The study describes essential genetic roles of Smad4, a key signaling mediator of the TGF-β/BMP signaling pathways, in retinal growth, retinal spatial patterning, and axon guidance during mouse eye development.
Abstract
Purpose.
The present study was aimed at defining developmental roles of Smad4, a key mediator of the TGF-β superfamily signaling system, in the embryonic mouse retina.
Methods.
Using a Cre/loxP-mediated conditional gene targeting approach, Smad4 gene function was deleted from the embryonic mouse retina. Mutant phenotypes were morphologically and molecularly examined.
Results.
Loss of Smad4 in the developing retina led to varying degrees of microphthalmia at birth, presumably because of elevated apoptosis observed transiently at embryonic day 12.5 in the developing retina. This was also associated with an apparent delay in accumulation of retinal ganglion cells. Smad4 conditional mutants also exhibited alterations of retinal spatial patterning along the dorsal-ventral axis, consistent with a known function of BMP signaling in the embryonic retina. However, despite a known role for BMP signaling in retinal cell survival, proliferation, and differentiation, Smad4 mutant retinal progenitor cells were capable of maintaining growth and neurogenesis throughout embryonic development. We also found that the loss of Smad4 led to abnormal targeting of retinal ganglion cell axons to the optic nerve head, a phenotype consistent with reduced BMP signaling in the developing retina.
Conclusions.
These results suggest that Smad4 is essential for a subset of, but not all, TGF-β/BMP–dependent developmental processes in the embryonic retina. In addition, genetic requirements for Smad4 in the embryonic retina are evident predominantly in the developmental events regulated by the BMP branch of the TGF-β signaling pathway.
Development of the vertebrate retina involves the concerted action of numerous cell-intrinsic and -extrinsic factors for retinal progenitor cell growth, elaboration of distinct neural cell types, spatial patterning, and axonal connectivity.1 Among a number of evolutionarily conserved signaling pathways, transforming growth factor-β (TGF-β) superfamily of secreted ligands are known to mediate important functions during retinal development.2,3
One of two major branches of the TGF-β signaling system is the bone morphogenetic protein (BMP) pathway.4 Experiments using various vertebrate model organisms have shown that BMP signaling is essential for a wide spectrum of retinal development, starting from early morphogenesis of the retina.5–9 BMP signaling is also known to control apoptotic cell death,10,11 retinal growth and regeneration,12 initiation of neurogenesis,13 retinal pigment epithelium fate determination,14 and retinal ganglion cell (RGC) axon guidance.15 Another important genetic influence by BMP signaling on retinal development is spatial patterning, ultimately impacting visual function.16 The dorsal retinal identity is maintained by a signal involving dorsally restricted Bmp4 and its apparent target Tbx5,10,13,17 which, in turn, regulates the establishment of the retinocollicular map.17,18 In addition, recent studies19 have shown that dorsal BMP signaling is also essential to establish expression gradients of visual pigments (S- and M-opsins) in cone photoreceptor cells. The other branch of the TGF-β signaling system is the canonical TGF-β/activin pathway mediated by founding TGF-β members (TFG-β1, TFG-β2, and TFG-β3), activin, nodal, and growth differentiation factors (GDFs).20 During retinal development, TGF-βs are known to control retinal cell death and cell density2 and the regulation of retinal progenitor competence for neuronal fate allocation.21
Well-characterized core pathways of the TGF-β superfamily signaling system involve the intracellular signaling mediators Smads.22,23 In particular, the current model for Smad-mediated gene regulation dictates a central role for Smad4—the only common Smad class member in vertebrates—which is involved in both branches of the TGF-β/BMP signaling system.24 However, the evidence for Smad4 as an obligate component of Smad-mediated transcriptional regulation is elusive.22 Furthermore, TGF-β/BMP signaling is also mediated by Smad-independent signaling mechanisms.25 In the context of retinal development, however, the relative contribution of Smad4 in TGF-β/BMP signaling-mediated processes is not known.
Here we report the generation of retina-specific Smad4 mutant mice to investigate the functional contribution of Smad4 during retinal development. We demonstrate that Smad4 is essential for a subset of, but not all, TGF-β/BMP–dependent developmental processes in the embryonic mouse retina. The genetic requirement for Smad4 is most evident in retinal spatial patterning and RGC axon behavior, both known to be under the control of BMP signaling. These results provide genetic evidence that Smad4 is predominantly required for a subset of developmental processes dependent on the BMP branch of the TGF-β signaling pathway in the embryonic retina.
Methods
Mice
Mice carrying a conditional mutant allele for Smad426 (referred to as Smad4fx), those with a series of Bmpr1a and Bmpr1b mutant alleles13 (referred to as Bmpr1a−, BmprIafx, and BmprIb−), and a retina-specific Cre transgene, TgN(Six3Cre)69Frty27 (referred to as Tg-Six3Cre), have previously been described. Retina-specific Smad4 conditional knock-out mutants (Smad4fx/fx;Tg-Six3Cre [referred to as Smad4-CKO]) were obtained by crossing animals carrying Smad4fx/+;Tg-Six3Cre and Smad4fx/fx genotypes. Littermate conditional heterozygous mice (Smad4fx/+;Tg-Six3Cre) were used as controls. Established PCR protocols were used to genotype DNA samples extracted from the tail or embryo yolk sac with appropriate allele-specific primer sets. Housing and handling of animals were performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the guidelines established by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center.
Tissue Processing for Marker Analyses
Tissues were fixed in 4% paraformaldehyde in PBS, followed by standard histologic sectioning. Sections were processed for hematoxylin and eosin staining, in situ hybridization, or immunohistochemistry. In situ hybridization using digoxygenin- or 35S-labeled riboprobes was performed essentially as described.28 Templates for Atoh7, Crx, Dcc, Efnb2, Fgf15, Ntn1, Rax, Tbx5, Vax2, Vsx2, and Zic2 RNA probes have previously been described.13,29–36 Images for digoxygenin in situ sections were photographed under DIC contrast illumination on a compound microscope. Pseudocolor images of radioactive in situ sections were generated as previously described.6 Antibodies used for immunohistochemical detection were anti-BrdU monoclonal antibody (mAb) (Developmental Studies Hybridoma Bank [DSHB], University of Iowa, Iowa City, IA), anti-Brn3 polyclonal antibody (pAb) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Isl1/2 mAb (DSHB; Iowa City, IA), anti-Nr2f2 (COUP-TFII) mAb (Perseus Proteomics, Tokyo, Japan), anti-Smad4 mAb (Santa Cruz Biotechnology), and anti-phospho-Smad1/5/8 pAb (Cell Signaling Technology, Beverly, MA). Secondary antibodies conjugated with Cy2, Cy3, biotin (Jackson ImmunoResearch Laboratories, West Grove, PA), or Alexa Fluor (Alexa 488 and Alexa 555) (Molecular Probes/Invitrogen, Carlsbad, CA) were selected from appropriate animal species based on primary antibodies. Propidium iodide (Molecular Probes/Invitrogen, Carlsbad, CA) was used for nuclear counterstaining. Fluorescent signals were visualized and photographed on a confocal microscope (Olympus, Center Valley, PA). Immunoreactivity of biotin-conjugated antibodies was visualized by peroxidase staining (Vectastain ABC Kit; Vector Laboratories, Burlingame, CA) in accordance with the manufacturer's instructions. For cell proliferation assays, pregnant mice were injected intraperitoneally with 1 mL/100 g body weight of BrdU solution (Cell Proliferation Labeling Reagent; GE Healthcare, Piscataway, NJ) 2 hours before embryo dissection. Apoptotic cells were detected by TUNEL staining (ApopTag Cell Death Detection Kit; Millipore, Billerica, MA) essentially according to the manufacturer's instructions. Three to eight independent samples from both control and mutant groups were analyzed for individual markers.
Quantification of Tissue Morphology and Cellular and Molecular Marker Expression
For quantification of retinal proliferation, differentiation, and cell death markers, the relative population of signal-positive nuclei was assessed on sections through the ocular dorsal-ventral axis. Six to 10 distant sections through and near the optic nerve head (ONH) from each eye were measured. Central depression of the ONH area was used as a landmark to demarcate dorsal and ventral retinal areas. Fluorescent nuclear stainings of PI and Pou4f2 signals were digitally counted with ImageJ software (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html). TUNEL-positive nuclei were manually counted on collections of high-magnification immunofluorescent images. Measurement of diameters of the optic nerves was performed on a series of images from histologic sections. Statistical evaluations between control and mutant samples were performed using the unpaired Student's t-test (two-tailed).
Retinal Ganglion Cell Axon Labeling
Embryonic retinas were isolated by surgically removing the sclera, choroid, cornea, and lens from enucleated eyes; incisions were then made to prepare a retinal flat mount. Pellets of DiI (Molecular Probes/Invitrogen; Carlsbad, CA) suspended in dimethyl formamide were placed on the periphery of the flattened retina using a glass needle. Retinal preparations were incubated in 1% paraformaldehyde in phosphate-buffered saline at 37°C for 2 days. Fluorescent signals were observed and photographically documented using an epifluorescence stereoscope (Olympus, Center Valley, PA).
Results
Loss of Smad4 in the Embryonic Retina Causes Varying Degrees of Microphthalmia
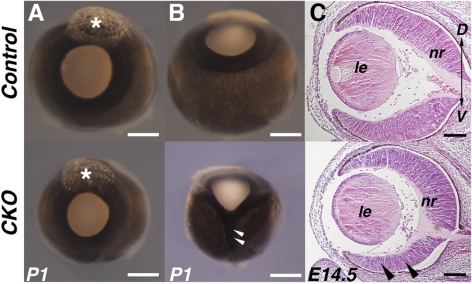
Homozygous null mutations of the Smad4 gene lead to early embryonic lethality before the initiation of ocular development.37,38 In the present study, a Cre-lox strategy was used to investigate the genetic role of Smad4 during embryonic mouse retinal development. Embryos that lack retinal Smad4 function (Smad4-CKO; see Methods) developed apparently normally to term, and all the possible genotypes were segregated at expected Mendelian frequencies. Eyes of Smad4-CKO embryos were morphologically indistinguishable from control littermates up to around embryonic day (E) 12.5. By birth, however, Smad4-CKO mutants exhibited varying degrees of microphthalmia with occasional coloboma (Figs. 1A, 1B). Histologic examination of the mid-gestation retina revealed that, though Smad4-CKO embryonic eyes appeared to be patterned near normally, the retinal neuroepithelium was reduced in thickness, most notably in the ventral quadrant (Fig. 1C, arrowheads).
Figure 1.
Microphthalmia and ventral retinal thinning in Smad4-CKO mutants. (A, B) Newborn eyes from control (top) and Smad4-CKO mutants (bottom) viewed from anterior (A) and ventral (B) orientations. The mutant eye (A) is a representative of mild microphthalmia, whereas a more severe case with concomitant coloboma is shown (B). Asterisks: resin marking dorsal side of the eye. (C) Embryonic eyes sectioned through the dorsal-ventral axis showing apparent thinning in the ventral quadrant of the mutant retina (arrowheads). D, dorsal; le, lens; nr, neural retina; V, ventral. Scale bars: 1 mm (A, B); 100 μm (C).
Increased Apoptosis in the Smad4-CKO Embryonic Retina
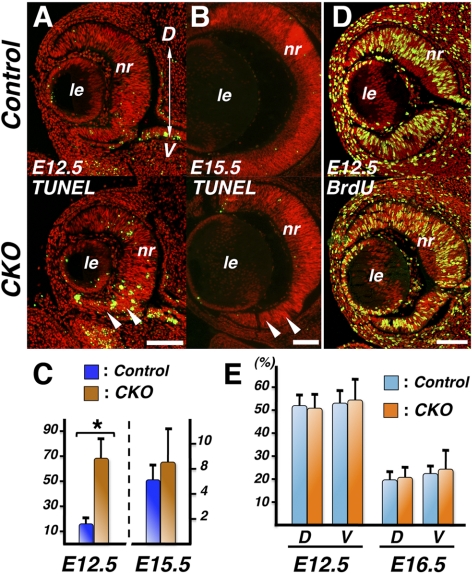
TUNEL assays revealed that retinal apoptosis was rapidly increased at around E12.5, most notably in the ventral aspects of the retina (Figs. 2A, 2C). Soon thereafter, however, this excessive cell death in CKO mutants appeared to subside, even in thinned ventral neural retinas, where elevated cell death was evident earlier in development (Fig. 2B, arrowheads). By E15.5, no statistically significant difference in retinal cell death was detectable compared with controls. Cell proliferation assays at E12.5 and E16.5 revealed no significant difference in Smad4-CKO retinas compared with controls, even in the thin ventral neural retina, where apoptosis was transiently elevated (Figs. 2D, 2E). These observations suggested that transiently increased apoptosis at around E12.5, predominantly in the ventral retina (Fig. 2A), is one main causative factor for microphthalmia phenotypes in embryonic and neonatal Smad4-CKO eyes.
Figure 2.
Embryonic retinal growth in Smad4-CKO mutants. (A) TUNEL staining on coronal sections showing elevated apoptotic cell death predominantly in the ventral retina of a Smad4-CKO mutant at E12.5 (arrowheads). (B) Later in development at E15.5, the Smad4-CKO retina no longer shows evidence of increased apoptosis, even in the thinned ventral retina (arrowheads). (C) Quantification of TUNEL-positive nuclei (per 1000 retinal cells) at E12.5 (n = 6 for control; n = 8 for mutants; *P = 0.0045) and E15.5 (n = 4 for control; n = 6 for mutants). (D) Representative images from BrdU incorporation assays at E12.5. (E) Quantification of BrdU incorporation in the dorsal (D) and ventral (V) regions of the developing retinas at E12.5 (n = 4) and E16.5 (n = 6). No statistically significant differences between controls and mutants are detected. D, dorsal; le, lens; nr, neural retina; V, ventral. Scale bars, 100 μm.
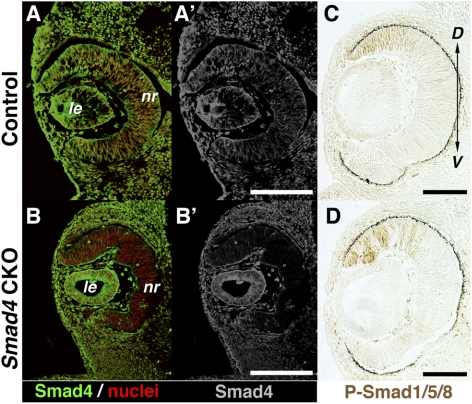
To assess the timing of Smad4 functional deletion in Smad4-CKO embryos, expression of the Smad4 protein was examined. At E10.5, Smad4 protein was ubiquitously distributed throughout the embryonic head, including the retina, and this pattern was unaltered in the Smad4-CKO mutants (not shown). By E11.5, Smad4 immunoreactivity became largely diminished from the mutant retina (Figs. 3A, 3B). This suggests that the loss of Smad4 protein expression in the CKO retina precedes elevated retinal cell death. During this period, BMP receptor-mediated signaling, as assessed by dorsal enrichment of phospo-Smad1/5/8 within the retina, appeared comparable to that of controls (Fig. 3C), suggesting the upstream signaling machinery from BMP ligands to the activation of BMP receptors and the phosphorylation of R-Smads is intact.
Figure 3.
Expression of Smad4 and phospho-Smad1/5/8 in the developing retina. (A, B′) Immunohistochemical detection of Smad4 in the developing retina at E11.5, revealing wide distribution of Smad4 in and around the eye, including the retina, in the control (A, A′). In a CKO littermate, Smad4 immunoreactivity is diminished specifically in the retina (B, B′). (C, D) Immunolocalization of phospho-Smad1/5/8 in the embryonic retina at E12.5, revealing high levels of signal in the dorsal compartment in the CKO mutant (D), comparable to the control (C). D, dorsal; le, lens; nr, neural retina; V, ventral. Scale bars, 100 μm.
Defects in Retinal Spatial Patterning in the Smad4-CKO Retina
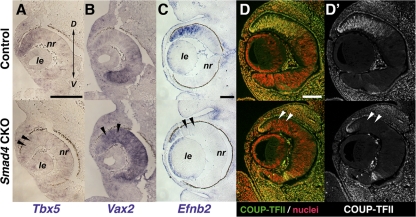
Next, the possible involvement of Smad4 in retinal spatial patterning was examined in light of the clear importance of BMP signaling in this process. In Smad4-CKO mutants, the expression of Tbx5, a dorsal retinal marker regulated by BMP signaling, appeared normal up to E10.5 (not shown). Concomitant with the loss of the Smad4 protein by E11.5 as mentioned above (Fig. 3B), Tbx5 transcripts became largely diminished in the mutant retina (Fig. 4A). In contrast, Vax2, normally expressed in the ventral aspect of the retina (Fig. 4B, top), was found expanded throughout the developing mutant retina (Fig. 4B, bottom). Furthermore, Efnb2, a dorsally expressed gene involved in axon guidance, and COUP-TFII (Nr2f2), an orphan nuclear receptor regulating cone opsin gene expression downstream of BMP signaling,19 were also lost in the mutant retina (Figs. 4C, 4D, 4D′), consistent with the suspected defects in dorsal retinal patterning in Smad4-CKO embryos.
Figure 4.
Defects in dorsal-ventral retinal patterning in Smad4-CKO mutants. (A, B) Expression of the dorsal marker Tbx5 (A) and ventral marker Vax2 (B) in E11.5 retinas. Arrowheads: loss of dorsal Tbx5 expression (A, bottom) and dorsal expansion of Vax2 expression domain (B, bottom), respectively. (C) Expression of Efnb2 in E14.5 retinas, showing diminished expression in the CKO mutant (arrowheads). (D, D′) Expression of COUP-TFII in E13.5 retinas. Although normally localized dorsally in the developing normal retina (top), its expression is lost in the Smad4-CKO retina (bottom, arrowheads). D, dorsal; le, lens; nr, neural retina; V, ventral. Scale bars, 100 μm.
Retinal Growth and Retinal Ganglion Cell Development in Smad4-CKO Mutants
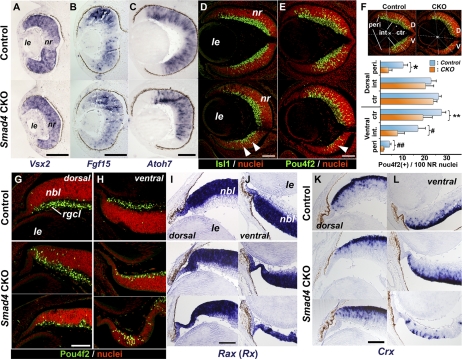
We previously showed that mutant mice lacking both Bmpr1a and Bmpr1b receptor genes in the developing retina fail to initiate retinal neurogenesis, associated with the loss of retinal predifferentiation markers Vsx2 (previously known as Chx10) and Fgf15.13 In Smad4-CKO mutants, both these genes were expressed at comparable levels in the retinal progenitor cells at early phases of neurogenesis (Figs. 5A, 5B). Normally, the initiation of retinal neurogenesis occurs in the central region of the retina at around E11.5, associated with the expression of a pro-neural gene, Atoh7 (previously known as Math5).39 In the Smad4-CKO retina, though the initiation of Atoh7 was unaffected in Smad4-CKO mutants at E11.5 (not shown), radial expansion of the Atoh7 expression domain appeared to be delayed (Fig. 5C). During later retinal differentiation, RGCs express Isl1 and Pou4f2 (previously known as Brn3b), key regulatory nuclear proteins for RGC development, with apparent central-to-peripheral gradients in the normal control retina (Figs. 5D, 5E, top).40 Expression of these RGC markers appeared to be grossly comparable in the dorsal aspect of the retina in Smad4-CKO mutants (Figs. 5D, 5E). In thinned areas of the ventral retina, however, cells expressing Isl1 and Pou4f2 were apparently reduced (Figs. 5D, 5E, arrowheads). Quantification of Pou4f2-expressing cells within the developing neural retina at E14.5 revealed that, though no statistical difference is detected in the proximal region of the dorsal retina (Fig. 5F; dorsal intermediate and central compartments), RGCs are significantly reduced in the dorsal periphery and the entire ventral neural retina (Fig. 5F).
Figure 5.
Expression of markers for retinal growth and differentiation in Smad4-CKO mutants. (A, B) Expression of Vsx2 (Chx10) at E11.5 (A) and Fgf15 at E12.5 (B), both implicated in retinal progenitor growth. (C) Expression of the proneural gene Atoh7 (Math5) in the developing retina at E12.5. (D, E) Distribution of differentiating RGC markers Isl1 (D) and Pou4f2 (Brn3b) (E) in the developing retina at E14.5. (F) Regional distribution of Pou4f2-expressing cells in E14.5 retinas. Dorsal and ventral retinas in reference to the central depression of the ONH were, respectively, divided into peripheral (peri), intermediate (int), and central (ctr) domains. These domains were demarcated in equal angles from the reference point (X) between the distal tip of the retina and the ONH. The reference point was defined as the midpoint between the distal tip of the lens and the lumen of the ONH. Numbers of RGCs were counted separately in each of these domains of the neural retina (n = 4 for control; n = 6 for mutants; *P = 0.011; **P = 0.046; #P = 0.029; ##P = 0.0037). (G–L) Expression of Pou4f2 (G, H), Rax (I, J), and Crx (K, L) in peripheral regions of the dorsal (G, I, K) and ventral (H, J, L) retinas at E18.5. For Smad4-CKO mutants, moderately thinned and more severely affected examples are shown in the middle and bottom rows, respectively. D, dorsal; le, lens; nbl, neuroblastic layer; nr, neural retina; rgcl, RGC layer; V, ventral. Scale bars: 100 μm (A–E); 50 μm (G–L).
At E18.5, however, Pou4f2-positive cells were present in the mutant peripheral neural retina both dorsally and ventrally to an extent comparable to that for the control (Figs. 5G, top and middle panels). Even in mutants that showed severe thinning of the peripheral retina, Pou4f2-expressing cells spread toward the distal margin, albeit at reduced numbers (Figs. 5G, 5H, middle and bottom panels). Retinal progenitor marker Rax was present in the periphery of moderately thinned or severely deformed ventral retinas (Fig. 5J) and in less affected dorsal retinas (Fig. 5I), suggesting sustained activity of neurogenesis in these areas. Crx is expressed in rod photoreceptor cells and their progenitors, whose generation peaks after RGCs.41 Expression of Crx was also present in the mutant retina at levels comparable to those for controls (Figs. 5K, 5L), although it was reduced in overtly deformed ventral retinas in some severely affected CKO mutant eyes (Fig. 5L). Our results failed to demonstrate a reverse correlation between the degrees of RGC and rod photoreceptor cell differentiation as was reported for GDF11 mutants.21 Rather, the general reduction of both cell type markers coincided with retinal thinning in the periphery, especially in ventral regions.
Intraretinal RGC Axon Guidance in Smad4- and BMP Type 1 Receptor-CKO Mutants
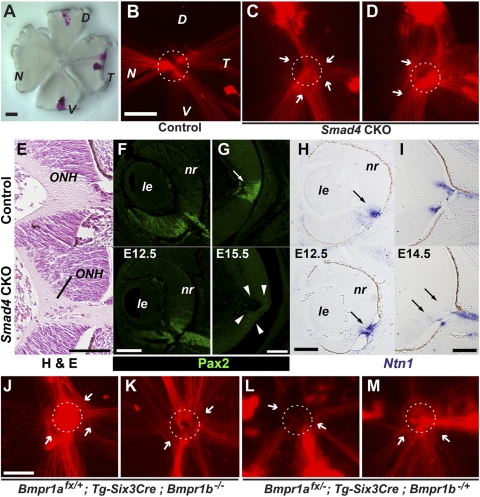
Previously, roles for BMP signaling in the guidance of RGC axons to the optic disc were reported.15,42 To examine whether Smad4 is potentially involved in this BMP–dependent process, the behavior of RGC axons was analyzed by anterograde labeling of the RGC axons with the lipophilic dye DiI (Fig. 6A). In the control retina, the RGC axons were traced from their site of origin to the optic disc in a highly stereotypic manner (Fig. 6B). In contrast, in Smad4-CKO embryos examined at E15.5 through E17.5, overt defects in optic disc axon trajectories were noted (Figs. 6C, 6D). These defects were not localized to axons arising from any particular retinal quadrant. In several instances, the number of misrouting axons was large enough to obscure the optic disc itself (Fig. 6D). At histologic levels, an apparent majority of RGC axons was fascicled to form the optic nerve at the ONH in CKO mutants at E14.5 (Fig. 6E). Earlier in development, at E12.5, Pax2 expression marking the ventral aspects of the developing retina, including the prospective ONH, was observed in patterns comparable to that of controls (Fig. 6F). However, by E15.5, Pax2 expression normally maintained in the ONH area (Fig. 6G, arrow) was almost diminished in the mutant retina (Fig. 6G, arrowheads). We also found abnormality in the expression of netrin1 (Ntn1), a key axon guidance gene, in the ONH (Figs. 6H, 6I). Although Ntn1 transcripts were properly initiated in the prospective ONH area at E12.5 in the Smad4-CKO retina (Fig. 6H, arrows), it was not upregulated and maintained within the retinal proper to define the ONH at E14.5 (Fig. 6I, arrows). These defects were associated with markedly hypoplastic optic nerves (Supplementary Fig. S1A, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental).
Figure 6.
Defects in RGC axon targeting to the optic disc in Smad4-CKO and Bmpr1a;Bmpr1b compound mutants. (A) A representative retinal flat mount preparation subjected to DiI labeling. DiI pellets have been placed on three peripheral positions of the retina to track orientation. (B) An E17.5 control embryonic retina showing labeled RGC axons from all labeled quadrants of the retina targeting the optic disc (dotted circle). (C, D) In Smad4-CKO mutants, though most RGC axons appeared to reach the optic disc region, several went astray locally around the disc (arrows), failing to appropriately target the optic disc and join the optic nerve. In severe cases, RGC axon mistargeting obscures demarcation of the optic disc (D). Note that RGC axon mistargeting does not appear to be biased to those from any particular quadrant in the retina. (E) Morphology of the ONH of control and mutant retinas at E14.5. (F, G) Expression of Pax2 in the central regions of the retina at E12.5 (F) and E15.5 (G). Note the loss of Pax2 expression in the ONH at E15.5 in the mutant (G, bottom, arrowheads). (H, I) Expression of Ntn1 (netrin 1) in the central regions of the retina at E12.5 (H) and E14.5 (I). Note the attenuated expression of Ntn1 in the mutant ONH at E14.5 (I, bottom, arrows). (J–M) Two representative samples each from Bmpr1afx/+;Tg-Six3Cre;Bmpr1b−/− (J, K) and Bmpr1afx/−;Tg-Six3Cre;Bmpr1b−/+ (L, M) mutants. Although peripheral RGC axons show directional extension toward the optic disc, they often go astray locally at the disc (arrows). Misrouting axons appear to arise from all directions. D, dorsal; le, lens; N, nasal; nr, neural retina; T, temporal; V, ventral. Scale bars: 50 μm (A–E, I); 100 μm (F–H).
The RGC axon defects observed in Smad4-CKO eyes appeared more severe than the phenotypes in the BmprIb−/− mutants previously reported by Liu et al.15 In these mutants, only ventral retinal axons were found affected, correlating with ventrally enriched expression of the Bmpr1b receptor in the developing retina.6 We next tested whether the loss of additional type I receptor alleles can exacerbate this RGC axon guidance phenotype. Compound mutants with the genotypes BmprIafx/+;Tg-Six3Cre;BmprIb−/−, and BmprIafx/−;Tg-Six3Cre;BmprIb−/+ differ from the BmprIb−/− mutants in terms of the dosage of type I receptor genes; each has only 1 of 4 functional Bmpr1 gene alleles, whereas both alleles of the Bmpr1a gene are intact in BmprIb−/− mutants. In every retina of compound mutant animals lacking 3 of 4 type I BMP receptor gene alleles examined at E16.5 to E17.5, a subset of axons frequently misrouted locally at the optic disc (Figs. 6J–M). Unlike the BmprIb−/− mutants,15 RGCs corresponding to the misrouted axons were not biased toward a particular region of the retina, resembling the defects observed in the Smad4-CKO mutant retina (Figs. 6C, 6D). Consistently, eyes of both BmprIafx/+; Tg-Six3Cre;BmprIb−/− and BmprIafx/−;Tg-Six3Cre;BmprIb−/+ mutants exhibited hypoplastic optic nerves (Supplementary Figs. S1B, S1C, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental) compared with control BmprIafx/+; Tg-Six3Cre; BmprIb−/+ (doubly heterozygous) mice with approximately 11.5% and 16.5% reductions in average optic nerve diameters, respectively (BmprIafx/+;Tg-Six3Cre;BmprIb−/+ [control: *, Supplementary Figs. S1B, S1C, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental], 155.7 μm ± 1.9 [n = 6]; BmprIafx/+;Tg-Six3Cre;BmprIb−/− [#, Supplementary Figs. S1B, S1C, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental], 131.6 ± 8.9 [P = 0.0058; n = 4]; BmprIafx/−;Tg-Six3Cre;BmprIb−/+ [∧, Supplementary Figs. S1B, S1C, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental], 137.8 ± 4.5 [P = 0.0045; n = 6]). Analyses of retinal molecular markers (Supplementary Fig. S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental), however, did not reveal apparent abnormalities. For example, Ntn1 was properly expressed in the ONH in these Bmpr1a;Bmpr1b compound mutants (Supplementary Figs. S2A–C, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental), in contrast to Smad4-CKO mutants as described (Fig. 6I).
Discussion
The present study has provided genetic evidence that Smad4, a key mediator of the TGF-β signaling system, is required for normal retinal development in the mouse. In addition, comparison of Smad4-CKO mouse phenotypes with the range of retinal defects previously observed in various TGF-β/BMP signaling component mutants provides a platform with which to evaluate the degree of Smad4 requirement in specific contexts of retinal development. For example, the comparison of retinal phenotypes between Smad4-CKO (the present study) and the allelic series of Bmpr1a;Bmpr1b compound mutants13 suggests that Smad4 function is essential only for a subset of BMP signal-regulated processes.
Smad4 and Retinal Growth and Differentiation
Mouse embryos that lack the function of both Bmpr1a and Bmpr1b in the embryonic retina, thus with no BMP receptor-mediated signaling, exhibit elevated retinal cell death, followed by deficits in cell proliferation.13 Thus, the ultimate consequence of a complete loss of BMP receptor-mediated signaling is degeneration of the embryonic retina. In addition, expression of the proneural gene Atoh7 is not detected in the Bmpr1a;Bmpr1b-double mutant retina.13 In contrast, the Smad4-CKO retina shows expression of Atoh7 and subsequent differentiation of RGCs, and further retinal growth and differentiation proceed throughout embryogenesis, albeit at reduced degrees. This suggests that Smad4 is genetically dispensable for certain aspects of BMP-dependent retinal growth and differentiation.
Elevated cell death in the Smad4-CKO retina observed predominantly in the ventral quadrant suggests a regional difference in the degrees of Smad4 dependency within the developing retina. In the developing mouse retina, a major BMP ligand, Bmp4, is concentrated in the dorsal retina.6 In contrast, a secreted BMP antagonist, Chdrl1 (formerly known as ventroptin), is ventrally localized.43 Thus, the overall BMP signaling levels may be significantly lower in the ventral region, as reflected by the lower levels of phosphorylated BMP R-Smads in the ventral retina (e.g., Fig. 3C). Ventrally localized cell death in the Smad4-CKO retina may suggest that ventral retinal cells receiving low levels of BMP signaling have a higher dependence on Smad4 than dorsal regions for BMP-mediated cell survival.
Smad4 and Retinal Spatial Patterning
In contrast to the observations that Smad4 is largely dispensable for retinal cell proliferation and differentiation, loss of Smad4 caused a clear disruption in retinal spatial patterning, consistent with the known function of BMP signaling in retinal D-V patterning.10,13,17 In epiblast-specific Smad4 mutants, the specification of primordial germ cells, a process known to require BMP signaling,44 failed to occur.45 However, in the same mutants, allantois formation, another BMP-dependent process in early mouse embryos, was not abolished.45,46 Genetic studies indicate that the specification of these distinct cell types may require different threshold levels of BMP signaling.44,45,47 Thus, in addition to aiding the maintenance of ventral retinal cell survival at a low BMP signaling input, Smad4 may also be required to maintain sufficient signaling levels to sustain dorsal retinal characteristics that require a higher BMP signaling input.
Smad4 and RGC Axon Guidance
Despite largely redundant genetic functions of Bmpr1a and Bmpr1b in multiple developmental contexts,13,48,49 Bmpr1b has been shown to play unique roles in intraretinal RGC axon guidance in the mouse15 and BMP-mediated chemo-repulsion of spinal commisural axons in the chick.50 In the present study, we have observed similar defects in the targeting of RGC axons to the optic disc in Smad4-CKO and Bmpr1a;Bmpr1b compound mutants. Decreasing the dosage of Bmpr1a in the Bmpr1b−/− mutant background appears to exacerbate this defect. Of note, these mutants show no overt retinal morphologic and patterning defects.13 Bmpr1a−/fx;Tg-Six3Cre;Bmpr1b−/+ mutants (retaining one functional copy of the Bmpr1b gene), which have a morphologically normal retina but show D-V patterning defects,13 also exhibit RGC axon targeting defects similar to those seen in Bmpr1b−/−, Bmpr1afx/+;Tg-Six3Cre;Bmpr1b−/−, and Smad4-CKO mutants. These results suggest a potential importance of the overall type I Bmp receptor levels rather than a specific requirement for Bmpr1b. In this context, Smad4 may be required to maintain a critical level of BMP signaling to control proper RGC axon behaviors.
Interpretation of these results, however, requires some precautions. At present, we cannot exclude the possibility that defects in RGC axon guidance in Smad4-CKO and Bmpr1a−/fx; Tg-Six3Cre;Bmpr1b−/+ mutants are secondary due to altered D-V patterning. Furthermore, several lines of evidence suggest that some aspects of BMP-mediated response of growth cone and axon extension are independent of conventional Smad-mediated mechanisms.51
One striking difference between Smad4-CKO mutants and Bmpr1a;Bmpr1b compound mutants with regard to RGC axon behaviors is that Smad4-CKO retinas fail to maintain some of the key factors for ONH specification and function, including Pax2 and Ntn1. Loss of these molecules in the ONH is likely to be an additional causative factor for RGC axon abnormality in Smad4-CKO mutants, which, together with generally reduced eye size, leads to severe optic nerve hypoplasia. Underlying molecular mechanisms by which Smad4 controls the expression of Pax2 and Ntl1 in the ONH are unclear at the moment. Recent studies have implicated an important role for Bmp7 to properly specify the ONH in concert with Shh,42 and Smad4 may make a functional contribution to this process.
TGF-β/Activin Signaling in the Embryonic Eye and Smad4 Loss of Function
Several TGF-β family ligands that signal through the canonical TGF-β/activin branch pathway have been shown to play roles during retinal development in vivo. Tgf-β2 and Tgf-β3 are expressed in the developing retina,52,53 and Tgfb2, or combinatorial Tgfb2 and Tgfb3 mutant mice exhibit hyperplastic retinas, associated with decreased apoptosis.53 Consistently, in the chick retina, application of a TGF-β–neutralizing antibody resulted in a marked decrease in apoptosis.52 These phenotypic characteristics of Tgfb2- and Tgfb2;Tgfb3 mutants are clearly distinct from those seen in the Smad4-CKO retina. GDF11 is another canonical TGF-β/activin pathway ligand expressed in the retinal ganglion cell layer.21 Loss of Gdf11 in the mouse causes an increase in retinal ganglion cells at the expense of rod photoreceptor and amacrine cell lineages, suggesting that GDF11 limits the time window for retinal progenitor cells to adopt an RGC fate.21 In contrast, Smad4-CKO mutants do not show evidence for an increased proportion of RGC number and a concomitant decrease in rod photoreceptor fate and thus fail to display common phenotypic characteristics with the Gdf11 mutants. Hypoplastic portions of the Smad4-CKO mutant retina tend to show reductions of both markers for RGC and rod photoreceptor lineages rather than presenting a reverse correlation of these cell types. In addition, our recent preliminary microarray studies indicated that the expression of multiple regulatory genes for photoreceptor cell lineages, including Crx and Rax, show no significant changes in the early postnatal retina (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5940/-/DCSupplemental). Taken together, these results suggest that the known in vivo functions for the TGF-β/activin family ligands can bypass the genetic requirement for Smad4 during embryonic retinal development.
Smad4 Requirement and Alternative Pathways Downstream of TGF-β/BMP Receptors
The present study provided evidence that Smad4 is not necessarily required for all the aspects of TGF-β/BMP signaling in the developing retina, consistent with a growing body of similar evidence from other developmental contexts.45,54,55 A large body of research primarily using in vitro cell culture studies has shown that TGF-β/BMP signaling is also transduced through Smad-independent pathways, including those involving JNK, p38/MAPK, and ERK.25,56 Consistently, in mouse craniofacial development, Smad4 has been shown to function in parallel with the p38/MAPK pathway, constituting redundant mechanisms for palate fusion.57 On the contrary, TGF-β–activated kinase 1 (TAK1), one of the best-characterized downstream effectors for TGF-β/BMP receptors involved in p38/MAPK activation, has been shown to be dispensable for mouse sympathetic neuron development in vivo.54 More recently, R-Smad-dependent, but Smad4-independent, intracellular signaling mechanisms have also been identified.58,59 The Smad4-CKO mouse models developed in the present study will provide a useful platform for further genetic studies to better understand the involvement of unconventional TGF-β/BMP signaling mechanisms during retinal development.
Supplementary Material
Acknowledgments
The authors thank Mathew Chempanal, Heather Smith, Eun Young Kim, and Jody Polan for their skillful technical assistance; Takahisa Furukawa for plasmids; and Mini Kapoor, Li Zhang, and their research staff for microarray gene expression profiling studies.
Footnotes
Supported by National Institutes of Health Grants EY013128 (YF), CA16672 (MD Anderson Cancer Center), and EY010608 (UT Health Sciences Center at Houston); American Heart Association Grant 055037Y (YF); and Center for Stem Cells and Developmental Biology pilot grant of UT MD Anderson Cancer Center (YF).
Disclosure: D. Murali, None; M. Kawaguchi-Niida, None; C.-X. Deng, None; Y. Furuta, None
References
- 1. Harada T, Harada C, Parada LF. Molecular regulation of visual system development: more than meets the eye. Genes Dev. 2007;21:367–378 [DOI] [PubMed] [Google Scholar]
- 2. Duenker N. Transforming growth factor-beta (TGF-beta) and programmed cell death in the vertebrate retina. Int Rev Cytol. 2005;245:17–43 [DOI] [PubMed] [Google Scholar]
- 3. Wordinger RJ, Clark AF. Bone morphogenetic proteins and their receptors in the eye. Exp Biol Med (Maywood). 2007;232:979–992 [DOI] [PubMed] [Google Scholar]
- 4. Mishina Y. Function of bone morphogenetic protein signaling during mouse development. Front Biosci. 2003;8:d855–869 [DOI] [PubMed] [Google Scholar]
- 5. Adler R, Belecky-Adams TL. The role of bone morphogenetic proteins in the differentiation of the ventral optic cup. Development. 2002;129:3161–3171 [DOI] [PubMed] [Google Scholar]
- 6. Furuta Y, Hogan BL. BMP4 is essential for lens induction in the mouse embryo. Genes Dev. 1998;12:3764–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hammerschmidt M, Kramer C, Nowak M, Herzog W, Wittbrodt J. Loss of maternal Smad5 in zebrafish embryos affects patterning and morphogenesis of optic primordia. Dev Dyn. 2003;227:128–133 [DOI] [PubMed] [Google Scholar]
- 8. Hanel ML, Hensey C. Eye and neural defects associated with loss of GDF6. BMC Dev Biol. 2006;6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wawersik S, Purcell P, Rauchman M, Dudley AT, Robertson EJ, Maas R. BMP7 acts in murine lens placode development. Dev Biol. 1999;207:176–188 [DOI] [PubMed] [Google Scholar]
- 10. Behesti H, Holt JK, Sowden JC. The level of BMP4 signaling is critical for the regulation of distinct T-box gene expression domains and growth along the dorso-ventral axis of the optic cup. BMC Dev Biol. 2006;6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trousse F, Esteve P, Bovolenta P. Bmp4 mediates apoptotic cell death in the developing chick eye. J Neurosci. 2001;21:1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haynes T, Gutierrez C, Aycinena JC, Tsonis PA, Del Rio-Tsonis K. BMP signaling mediates stem/progenitor cell-induced retina regeneration. Proc Natl Acad Sci U S A. 2007;104:20380–20385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murali D, Yoshikawa S, Corrigan RR, et al. Distinct developmental programs require different levels of Bmp signaling during mouse retinal development. Development. 2005;132:913–923 [DOI] [PubMed] [Google Scholar]
- 14. Muller F, Rohrer H, Vogel-Hopker A. Bone morphogenetic proteins specify the retinal pigment epithelium in the chick embryo. Development. 2007;134:3483–3493 [DOI] [PubMed] [Google Scholar]
- 15. Liu J, Wilson S, Reh T. BMP receptor 1b is required for axon guidance and cell survival in the developing retina. Dev Biol. 2003;256:34–48 [DOI] [PubMed] [Google Scholar]
- 16. McLaughlin T, O'Leary DD. Molecular gradients and development of retinotopic maps. Annu Rev Neurosci. 2005;28:327–355 [DOI] [PubMed] [Google Scholar]
- 17. Koshiba-Takeuchi K, Takeuchi JK, Matsumoto K, et al. Tbx5 and the retinotectum projection. Science. 2000;287:134–137 [DOI] [PubMed] [Google Scholar]
- 18. Plas DT, Dhande OS, Lopez JE, et al. Bone morphogenetic proteins, eye patterning, and retinocollicular map formation in the mouse. J Neurosci. 2008;28:7057–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Satoh S, Tang K, Iida A, et al. The spatial patterning of mouse cone opsin expression is regulated by bone morphogenetic protein signaling through downstream effector COUP-TF nuclear receptors. J Neurosci. 2009;29:12401–12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu HJ, Burgess AW. Regulation of transforming growth factor-beta signaling. Mol Cell Biol Res Commun. 2001;4:321–330 [DOI] [PubMed] [Google Scholar]
- 21. Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930 [DOI] [PubMed] [Google Scholar]
- 22. Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810 [DOI] [PubMed] [Google Scholar]
- 23. Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408 [DOI] [PubMed] [Google Scholar]
- 24. Schmierer B, Hill CS. TGFβ-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982 [DOI] [PubMed] [Google Scholar]
- 25. Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584 [DOI] [PubMed] [Google Scholar]
- 26. Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81 [DOI] [PubMed] [Google Scholar]
- 27. Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26:130–132 [PubMed] [Google Scholar]
- 28. Nagy A, Gertsenstein M, Vinterstein K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd ed Cold Spring Harbor, NY: Cold Spring Haobor Laboratory Press; 2003 [Google Scholar]
- 29. Barbieri AM, Broccoli V, Bovolenta P, et al. Vax2 inactivation in mouse determines alteration of the eye dorsal-ventral axis, misrouting of the optic fibres and eye coloboma. Development. 2002;129:805–813 [DOI] [PubMed] [Google Scholar]
- 30. Chapman DL, Garvey N, Hancock S, et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390 [DOI] [PubMed] [Google Scholar]
- 31. Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci U S A. 1997;94:3088–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541 [DOI] [PubMed] [Google Scholar]
- 33. Vincentz JW, McWhirter JR, Murre C, Baldini A, Furuta Y. Fgf15 is required for proper morphogenesis of the mouse cardiac outflow tract. Genesis. 2005;41:192–201 [DOI] [PubMed] [Google Scholar]
- 34. Williams SE, Mann F, Erskine L, et al. Ephrin-B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron. 2003;39:919–935 [DOI] [PubMed] [Google Scholar]
- 35. Bertuzzi S, Hindges R, Mui SH, O'Leary DD, Lemke G. The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev. 1999;13:3092–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gad JM, Keeling SL, Wilks AF, Tan SS, Cooper HM. The expression patterns of guidance receptors, DCC and Neogenin, are spatially and temporally distinct throughout mouse embryogenesis. Dev Biol. 1997;192:258–273 [DOI] [PubMed] [Google Scholar]
- 37. Sirard C, de la Pompa JL, Elia A, et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12:107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang X, Li C, Xu X, Deng C. The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proc Natl Acad Sci U S A. 1998;95:3667–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833 [DOI] [PubMed] [Google Scholar]
- 40. Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A. 2008;105:6942–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–38 [DOI] [PubMed] [Google Scholar]
- 42. Morcillo J, Martinez-Morales JR, Trousse F, Fermin Y, Sowden JC, Bovolenta P. Proper patterning of the optic fissure requires the sequential activity of BMP7 and SHH. Development. 2006;133:3179–3190 [DOI] [PubMed] [Google Scholar]
- 43. Sakuta H, Suzuki R, Takahashi H, et al. Ventroptin: a BMP-4 antagonist expressed in a double-gradient pattern in the retina. Science. 2001;293:111–115 [DOI] [PubMed] [Google Scholar]
- 44. McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15 [DOI] [PubMed] [Google Scholar]
- 45. Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512 [DOI] [PubMed] [Google Scholar]
- 46. Lawson KA, Dunn NR, Roelen BA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Loebel DA, Watson CM, De Young RA, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14 [DOI] [PubMed] [Google Scholar]
- 48. Wine-Lee L, Ahn KJ, Richardson RD, Mishina Y, Lyons KM, Crenshaw EB., 3rd Signaling through BMP type 1 receptors is required for development of interneuron cell types in the dorsal spinal cord. Development. 2004;131:5393–5403 [DOI] [PubMed] [Google Scholar]
- 49. Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:5062–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamauchi K, Phan KD, Butler SJ. BMP type I receptor complexes have distinct activities mediating cell fate and axon guidance decisions. Development. 2008;135:1119–1128 [DOI] [PubMed] [Google Scholar]
- 51. Sanchez-Camacho C, Bovolenta P. Emerging mechanisms in morphogen-mediated axon guidance. Bioessays. 2009;31:1013–1025 [DOI] [PubMed] [Google Scholar]
- 52. Dunker N, Schuster N, Krieglstein K. TGF-beta modulates programmed cell death in the retina of the developing chick embryo. Development. 2001;128:1933–1942 [DOI] [PubMed] [Google Scholar]
- 53. Dunker N, Krieglstein K. Reduced programmed cell death in the retina and defects in lens and cornea of Tgfbeta2(−/−) Tgfbeta3(−/−) double-deficient mice. Cell Tissue Res. 2003;313:1–10 [DOI] [PubMed] [Google Scholar]
- 54. Morikawa Y, Zehir A, Maska E, et al. BMP signaling regulates sympathetic nervous system development through Smad4-dependent and -independent pathways. Development. 2009;136:3575–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rajagopal R, Huang J, Dattilo LK, et al. The type I BMP receptors, Bmpr1a and Acvr1, activate multiple signaling pathways to regulate lens formation. Dev Biol. 2009;335:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu X, Han J, Ito Y, Bringas P, Jr, Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15:322–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31:918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.