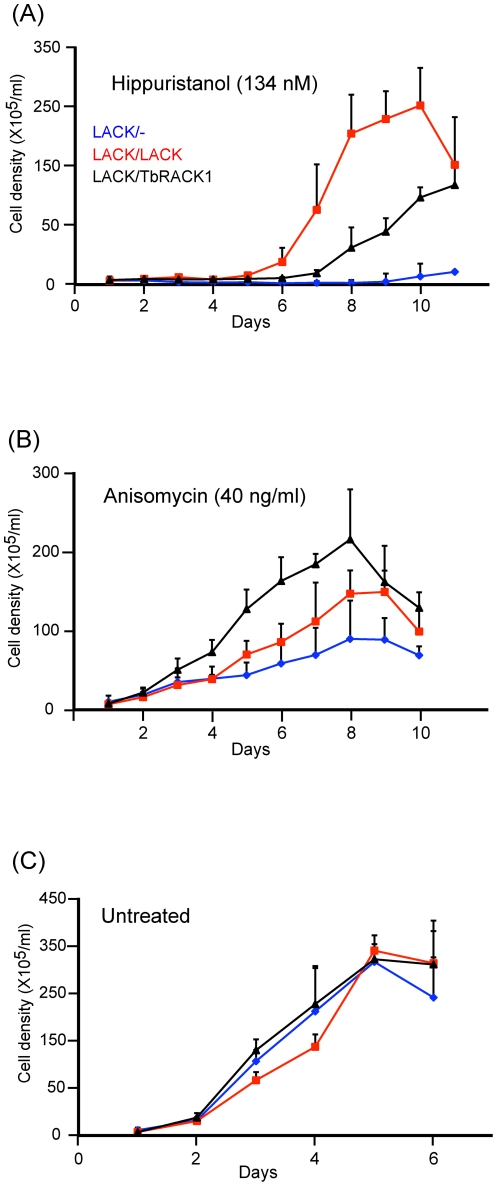
Because recent evidence indicates roles for LACK in translation elongation [18] and initiation [11], [16], we tested the effect of the elongation inhibitor, anisomycin [25], [26], and the initiation inhibitor, hippuristanol [27], on the Leishmania strains used in this report. By occupying the A-site, anisomycin inhibits translation elongation via interfering with the peptidyl transferase activity of the 80S ribosome. Hippuristanol binds the eukaryotic translation initiation factor 4 alpha (eIF4A) and inhibits its essential function as an RNA helicase to block initiation.
As shown in Figure 7A, growth of LACK/- parasites was severely affected by exposure to hippuristanol. This growth defect was somewhat ameliorated in LACK/TbRACK1 parasites, and to a significantly greater extent in LACK/LACK parasites. This result indicates that either LACK/LACK parasites are more resistant to translation inhibitors, or that the functions of LACK and TbRACK1 differ in translation initiation.
Figure 7. Effects of translation inhibitors on parasite growth.
The indicated parasite lines were seeded in medium 199/10% FBS at 5×105/ml and incubated at 27°C in the presence of the indicated concentrations of the translation inhibitors hippuristanol (A) and anisomycin (B), or no inhibitor (C). Cell densities were determined daily for seven days by enumeration, using a hemocytometer for two independent sets of clones performed at least twice. The data were averaged and plotted with standard error bars as indicated.
Next, we tested the effect of anisomycin on the growth of LACK/-, LACK/TbRACK1 and LACK/LACK parasites. Once again, as shown in Figure 7B, LACK/- parasites were more susceptible to anisomycin than the other strains. However, LACK/TbRACK1 parasites were more resistant to anisomycin than LACK/LACK parasites. Therefore it is unlikely that the differential effect of hippuristanol on LACK/LACK and LACK/TbRACK1 parasites represents a general property of the former. Rather, it is simpler to interpret that the function of LACK in translation subtly differs from the function of TbRACK1. Both proteins function in translation with differential emphases on initiation and elongation.
TbRACK1 restores virulence to LACK/- parasites
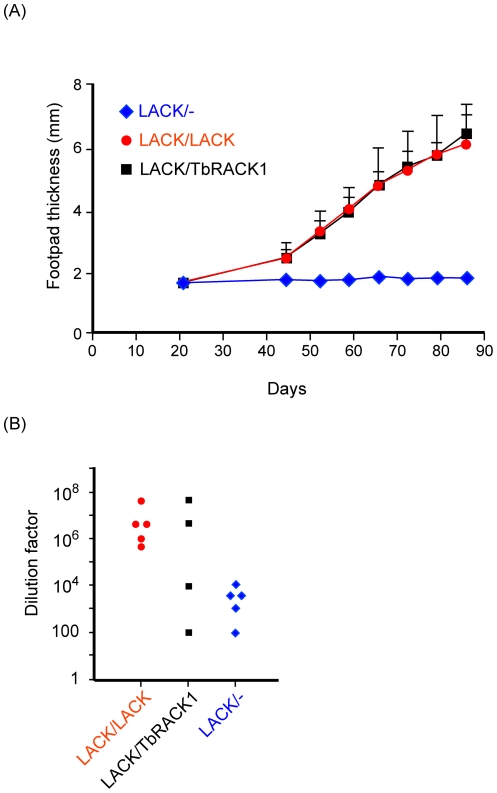
The preceding data, demonstrate that although introduction of TbRACK1 into LACK-deficient parasites can restore viability and increased resistance to translation inhibitors, there are differences between LACK and TbRACK1. We wondered whether these differences would impinge on parasite virulence in vivo. Therefore, we compared the LACK/LACK and LACK/TbRACK1 strains for their capacity to induce disease in the susceptible BALB/c mouse strain. Hind footpads of BALB/c mice were injected sub-cutaneously with parasites as described in the methods, after which the development of footpad lesions was monitored (Figure 8). Footpad lesions developed comparably in LACK/LACK and LACK/TbRACK1 infected mice (Figure 8A), in contrast to LACK/- infections that resulted in negligible disease as reported previously [20].
Figure 8. Disease development and parasite burden following infection with LACK/TbRACK1 L. major.
(A) BALB/c mice were inoculated in the left hind footpad with 4×106 of each of the indicated L. major strains. Disease progression in the hind footpad was monitored with vernier-reading calipers (Mitutoyo, Japan) as indicated. (B) For parasite burden determinations, parasites were isolated from popliteal lymph nodes at the experimental endpoints for each parasite line, dispersed in medium 199 and decimally diluted. Parasite burdens for each strain, as indicated in the Figure, were determined by identifying the highest dilution from which viable parasites were recovered.
Similarly, analyses of parasite burdens from suspensions of popliteal lymph nodes that drain the infected footpad indicated that comparable levels of parasitemia were attained in mice infected with either LACK/LACK or LACK/TbRACK1 parasites (Figure 8B).
Therefore, although there are biochemical differences between LACK and TbRACK1, they are not critical to the development of disease in this animal model.
Discussion
RACK1 proteins are conserved in all eukaryotes and have multiple physiological roles as adaptor proteins, regulating cellular events including signaling and translation. Although ostensibly, RACK1 proteins show a remarkable degree of overall sequence conservation amongst eukaryotes, recent studies have uncovered functionally important motifs that are not universally conserved amongst RACK1s [21],[28], [29]. For example, RACK1, a substrate and regulator of mammalian Src kinase is phosphorylated on Y228 and Y246 by Src [21], [22], whereas L. major LACK has phenylalanine residues (F230 and F248) at the equivalent positions. Such observations indicate that throughout eukaryote evolution, RACK1 proteins have diverged in structurally subtle yet functionally important ways.
Our interest in the constitutively expressed Leishmania RACK1 ortholog, LACK [30], was piqued by the observation that diploid Leishmania and other trypanosomatids have four identical copies of this gene. In contrast, other eukaryotes ranging from mammals to yeast have two copies of RACK1 per diploid genome [31]. Indeed, RACK1 can be deleted completely from budding and fission yeast without impacting cell survival and proliferation [7], [10]. It is also likely that RACK1 is essential for trypanosomatid survival and proliferation. Depletion of RACK1 by RNA interference in T. brucei results in a dramatic defect on cytokinesis and disruption of the cell cycle [19]. While it is not possible to deplete proteins by RNA interference in L. major, our gene knockout studies indicate that survival of L. major requires at least one copy of LACK [20]. Further, strains of L. major with a single copy of LACK are considerably impaired in their survival at mammalian host temperatures, and severely impaired in pathogenesis. Parallel to observations in T. brucei, our findings indicate that Leishmania strains with a single copy of LACK are impaired in progression through the G2/M phases of the cell cycle, particularly at mammalian body temperature.
In light of studies highlighting a role for RACK1 proteins in regulating eukaryote translation [7], [8], [13], and the emerging dogma that trypanosomatid gene expression relies heavily upon translation regulation [32], [33], we sought to further elucidate the important role of LACK in L. major with respect to parasite viability, protein synthesis and virulence.
RACK1 proteins have been shown directly, or indirectly, to interact with ribosomes to modulate translation [13], [18]. This interaction requires a conserved RDK motif present within the first WD40 domain of these RACK1 proteins. This motif is not conserved in the RACK1 orthologs of Leishmania or T. cruzi; curiously, while biochemical and structural studies indicate that RACK1 directly associates with S. cerevisiae ribosomes [12], [13], other studies indicate absence of a direct RACK1 association with T. cruzi ribosomes [34]. We interpret this difference to indicate that a single positively charged residue with the RDK motif is insufficient for direct ribosome association. In contrast, co-sedimentation analysis indicates that T. cruzi RACK1 co-sediments with T. cruzi ribosomes [18], suggesting an indirect association rather than the direct association observed in S. cerevisiae. Taken together, these data suggest that two positively charged amino acids in the putative ribosomal binding domain are not an absolute requirement for the association of RACK1s with trypanosomatid ribosomes, or that the association between RACK1 proteins and ribosomes in trypanosomatids is indirect. We interpret our findings to indicate that RACK1 proteins from different species may associate with the translation apparatus in different ways that are likely dependent on the intracellular milieu. For example, in T. brucei, RACK1 interacts with eEF1A [18], however our concerted efforts indicate this interaction may not occur in L. major. Nevertheless, T. brucei RACK1 can complement LACK/- L. major strains to restore their growth properties and virulence. Differences between the RACK1 proteins are also indicated by a differential susceptibility to various translation inhibitors by LACK/LACK and LACK/TbRACK1 strains of L. major.
Both LACK and heterologously-expressed TbRACK1 co-sediment in monosome and polysome sucrose gradient fractions from L. major, but are absent from non-ribosomal fractions. Such profiles are comparable to those observed for RACK1 in humans, mice, Drosophila, C. elegans and S. cerevisiae [10]. In contrast, TbRACK1 is detected in T. brucei ribosome-containing, and non-ribosomal fractions, similar to findings with S. pombe RACK1 [7], [35]. Although the functional consequences of such species-specific differences in cellular compartmentalization of RACK1 proteins are unclear, our findings may explain why eEF1A, localized predominantly in low molecular weight sucrose gradient fractions, was not detected in L. major complexes immunoprecipitated with anti-HA or anti-TbRACK1 antisera. The presence of a minor population of eEF1A in specific polysome fractions is a novel observation and may represent association of eEF1A with non-ribosomal high-order protein complexes [36].
Consistent with roles for RACK1 orthologs in Leishmania protein synthesis, LACK/LACK and LACK/TbRACK1 lines were more resistant to translation inhibitors than LACK/- L. major. Interestingly, LACK/LACK and LACK/TbRACK1 lines can be distinguished by their resistance to the translation inhibitors hippuristanol and anisomycin. Hippuristanol interferes with translation initiation by inhibiting initiation factor eIF4A, a DEAD-box family RNA helicase that unwinds 5′ secondary structures in mRNAs to promote translation initiation [27]. Conversely, anisomycin inhibits peptidyl transferase activity in translation elongation [25], [26]. Previous studies indicate that RACK1 may modulate eIF4A [11]; it is reported that phosphorylation of eIF4A may negatively regulate translation [37] [38]; pertinently, S. cerevisiae RACK1 decreases phosphorylation of eIF4A [11]. Our findings indicate that L. major strains with a single copy of LACK, either LACK/- or LACK/TbRACK1, display greater sensitivity to the eIF4A inhibitor hippuristanol. These findings imply that abundant LACK levels positively impact the function of eIF4A. Given the well-established roles for RACK1 proteins in regulating protein phosphorylation, one speculative scenario that may account for our findings is that LACK may augment Leishmania protein synthesis by down-modulating eIF4A phosphorylation. Further, it appears to do this more efficiently than TbRACK1. Our future investigations will be directed toward understanding the mechanism by which LACK impacts eIF4A function.
Although LACK/TbRACK1 and LACK/LACK strains are distinguished by their sensitivity to translation inhibitors, both strains displayed equivalent virulence in mice. Both strains replicated equivalently at mammalian host temperatures and were recovered comparably from infected mouse footpad lesions. Therefore, although these proteins likely have different biochemical properties, each of them can restore growth and virulence to LACK/- strains. Our future studies will explore functions of these proteins necessary for axenic growth, and functions necessary for virulence.
In conclusion, our data are consistent with a model where LACK participates in protein synthesis similar to the function proposed for TbRACK1 in T. brucei [18]. The effect of LACK and TbRACK1 on cell cycle progression and cytokinesis raises the possibility that RACK1 proteins may selectively impact translation of specific mRNAs, as proposed previously [17]. Because of the multi-functional nature of RACK1 proteins, however, it is also possible that the role of these proteins in the Leishmania cell cycle could involve processes additional to translation. Further, given the recent approval of the translation inhibitor, paromomycin as a treatment against leishmaniasis [39] the differential sensitivities of L. major lines, expressing distinct RACK1 orthologs, to the specific translation inhibitors used in our studies, suggest RACK1 proteins as potential species-specific targets for parasite-selective therapies.
Methods
Parasite culture
All parasite lines used were derived from L. major strain WHOM/IR/-/173 and cultured at either 27°C or 35°C in medium 199,10% heat-inactivated FBS as previously described [20].
Microscopy
Parasites from 4-day old cultures incubated at either 27°C or 35°C were washed twice in PBS, fixed in 4% formaldehyde/PBS, and applied to poly-lysine-coated glass slides using a Cytospin instrument (Shandon/Thermo Fisher Scientific) in accordance with manufacturers' instructions. The slides were then washed in PBS/0.5% Triton-X100 and mounted in ProLong Antifade with DAPI (Invitrogen). Differential interference contrast (DIC) and DAPI-fluorescent images of the parasites were then obtained using a Leica DM RA2 epifluorescence microscope (Leica Microsystems) equipped with the appropriate filters, excitation sources and motorized z-stage controlled by Slide Book software (Intelligent Imaging Innovations).
Image analysis
DIC microscopic images were analysed to quantify promastigote flagellar lengths using open source ImageJ software, available free of charge at http://rsb.info.nih.gov/ij/.
Cell cycle analysis
Parasites cultured for four days at 27°C or 35°C were fixed at −20°C with 90% ice cold methanol for 1 hr, then stained with propidium iodide as previously described [40] and analysed following doublet exclusion, using a FACSCalibur flow cytometer and Cellquest software (BD Scientific).
Polysome analyses
Sucrose gradient fractionation analyses of Leishmania polysomes were performed as described previously [41]. Briefly, 0.5–1×109 cells were pelleted, resuspended in 5 ml medium 199 containing 100 µg/ml cycloheximide for 5 min at room temperature. The cells were then washed twice in 50 ml ice-cold PBS containing 100 µg/ml cycloheximide. The cells were then resuspended in 50 ml lysis buffer (15 mM Tris-HCl (pH 7.4), 0.3 M KCl, 5 mM MgCl2) containing 0.5 mM DTT, 100 µg/ml cycloheximide, 1 mg/ml heparin, 1× Complete Mini protease inhibitor cocktail (Roche), 120 units of RNAsin (Promega Inc.) and incubated on ice for 10 min. The cells were then lysed by addition of 25 µl of 20% Triton-X 100. Lysates were then centrifuged at 12,000× g for 15 min at 4°C. Lysate supernatants were then layered onto 11 ml 10–50% sucrose step gradients made up in polysome buffer (20 mM Tris-HCl (pH 8.0), 140 mM KCl, 5 mM MgCl2) and centrifuged for 160 min at 35,000 rpm in a SW-40 rotor (Beckman Instruments). 0.5 ml fractions were then collected from the top of the gradient using a Foxy Jr. fraction collector (Teledyne Isco, Inc.). Proteins were isolated from each fraction by methanol/chloroform precipitation as previously described [18] and submitted for Western analysis.
Immunoblotting
Lysates from 2×107 parasites per lane were prepared using 2× Laemmli buffer and run on 12% SDS-PAGE gels and blotted onto PVDF membranes (ImmunBlot, BioRad) according to the manufacturer's instructions. After overnight blocking in 5% milk powder [20] in TBS with 0.05% Tween 20 (TBS/T) at 4°C, blots were incubated with 1∶1,000 rabbit anti–LACK antiserum, 1∶2000 rabbit anti-TbRACK1 (kindly provided by Dr. Larry Ruben, Southern Methodist University, Dallas), 1∶2500 rabbit anti-TcPO antiserum (a generous gift from Drs. Ajay Bhatia and Steven Reed, Infectious Disease Research Institute, Seattle) and 1∶3000 mouse anti-EF1A (Upstate Biotechnology, Lake Placid) for 1 h at 37°C. After washing in TBS/T, blots were incubated with 1∶3000 goat anti–rabbit-Ig conjugated with horseradish peroxidase for 30 min. The blots were washed and developed using ECL chemiluminescence reagent (GE Healthcare) according to the manufacturer's instructions.
Construction of the TbRACK1 and HA-LACK targeting plasmids
The TbRACK1 coding sequence was obtained by PCR amplification from genomic DNA isolated from T. brucei strain 427 procyclic stages, kindly provided by Dr. Zachary Mackey, University of California, San Francisco.
The TbRACK1-targeting construct, pL2PD-TbRK-L2, was created as described previously for pL2PD-LKΔ-L2 [20], except that the TbRACK1 coding sequence was inserted between the puromycin selection cassette and the 0.6 kb lack2 3′ flank in pL2PDL2.The HA-LACK-targeting plasmid was created in similar fashion to pL2PD-TbRK-L2, except the HA-LACK coding sequence was obtained by PCR amplification of the LACK gene, using a 5′ primer encoding methionine, followed by three HA epitope sequences, separated by serine-glycine dipeptide linkers, followed by LACK codons two, to nine.
Replacement of lack2 with TbRACK1 and HA-LACK
The downstream lack2 gene from lack++/−− L. major was replaced with a cassette containing either the TbRACK1 or the HA-LACK open reading frame, by transfecting the lack++/−− line [20] with the linear insert purified from pL2PD-TbRK-L2 or pL2PD-HALK-L2 (Fig. 4B), using methods previously described [20]. Generation of LACK/LACK lines, previously termed lack++ Δ /−−, has been described previously [20].
Southern blot analysis
Genomic DNAs isolated from control L. major transfectant clones previously described [20] and LACK/TbRACK1 L. major were digested with Stu I and size fractionated by agarose gel electrophoresis. The nucleic acids were blotted onto Hybond N+ nylon membranes (GE Healthcare) and hybridized using Rapid-Hyb (GE Healthcare) with DNA fragments g or h corresponding to the lack1 coding region and the 3′-most region of the lack allele, respectively (Fig. 4 B), according to the manufacturer's instructions (Phototope, NEB). Blots were washed at 65°C with 2X SSC-0.2X SSC/0.5% SDS, as described earlier [20]. Hybridizing fragments were visualized by chemiluminescent detection according to manufacturer's protocols (Phototope, New England biolabs). A similar strategy was used to create an L. major line where the downstream lack2 gene was replaced with a sequence encoding an HA-epitope-tagged version of LACK (data not shown).
Co-immunoprecipitation assays
HA-tagged LACK or TbRACK1 was immunoprecipitated from LACK/HA-LACK and LACK/TbRACK1 parasite lysates, respectively, using procedures and conditions described previously [18]. HA-LACK was immunoprecipitated under these conditions using ProFound anti-HA agarose beads (ThermoScientific/Pierce), in accordance with manufacturers' protocols. TbRACK1 was immunoprecipitated using anti-TbRACK1 anti-serum, kindly provided by L. Ruben, Southern Methodist University, Dallas, with protein A/G PLUS-Agarose (Santa Cruz Biotechnology) in accordance with manufacturer's directions.
Translation inhibitors and growth assays
For growth assays, parasites were diluted to 5×105/ml in fresh medium and incubated at 27°C or 35°C. Parasites were fixed in 0.4% formaldehyde/PBS then enumerated daily using a hemocytometer. Inhibitory concentrations of translation inhibitors anisomycin (40 ng/ml) (Sigma-Aldrich, St. Louis, MO), and hippuristanol (134 nM) (kindly provided by Dr. Jerry Pelletier, McGill University, Montreal) that were sub-lethal to LACK/LACK L. major were determined empirically such that LACK/LACK parasite cell densities peaked at approximately 1–2.5×107/ml.
Parasite infections
Cohorts of five female BALB/c mice (The Jackson Laboratory, ME) were inoculated subcutaneously in the left hind footpad with 4×106 stationary phase promastigotes, prepared as previously described [20]. Lesions were measured weekly using a Vernier-reading caliper. At the experiment termination point, popliteal lymph nodes were collected from infected mice, resuspended in M199/10% FCS, and diluted decimally into duplicate microtiter wells. After 7 d of culture at 27°C, parasites were quantified by microscopic evaluation of the highest dilutions containing viable organisms.
Animal ethics statement
All animal infection experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The infection protocol was approved by the Louisiana State University Health Sciences Center Institutional Animal Care and Use Committee (IACUC), IACUC protocol number 2584. All infections were performed under isoflurane anesthesia, and all efforts were made to minimize discomfort.
Supporting Information
Effects of elevated temperature and translation inhibitors on LACK/- L. major complemented with a LACK expression plasmid. (A) Western analysis of LACK/-/pXGLACK transfectants. Extracts from 2×107 LACK/- L. major and LACK/- L. major, complemented by expression of epitope-tagged LACK from the expression plasmid pXGLACK, as previously described [24], were size-fractionated, blotted and probed with anti-LACK antisera as described in Materials and Methods. Upper arrowed band denotes the epitope-tagged LACK protein; middle band indicates endogenous LACK, faint lower band represents an unknown cross-reacting protein. (B) Growth of LACK/-/pXGLACK transfectants at host temperatures. Cell densities of the indicated parasite lines were determined daily for seven days, by enumeration as described in the legend to Figure 4. The LACK/LACK line is included as a positive control. (C) Effect of hippuristanol on LACK/-/pXGLACK transfectants. Cell densities of the indicated parasite lines incubated in medium containing 134 nM hippuristanol were determined by enumeration daily for seven days, as described in the legend to Figure 7. The LACK/LACK line is included as a positive control.
(TIF)
Acknowledgments
We thank Connie Poretta and the Louisiana State University Health Sciences Center Vaccine Immunology Core for assistance with flow cytometry experiments, and our colleagues Nick Lapara, Doug Johnston, Glen Palmer and Joy Sturtevant for their suggestions during the course of this work. We thank Larry Ruben for generously providing antisera against TbRACK1 and suggestions on experimental protocols. We thank Steve Reed and Ajay Bhatia for antisera against TcPO.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded in part by National Institutes of Health Award AI055172 awarded to Ben Kelly and Ashok Aiyar. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 2.Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, et al. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 2008;17:1771–1780. doi: 10.1110/ps.035121.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goncalves KA, Borges JC, Silva JC, Papa PF, Bressan GC, et al. Solution structure of the human signaling protein RACK1. BMC Struct Biol. 2010;10:15. doi: 10.1186/1472-6807-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 5.Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 6.Vomastek T, Iwanicki MP, Schaeffer HJ, Tarcsafalvi A, Parsons JT, et al. RACK1 targets the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway to link integrin engagement with focal adhesion disassembly and cell motility. Mol Cell Biol. 2007;27:8296–8305. doi: 10.1128/MCB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shor B, Calaycay J, Rushbrook J, McLeod M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J Biol Chem. 2003;278:49119–49128. doi: 10.1074/jbc.M303968200. [DOI] [PubMed] [Google Scholar]
- 8.Baum S, Bittins M, Frey S, Seedorf M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem J. 2004;380:823–830. doi: 10.1042/BJ20031962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLeod M, Shor B, Caporaso A, Wang W, Chen H, et al. Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase To regulate cell cycle progression and meiotic development. Mol Cell Biol. 2000;20:4016–4027. doi: 10.1128/mcb.20.11.4016-4027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol. 2004;24:8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valerius O, Kleinschmidt M, Rachfall N, Schulze F, Lopez Marin S, et al. The Saccharomyces homolog of mammalian RACK1, Cpc2/Asc1p, is required for FLO11-dependent adhesive growth and dimorphism. Mol Cell Proteomics. 2007;6:1968–1979. doi: 10.1074/mcp.M700184-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Coyle SM, Gilbert WV, Doudna JA. Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol. 2009;29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, et al. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- 14.Grosso S, Volta V, Vietri M, Gorrini C, Marchisio PC, et al. Eukaryotic ribosomes host PKC activity. Biochem Biophys Res Commun. 2008;376:65–69. doi: 10.1016/j.bbrc.2008.08.118. [DOI] [PubMed] [Google Scholar]
- 15.Grosso S, Volta V, Sala LA, Vietri M, Marchisio PC, et al. PKCbetaII modulates translation independently from mTOR and through RACK1. Biochem J. 2008;415:77–85. doi: 10.1042/BJ20080463. [DOI] [PubMed] [Google Scholar]
- 16.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, et al. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regmi S, Rothberg KG, Hubbard JG, Ruben L. The RACK1 signal anchor protein from Trypanosoma brucei associates with eukaryotic elongation factor 1A: a role for translational control in cytokinesis. Mol Microbiol. 2008;70:724–745. doi: 10.1111/j.1365-2958.2008.06443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothberg KG, Burdette DL, Pfannstiel J, Jetton N, Singh R, et al. The RACK1 homologue from Trypanosoma brucei is required for the onset and progression of cytokinesis. J Biol Chem. 2006;281:9781–9790. doi: 10.1074/jbc.M600133200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly BL, Stetson DB, Locksley RM. Leishmania major LACK antigen is required for efficient vertebrate parasitization. J Exp Med. 2003;198:1689–1698. doi: 10.1084/jem.20031162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang BY, Harte RA, Cartwright CA. RACK1: a novel substrate for the Src protein-tyrosine kinase. Oncogene. 2002;21:7619–7629. doi: 10.1038/sj.onc.1206002. [DOI] [PubMed] [Google Scholar]
- 22.Mamidipudi V, Chang BY, Harte RA, Lee KC, Cartwright CA. RACK1 inhibits the serum- and anchorage-independent growth of v-Src transformed cells. FEBS Lett. 2004;567:321–326. doi: 10.1016/j.febslet.2004.03.125. [DOI] [PubMed] [Google Scholar]
- 23.Folgueira C, Quijada L, Soto M, Abanades DR, Alonso C, et al. The translational efficiencies of the two Leishmania infantum HSP70 mRNAs, differing in their 3′-untranslated regions, are affected by shifts in the temperature of growth through different mechanisms. J Biol Chem. 2005;280:35172–35183. doi: 10.1074/jbc.M505559200. [DOI] [PubMed] [Google Scholar]
- 24.Kelly BL, Locksley RM. The Leishmania major LACK antigen with an immunodominant epitope at amino acids 156 to 173 is not required for early Th2 development in BALB/c mice. Infect Immun. 2004;72:6924–6931. doi: 10.1128/IAI.72.12.6924-6931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Dyke N, Pickering BF, Van Dyke MW. Stm1p alters the ribosome association of eukaryotic elongation factor 3 and affects translation elongation. Nucleic Acids Res. 2009;37:6116–6125. doi: 10.1093/nar/gkp645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gay DA, Sisodia SS, Cleveland DW. Autoregulatory control of beta-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci U S A. 1989;86:5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 28.Liu YV, Hubbi ME, Pan F, McDonald KR, Mansharamani M, et al. Calcineurin promotes hypoxia-inducible factor 1alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J Biol Chem. 2007;282:37064–37073. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buensuceso CS, Woodside D, Huff JL, Plopper GE, O'Toole TE. The WD protein Rack1 mediates protein kinase C and integrin-dependent cell migration. J Cell Sci. 2001;114:1691–1698. doi: 10.1242/jcs.114.9.1691. [DOI] [PubMed] [Google Scholar]
- 30.Mougneau E, Altare F, Wakil AE, Zheng S, Coppola T, et al. Expression cloning of a protective Leishmania antigen. Science. 1995;268:563–566. doi: 10.1126/science.7725103. [DOI] [PubMed] [Google Scholar]
- 31.Chou YC, Chou CC, Chen YK, Tsai S, Hsieh FM, et al. Structure and genomic organization of porcine RACK1 gene. Biochim Biophys Acta. 1999;1489:315–322. doi: 10.1016/s0167-4781(99)00213-4. [DOI] [PubMed] [Google Scholar]
- 32.Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Gale M, Jr, Carter V, Parsons M. Cell cycle-specific induction of an 89 kDa serine/threonine protein kinase activity in Trypanosoma brucei. J Cell Sci. 1994;107(Pt 7):1825–1832. doi: 10.1242/jcs.107.7.1825. [DOI] [PubMed] [Google Scholar]
- 34.Gao H, Ayub MJ, Levin MJ, Frank J. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc Natl Acad Sci U S A. 2005;102:10206–10211. doi: 10.1073/pnas.0500926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nunez A, Franco A, Madrid M, Soto T, Vicente J, et al. Role for RACK1 orthologue Cpc2 in the modulation of stress response in fission yeast. Mol Biol Cell. 2009;20:3996–4009. doi: 10.1091/mbc.E09-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateyak MK, Kinzy TG. eEF1A: thinking outside the ribosome. J Biol Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le H, Browning KS, Gallie DR. The phosphorylation state of the wheat translation initiation factors eIF4B, eIF4A, and eIF2 is differentially regulated during seed development and germination. J Biol Chem. 1998;273:20084–20089. doi: 10.1074/jbc.273.32.20084. [DOI] [PubMed] [Google Scholar]
- 38.Rogers GW, Jr, Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 39.Mondal S, Bhattacharya P, Ali N. Current diagnosis and treatment of visceral leishmaniasis. Expert Rev Anti Infect Ther. 2010;8:919–944. doi: 10.1586/eri.10.78. [DOI] [PubMed] [Google Scholar]
- 40.Selvapandiyan A, Duncan R, Debrabant A, Bertholet S, Sreenivas G, et al. Expression of a mutant form of Leishmania donovani centrin reduces the growth of the parasite. J Biol Chem. 2001;276:43253–43261. doi: 10.1074/jbc.M106806200. [DOI] [PubMed] [Google Scholar]
- 41.Yoffe Y, Zuberek J, Lerer A, Lewdorowicz M, Stepinski J, et al. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot Cell. 2006;5:1969–1979. doi: 10.1128/EC.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of elevated temperature and translation inhibitors on LACK/- L. major complemented with a LACK expression plasmid. (A) Western analysis of LACK/-/pXGLACK transfectants. Extracts from 2×107 LACK/- L. major and LACK/- L. major, complemented by expression of epitope-tagged LACK from the expression plasmid pXGLACK, as previously described [24], were size-fractionated, blotted and probed with anti-LACK antisera as described in Materials and Methods. Upper arrowed band denotes the epitope-tagged LACK protein; middle band indicates endogenous LACK, faint lower band represents an unknown cross-reacting protein. (B) Growth of LACK/-/pXGLACK transfectants at host temperatures. Cell densities of the indicated parasite lines were determined daily for seven days, by enumeration as described in the legend to Figure 4. The LACK/LACK line is included as a positive control. (C) Effect of hippuristanol on LACK/-/pXGLACK transfectants. Cell densities of the indicated parasite lines incubated in medium containing 134 nM hippuristanol were determined by enumeration daily for seven days, as described in the legend to Figure 7. The LACK/LACK line is included as a positive control.
(TIF)