The authors report the identification of novel gene targets of miR-204 and show that this microRNA may play an important role in the regulation of multiple functions in human trabecular meshwork cells, including apoptosis, accumulation of damaged proteins, endoplasmic reticulum stress response, and expression of inflammatory mediators.
Abstract
Purpose.
To investigate the biological functions of miR-204 in human trabecular meshwork (HTM) cells.
Methods.
Changes in gene expression induced by miR-204 in HTM cells were evaluated by gene array analysis using arrays and confirmed by quantitative-PCR (Q-PCR). Direct targeting of miR-204 to 12 potential novel targets was confirmed using a luciferase system, and five of them were verified by Western blot analysis. Effects of miR-204 on apoptosis, cell viability, and accumulation of carbonylated proteins were evaluated in HTM cells treated with H2O2. Induction of endoplasmic reticulum (ER) stress markers by tunicamycin was analyzed by Q-PCR, and expression of IL-8 and IL-11 was analyzed by ELISA.
Results.
MiR-204 decreased the expression of multiple genes in HTM cells. Twelve genes (AP1S2, Bcl2l2, BIRC2, EDEM1, EZR, FZD1, M6PR, RAB22A, RAB40B, SERP1, TCF12, and TCF4) were validated as direct targets of miR-204. Downregulation of expressions at protein levels of Bcl2l2, BIRC2, EZR, M6PR, and SERP1 were confirmed by Western blot analysis. HTM cells transfected with miR-204 showed increased levels of apoptosis, decreased viability, increased accumulation of oxidized proteins after H2O2 treatment, decreased induction of ER stress response markers, and reduced expression of inflammatory mediators IL-8 and IL-11.
Conclusions.
MiR-204 potentially plays an important role in the regulation of multiple functions in HTM cells including apoptosis, accumulation of damaged proteins, ER stress response, and expression of inflammatory mediators.
Increased presence of senescent cells in aging tissues has been hypothesized to contribute to pathophysiological changes associated with several age-related conditions.1–3 Specifically, senescence of human trabecular meshwork (HTM) cells has been proposed to play a role in the functional alterations of this tissue in primary open angle glaucoma.4 We have previously reported that senescence of HTM cells is associated with significant changes of several microRNAs (miRNAs) and that miRNAs might contribute to the regulation of the phenotypic alterations characteristic of senescent cells. One of the miRNAs significantly downregulated in senescent HTM cells was miR-204.5,6
MiR-204 has been proposed to be involved in the regulation of multiple functions in different cell types. It is expressed at relatively high levels in retinal pigment epithelium (RPE), where it has been demonstrated to be the target of TGF-beta receptor 2 (TGFβR2) and SNAIL-2, leading to a decrease in transepithelial resistance associated with reduced expression of claudins 10, 16, and 19.7 MiR-204 has also been found to be highly abundant in distal axons compared with the cell bodies of primary sympathetic neurons, suggesting some potential role in the maintenance of axonal structure and function as well as neuronal growth and development.8 Expression of miR-204 is regulated by different light levels in the mouse retina, suggesting a potential role in adaptation to different levels of illumination.9 Finally, miR-204 has been found to be downregulated in several types of tumors,10,11 and it has been proposed that such downregulation could contribute to tumor growth through de-repression of the validated targets HOXA10 and MEIS111 and some predicted targets such as the antiapoptotic protein Bcl212 and the member of the RAS oncogene family RAB22.13 However, there is still little information about the genes regulated by miR-204 and the biological functions modulated by this miRNA.
To gain insight on the biological roles of miR-204 in the TM, we analyzed the changes in gene expression induced by this miRNA in HTM cells and identified 12 novel targets. Based on the genes downregulated by miR-204, we evaluated its role in the regulation of ER stress response, accumulation of oxidized proteins, and apoptosis in HTM cells.
Methods
Cell Culture of Primary HTM Cells
Postmortem human eyes or cornea rings were obtained from the New York Eye Bank within 7 days of death in accordance with the tenets of the Declaration of Helsinki. Primary cultures of HTM cells were generated and maintained following the methods previously described.14 All reagents were obtained from Invitrogen Corporation (Carlsbad, CA).
Transfection
Transfection of miRNAs was performed with a transfection system (Nucleofector; Amaxa Inc. Gaithersburg, MD) in accordance with the manufacturer's instructions. MiR-204 mimic (204M) or negative miRNA control mimic (ConM) (Dharmacon, Inc., Chicago, IL) (120 pmol per 5 × 105 cells) were transfected into HTM cells using the Amaxa program T23. The culture medium was replaced with fresh Dulbecco's modified Eagle's medium (DMEM) growth medium 24 hours after transfection, and cell culture supernatant or cells were collected 72 hours after transfection.
Microarray and Data Analysis
HTM cell cultures (HTM1073 passage 3) were transfected with 204M or ConM. Three days after transfection, total RNAs were isolated and hybridized to a gene expression array (Human Genome U133 2.0 Array, including Human Genome U133 Set; Affymetrix, Santa Clara, CA) at the Duke University Microarray facility (Durham, NC). This array includes 6500 additional genes for analysis of more than 47,000 transcripts. Raw data were normalized and analyzed (GeneSpring 10; Silicon Genetics, Wilmington, DE). Genes were filtered by intensity compared with the control channel, and P ≤ 0.05 of a paired Student's t-test was considered significant. The list of genes significantly downregulated was compared with three databases that predicted targets for miRNAs: Microcosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), TargetScan (http://www.targetscan.org), and PicTar-Vert (http://pictar.mdc-berlin.de/).
Q-PCR Analysis of Gene Expression
Total RNA was isolated (RNeasy kit; Qiagen Inc., Valencia, CA). RNA yields were measured using fluorescent dye (RiboGreen; Molecular Probes, Eugene, OR). First-strand cDNA was synthesized from total RNA (1 μg) by reverse transcription using oligo-dT and reverse transcriptase (Superscript II; Invitrogen Corporation) in a 20-μL volume. Q-PCR was performed in a 20-μL mixture that contained 1 μL cDNA preparation and 1× real-time PCR mix (iQ SYBR Green SuperMix; Bio-Rad, Hercules, CA), using the following PCR parameters: 95°C for 3 minutes followed by 40 cycles of 95°C for 10 seconds, 60°C for 30 seconds followed by melt curve (65°C–95°C in increments of 0.5°C for 5 seconds). Each quantification was conducted in triplicate, and the experiments were conducted in triplicate using two cell lines. The fluorescence threshold value (Ct) was calculated using real-time detection system software (iCycle; Bio-Rad). The absence of nonspecific products was confirmed by both the analysis of the melt curves to exclude primer-dimer and by electrophoresis in gels (3% Super AcrylAgarose; Bio-Rad) to verify the correct product size. β-Actin was used as an internal standard of mRNA expression to normalize the individual gene expression level. The specific primer pairs used to amplify genes were shown in Table 1.
Table 1.
Primer Pairs Used to Quantify Gene Expressions in HTM Cells
| Gene Name | GenBank Accession No. | Primer Pairs |
|---|---|---|
| AKAP1 | NM_003488.3 | For: 5′-AGA ACT GGG CAA TGA GGA GAG CTT |
| Rev: 5′-ATT ATC TGG AAG GCA GCC CGC TTA | ||
| AP1S1 | NM_001283 | For: 5′-TGT TCA GGG CAC TTC TTG TCC TCT |
| Rev: 5′-TTG TTC CTC CCT GGC ACT GGA TAA | ||
| AP1S2 | NM_003916.3 | For: 5′-GTC AGG GAA AGC TTC GAC TGC AAA |
| Rev: 5′-GCC ACT CAA GGA AGC TGC ACA TTT | ||
| ARHGAP29 | NM_004815.3 | For: 5′-TGT CTC ATC ACA CCC AGT CCC AAA |
| Rev: 5′-AGG ATT CCA ATG GTG TGC TCA GGA | ||
| ATP2B1 | NM_001682.2 | For: 5′-TAC CCA AGC ATG GTC CTG AAC AGT |
| Rev: 5′-TCA ACG ATC CAG GGC TCT TTG TCT | ||
| BCL2 | NM_000657 | For: 5′-ATT TCC TGC ATC TCA TGC CAA GGG |
| Rev: 5′-TGT GCT TTG CAT TCT TGG ACG AGG | ||
| Bcl2l2 | NM_004050.3 | For: 5′-GTT GTG GGC TTT GGT TCG GCT TTA |
| Rev: 5′-TAA CAC CCA TGC AAA CAG TGT GGC | ||
| BIRC2 | NM_001166.3 | For: 5′-TTG TTG TGA TGG TGG CTT GAG GTG |
| Rev: 5′-CAC ACC TTG GAA ACC ACT TGG CAT | ||
| CDH2 | NM_001792.3 | For: 5′-TGT GGG AAT CCG ACG AAT GGA TGA |
| Rev: 5′-TGG AGC CAC TGC CTT CAT AGT CAA | ||
| COL5A3 | NM_015719.3 | For: 5′-TTC AGC TCT TCT CGA GCG GGA TTT |
| Rev: 5′-TCA AAG CCT CAG CAC CAA ATG CAC | ||
| CXCL3 | NM_002090 | For: 5′-ACC GAA GTC ATA GCC ACA CTC AAG |
| Rev: 5′-ACT TCT CTC CTG TCA GTT GGT GCT | ||
| EDEM1 | NM_014674.2 | For: 5′-ACC AGA CCT TAA CGA CCA AAC CCA |
| Rev: 5′-ATT CCT ACA CCA CCA TGG ACA GCA | ||
| EZR | NM_003379.4 | For: 5′-AGC ACA AAC TTA CCA GGG ACC AGT |
| Rev: 5′-CAA GGG CAT CAA CTC CAA GCC AAA | ||
| FARP1 | NM_005766.2 | For: 5′-TCA GCA TCG CAC TGG AGA ATC AGT |
| Rev: 5′-AAG TTT GTG AAC ACC ACC CAC AGC | ||
| FZD1 | NM_003505.1 | For: 5′-GCA TGA AGC TTT GTG TGG GTT GGA |
| Rev: 5′-TAA GCG CAG GCT GTC TCA TTC TCA | ||
| HAS2 | NM_005328 | For: 5′-TGT CGA GTT TAC TTC CCG CCA AGA |
| Rev: 5′-AAT CAC ACC ACC CAG GAG GAT TGT | ||
| IL11 | NM_000641 | For: 5′-AGA TAT CCT GAC ATT GGC CAG GCA |
| Rev: 5′-TTG GAC TTC AGT GAT CCA CTC GCT | ||
| IL1B | NM_000576.2 | For: 5′-AAC AGG CTG CTC TGG GAT TCT CTT |
| Rev: 5′-ATT TCA CTG GCG AGC TCA GGT ACT | ||
| IL1RAP | NM_002182.2 | For: 5′-AAG GTG AAA GAG CTG AAG AGG GCT |
| Rev: 5′-ACC TGC CCT GTG GAT ACT TGG ATT | ||
| IL8 | NM_000584 | For: 5′-AGA AAC CAC CGG AAG GAA CCA TCT |
| Rev: 5′-CAC CTT CAC ACA GAG CTG CAG AAA | ||
| ITGB3 | NM_000212.2 | For: 5′-TGG ACA AGC CTG TGT CAC CAT ACA |
| Rev: 5′-TTG TAG CCA AAC ATG GGC AAG CAG | ||
| JARID2 | NM_004973.2 | For: 5′-ACC TCA AAT CTC AGG GCA TCA CCA |
| Rev: 5′-ATG CGC AGC ATG TCT GCT AGT TTG | ||
| M6PR | NM_002355.2 | For: 5′-AAT CGA CAC ACC CTA GCG GAC AAT |
| Rev: 5′-CAG CAA CCA GTG ATG CAA ACG TGA | ||
| MAPRE2 | NM_014268.2 | For: 5′-ATG TTC CGT GGT GTT TGG TTT CCC |
| Rev: 5′-GAA GGG CAA CAC GCT GTT AAG CAT | ||
| MYOC | NM_000261 | For: 5′-TGT ACA GGC AAT GGC AGA AGG AGA |
| Rev: 5′-ACT TGG AAA GCA GTC AAA GCT GCC | ||
| PLAT | NM_000930 | For: 5′-GCA TGA CTT TGG TGG GCA TCA TCA |
| Rev: 5′-AGA AGA GGC GGG ATC TCA TTT GCT | ||
| PLAUR | NM_002659 | For: 5′-TGT GGC TCA TCA GAC ATG AGC TGT |
| Rev: 5′-TTG TTG TGG AAA CCA TTG GAG CCC | ||
| RAB22A | NM_020673.2 | For: 5′-TAC CAA AGA GGC AAA GCA TGT GCG |
| Rev: 5′-AGA CAC CAT GCA ATC ACC AAC AGC | ||
| RAB40B | NM_006822.2 | For: 5′-GCA CTG AAA GAA ACA CGG CGG AAT |
| Rev: 5′-ACA AGA GCT TCA TGC ACA TCC ACG | ||
| SERINC3 | NM_006811.2 | For: 5′-TAC CAC CGT GTT AGA AAG CAG CCT |
| Rev: 5′-TAA ATG AGG CGA GTC ACC GTG GAA | ||
| SERP1 | NM_014445.3 | For: 5′-AAA TCT AGG GCG ACG CTT GAC AGA |
| Rev: 5′-AAG AGG AAG GAA ACG CAA CGC AAC | ||
| SERPINE1 (PAI-1) | NM_000602 | For: 5′-AAT GTG TCA TTT CCG GCT GCT GTG |
| Rev: 5′-ACA TCC ATC TTT GTG CCC TAC CCT | ||
| TCF4 | NM_003199 | For: 5′-TCC TCC AGG TTT GCC ATC TTC AGT |
| Rev: 5′-AAG GAG CTA GGG AAA GTG CTG GTT | ||
| TCF12 | NM_207036 | For: 5′-ACA CGA GTC TTC CAC CAA TGT CCA |
| Rev: 5′-TCC AAG TGC ATC ACC TGT CTG TGA | ||
| TGFBR2 | NM_003242.5 | For: 5′-TGT TGA GTC CTT CAA GCA GAC CGA |
| Rev: 5′-ACT TCT CCC ACT GCA TTA CAG CGA | ||
| β-actin | NM_001101.3 | For: 5′-CCT CGC CTT TGC CGA TCC G |
| Rev: 5′-GCC GGA GCC GTT GTC GAC G |
For, forward; Rev, reverse.
Luciferase Reporter Assay
The 3′UTR fragments containing miR-204 target sites of AP1S2, Bcl2l2, BIRC2, EDEM1, EZR, FZD1, M6PR, RAB22A, RAB40B, SERP1, TCF12, and TCF4 genes were amplified by PCR from human cDNA using forward and reverse primers listed in Table 2, which create XhoI and NotI sites, respectively, and ligate to pCR2.1 vector. The XhoI-NotI–digested products were then cloned into a vector (psiCheck2; Promega Corporation, Madison, WI) that included two luciferase reporter genes, Renilla (can be combined with 3′UTR insert) and firefly. The H293 cells were cotransfected in 12-well plates using reagent (Effectene, 301427; Qiagen, Germantown, MD) with 300 ng of the 3′UTR-luciferase report vector and 7 ng 204M or ConM (Dharmacon, Inc.). Twenty-four hours after transfection, firefly and Renilla luciferase activities were measured consecutively by using dual-luciferase assays (Promega Corporation) according to the manufacturer's protocol. Negative control vectors were generated by cloning the same 3′UTRs in reverse orientation of the individual gene.
Table 2.
Primer Pairs Used to Amplify 3′UTR of Target Genes
| Gene Name | GenBank Accession No. | Primer Pairs |
|---|---|---|
| AP1S2 3′UTR | NM_003916.3 | For: 5′-tgactcgagTGATCTACTGCAGGAGGAAGCTGA |
| Rev: 5′-tatgcggccgcATGTGTAGATAGGCTCTCAGTATGG | ||
| Bcl2l2 3′UTR | NM_004050.3 | For: 5′-tgactcgagTTTGCCCTCAAACAGAACAGCTCC |
| Rev: 5′-tatgcggccgcCAGTCCTTCTCATTAAACTTCTGGGC | ||
| BIRC2 3′UTR | NM_001166.3 | For: 5′-cgactcgagGAAATGCTGCGGCCAACATCTTCA |
| Rev: 5′-tatgcggccgcAAGCACCAAAGACAATTCGGCACC | ||
| EDEM1 3′UTR | NM_014674.2 | For: 5′-tgactcgagACAGTGCAGCAGGGTTGACTCTTA |
| Rev: 5′-tatgcggccgcATCACAGGCAGGAAGAACTCACCT | ||
| EZR 3′UTR | NM_003379.4 | For: 5′-cgactcgagACCCAACAGAAACATTCTGGGCTG |
| Rev: 5′-tatgcggccgcTGAAGGAGAAAGCAGTGCACGAGA | ||
| FZD1 3′UTR | NM_003505.1 | For: 5′-cgactcgagACAACTCTCTTTCGCAGGCTCCTT |
| Rev: 5′-tatgcggccgcTGTGGGATTGGCTATGAGACACCA | ||
| M6PR 3′UTR | NM_002355.2 | For: 5′-cgactcgagCCAAACCAAAGCTACACAGCCAGA |
| Rev: 5′-tatgcggccgcTGTGTCCAGTGGAGCCATGTTACT | ||
| RAB22A 3′UTR | NM_020673.2 | For: 5′-cgactcgagAATTCCATCCACTGACGCCAACCT |
| Rev: 5′-tatgcggccgcAGGAAAGCTCCTCAGTTGGTCTGA | ||
| RAB40B 3′UTR | NM_006822.2 | For: 5′-cgactcgagAAGCCACCTCAAGTCCTTCTCGAT |
| Rev: 5′-tatgcggccgcCGAAACTGTGCTGGCTGTGTTCAA | ||
| SERP1 3′UTR | NM_014445.3 | For: 5′-cgactcgagATGGGCATGTGAAGTGACTGACCT |
| Rev: 5′-tatgcggccgcGTCAAAGCCCATCACAATGCATCC | ||
| TCF12 3′UTR | NM_207036.1 | For: 5′-tgactcgagACCCATCCTGGGCTTAGTGAAACT |
| Rev: 5′-tatgcggccgcTTTCTCTCCCAGGGTTGTTGTGGT | ||
| TCF4 3′UTR | NM_003199.2 | For: 5′-tgactcgagGGAACTGGAAACACTCCCTGCATT |
| Rev: 5′-tatgcggccgcAACTCCAGCACACAACCTGTTTGG |
For, forward; Rev, reverse.
Protein Extraction and Immunoblot
Cells were washed twice in cold PBS. Total protein was extracted using RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 5 mM EDTA, pH 8.0) containing 1× protease inhibitor cocktail (Roche, Inc., Madison, WI). Protein concentration was determined (Micro BCA Protein Assay Kit; Pierce, Rockford, IL-). Total protein extracts (20–40 μg) were separated by 8% to 15% SDS-PAGE and were transferred to polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Membranes were blocked with 5% nonfat dry milk and incubated overnight with the primary antibodies anti-BIRC2, -Bcl2l2, -EZR (Cell Signaling, Inc., Danvers, MA), -SERP1 (GeneTex Inc., Irvine, CA), or anti-M6PR (Santa Cruz Biotechnology, Santa Cruz, CA). Then they were incubated with a secondary antibody conjugated to horseradish peroxidase for 1 hour at room temperature (RT). Immunoreactive proteins were visualized using chemiluminescence substrate (ECL Plus; GE Healthcare, Pittsburgh, PA). For detection of endogenous control, the membrane was stripped with stripping buffer (25 mM glycine, pH 3.0, plus 1% SDS) and then incubated with anti-β–tubulin (Sigma, St. Louis, MO).
Quantification of IL-8 and IL-11
HTM cells were transfected with 204M or ConM. Three days after transfection, cell culture supernatant was collected. The levels of IL-8 and IL-11 in the cell culture supernatant were detected using IL-8 and IL-11 ELISA kits (R&D Systems, Minneapolis, MN) in accordance with the manufacturer's instructions. Briefly, 50 μL (IL-8) or 100 μL (IL-11) supernatant was loaded onto each well pre-coated with IL-8 or IL-11 antibodies. After 2-hour incubation at RT, the wells were washed extensively with wash buffer, and then 100 μL (IL-8) or 200 μL (IL-11) conjugate was added to each well. After another 1-hour (IL-8) or 2.5-hour (IL-11) incubation at RT, the wells were washed again. Substrate solution (200 μL) was put into each well and incubated for 30 minutes at RT, and then 50 μL stop solution was added to each well. The optical density of each well was determined using a microplate reader with a setting of 450 nm and a correction setting of 540 nm.
Apoptosis and Cytotoxicity Assay
Apoptosis was measured with an assay kit (Vybrant Apoptosis Assay; Invitrogen). HTM cells were transfected with 204M or ConM (120 pmol per 5 × 105 cells) and seeded in a 96-well plate (7 × 103 cells/well). Twenty-four hours after transfection, the cell culture medium was changed; 48 hours later, the cells were treated with H2O2 0, 0.6, or 1.2 mM for 4 hours or with tunicamycin 1 μg/mL overnight. The cells were then incubated in 1 μL/mL iodide for 30 minutes at 37°C, 5% CO2. Fluorescence was measured using an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Cytotoxicity was determined by lactate dehydrogenase (LDH) release into cell culture supernatant using a nonradioactive cytotoxicity assay kit (CytoTox 96; Promega) in accordance with the manufacturer's instructions.
Detection of Oxidized Proteins
Accumulation of carbonylated proteins was determined using a protein oxidation detection kit (Oxyblot; Chemicon International, Temecula, CA) according to a slightly modified protocol. Briefly, cells were washed twice in cold PBS. Total protein was extracted using RIPA buffer containing 1× protease inhibitor cocktail and 1.5% β-mercaptoethanol (β-ME). Total protein extracts (5 μL) were mixed with 5 μL of 12% SDS and 10 μL of 1× DNPH (10 μL 1× derivatization control solution) to each Eppendof tube and were incubated at RT for 15 minutes. Neutralization solution (7.5 μL) was then added to the mixture. The proteins were separated by 10% SDS-PAGE and transferred to a PVDF membrane. Membranes were blocked with 5% nonfat dry milk and incubated for 1 hour with primary antibody. After washing, the membrane was incubated in secondary antibody for an additional 1 hour at RT. Immunoreactive proteins were visualized using chemiluminescence substrate. β-Tubulin antibody staining was used as an endogenous control.
Detection of ER Stress Markers by Real-Time Q-PCR
HTM1073/p10 and HTM682/p8 were transfected with 204M or ConM. Two days after transfection, the cells were incubated with 1 μg/mL tunicamycin (Sigma) or vehicle control in serum-free DMEM overnight. Total RNAs were then isolated, first-strand cDNA was synthesized, and Q-PCR was performed following the procedure mentioned. These specific primer pairs were used for Q-PCR: GRP94—forward, 5′-GTT TCT ATT CCG CCT TCC TTG; reverse, 5′-GAG TGT TTC CTC TTG GGT CAG; GRP78/BIP—forward, 5′-TGA TTC CAA GGA ACA CAG T; reverse, 5′-GTC AGA TCA AAT GTA CCC A; CHOP/DDIT3—forward, 5′-TTC TCT GGC TTG GCT GAC T; reverse, 5′-CTG GTT CTC CCT TGG TCT TC.
Statistical Analysis
The data were presented as mean ± SD. Statistical significance between groups was assessed by the Mann-Whitney U test. P < 0.05 was considered statistically significant.
Results
Changes in Gene Expression Induced by miR-204 in HTM Cells
To identify the genes regulated by miR-204, HTM cells were transfected with either 204M or ConM. Three days after transfection, RNAs were extracted and hybridized to gene arrays (Human Genome U133 2.0; Affymetrix). Forty-eight probe sets corresponding to 34 genes showed a significant downregulation higher than 1.5-fold (Table 3). Among these 34 genes, 28 showed consistently significant downregulation in two additional HTM cell lines by real-time Q-PCR. BcL2 was not affected by miR-204 in either of the cell lines (Table 4). As shown in Table 3, 23 downregulated genes were predicted targets of miR-204 in at least one of the following databases: Microcosm, TargetScan, and PicTar-Vert. Only one of these genes, TGFβR2, has been previously experimentally validated as an miR-204 target.7
Table 3.
Microarray Data in HTM Cells after 204M Transfection
| Probe Set ID | Gene Symbol | Fold Change ([204M] vs. [ConM]) | P | Unigene | Microcosm | TargetScan | PicTar-Vert |
|---|---|---|---|---|---|---|---|
| 201675_at | AKAP1 | −1.71555 | 0.017633 | Hs.463506 | T | T | T |
| 205195_at | AP1S1 | −1.575583 | 3.37E-05 | Hs.489365 | T | T | T |
| 205196_s_at | AP1S1 | −1.546286 | 0.001292 | Hs.489365 | T | T | T |
| 203299_s_at | AP1S2 | −3.228704 | 0.002527 | Hs.656471 | T | ||
| 203300_x_at | AP1S2 | −4.522286 | 0.0013 | Hs.656471 | T | ||
| 203910_at | ARHGAP29 | −1.75398 | 0.005955 | Hs.483238 | T | ||
| 212930_at | ATP2B1 | −2.281018 | 0.010067 | Hs.506276 | T | T | |
| 209311_at | Bcl2l2 | −2.116855 | 0.005594 | Hs.410026 | T | ||
| 202076_at | BIRC2 | −1.944986 | 2.29E-04 | Hs.696238 | T | ||
| 203440_at | CDH2 | −1.566819 | 0.006415 | Hs.464829 | T | T | |
| 52255_s_at | COL5A3 | −1.961802 | 0.009481 | Hs.235368 | T | T | |
| 207850_at | CXCL3 | −1.565648 | 0.004451 | Hs.89690 | |||
| 203279_at | EDEM1 | −2.205311 | 0.010543 | Hs.224616 | T | ||
| 208623_s_at | EZR | −1.613046 | 0.00998 | Hs.487027 | T | ||
| 201910_at | FARP1 | −1.533894 | 0.008742 | Hs.403917 | T | T | |
| 204451_at | FZD1 | −1.573639 | 0.008968 | Hs.94234 | T | ||
| 206432_at | HAS2 | −2.74259 | 0.03648 | Hs.159226 | |||
| 206924_at | IL11 | −2.432023 | 0.048955 | Hs.467304 | |||
| 205067_at | IL1B | −1.922176 | 0.015556 | Hs.126256 | T | ||
| 39402_at | IL1B | −1.871506 | 0.034115 | Hs.126256 | T | ||
| 210233_at | IL1RAP | −2.434636 | 0.009944 | Hs.478673 | |||
| 211506_s_at | IL8 | −1.825963 | 0.014736 | Hs.551925 | |||
| 204627_s_at | ITGB3 | −2.290012 | 0.003371 | Hs.218040 | |||
| 203297_s_at | JARID2 | −1.544999 | 0.004616 | Hs.630189 | T | T | |
| 203298_s_at | JARID2 | −2.019171 | 0.019289 | Hs.630189 | T | T | |
| 200900_s_at | M6PR | −2.616955 | 0.001963 | Hs.134084 | T | T | |
| 200901_s_at | M6PR | −2.73735 | 0.009838 | Hs.134084 | T | T | |
| 202501_at | MAPRE2 | −2.030328 | 0.006163 | Hs.532824 | T | T | |
| 210155_at | MYOC | −1.566554 | 0.026922 | Hs.436037 | |||
| 201860_s_at | PLAT | −3.632373 | 9.96E−04 | Hs.491582 | |||
| 210845_s_at | PLAUR | −1.995835 | 0.01926 | Hs.466871 | |||
| 211924_s_at | PLAUR | −1.951011 | 0.005251 | Hs.466871 | |||
| 213405_at | RAB22A | −4.138018 | 0.00481 | Hs.529044 | T | T | |
| 218360_at | RAB22A | −3.184726 | 0.004328 | Hs.529044 | T | T | |
| 204547_at | RAB40B | −1.917881 | 2.04E-04 | Hs.484068 | T | T | |
| 211769_x_at | SERINC3 | −2.8083 | 3.27E-04 | Hs.272168 | |||
| 221471_at | SERINC3 | −3.048062 | 0.00109 | Hs.272168 | |||
| 221472_at | SERINC3 | −2.798143 | 0.00244 | Hs.272168 | |||
| 221473_x_at | SERINC3 | −2.793758 | 0.002413 | Hs.272168 | |||
| 200969_at | SERP1 | −3.935548 | 0.003682 | Hs.518326 | T | ||
| 200970_s_at | SERP1 | −4.400927 | 0.003808 | Hs.518326 | T | ||
| 202627_s_at | SERPINE1 | −1.781277 | 1.31E-05 | Hs.414795 | |||
| 202628_s_at | SERPINE1 | −1.708365 | 0.002325 | Hs.414795 | |||
| 208986_at | TCF12 | −1.747276 | 0.00986 | Hs.511504 | T | T | |
| 212386_at | TCF4 | −1.51174 | 0.001397 | Hs.644653 | T | ||
| 212387_at | TCF4 | −1.58985 | 6.56E-04 | Hs.644653 | T | ||
| 213891_s_at | TCF4 | −1.647133 | 0.002283 | Hs.644653 | T | ||
| 208944_at | TGFBR2 | −2.066357 | 0.00198 | Hs.82028 | T |
Table 4.
Real-Time RT Q-PCR Results in Two HTM Cell Lines Transfected with 204M and Normalized by β-Actin
| Gene Name | HTM1073-07-26 (fold ± SD) | HTM681-09-27 (fold ± SD) |
|---|---|---|
| AKAP1 | −1.39 ± 0.23* | −1.42 ± 0.17 |
| AP1S1 | −2.08 ± 0.16 | −2.64 ± 0.27 |
| AP1S2 | −4.84 ± 0.15 | −4.42 ± 0.11 |
| ARHGAP | −1.32 ± 0.02 | −1.5 ± 0.03 |
| ATP2B1 | −1.29 ± 0.13* | −1.55 ± 0.11 |
| Bcl2 | 1.04 ± 0.53* | 1.97 ± 0.17* |
| Bcl2l2 | −2.87 ± 0.09 | −3.15 ± 0.11 |
| BIRC2 | −2.61 ± 0.05 | −3 ± 0.11 |
| CDH2 | −1.16 ± 0.13* | −1.73 ± 0.06 |
| COL5A3 | −4.86 ± 0.2 | −4.1 ± 0.19 |
| CXCL3 | −2.28 ± 0.11 | −3.13 ± 0.17 |
| EDEM1 | −2.62 ± 0.03 | −2.98 ± 0.07 |
| EZR | −1.53 ± 0.09 | −1.57 ± 0.14 |
| FARP1 | −1.01 ± 0.32* | −1.28 ± 0.14* |
| FZD1 | −2.57 ± 0.07 | −1.89 ± 0.17 |
| HAS2 | −2.67 ± 0.22 | −4.43 ± 0.22 |
| IL11 | −3.59 ± 0.09 | −2.18 ± 0.17 |
| IL-1β | −1.917 ± 0.0.16 | −2.06 ± 0.21 |
| IL1RAP | 1.09 ± 0.18* | −1.18 ± 0.17* |
| IL8 | −3.12 ± 0.22 | −4.19 ± 0.06 |
| ITGB3 | −2.54 ± 0.13 | −2.47 ± 0.2 |
| JARID2 | −1.73 ± 0.11 | −1.78 ± 0.31 |
| MAPRE2 | −1.81 ± 0.3 | −1.54 ± 0.25 |
| M6PR | −3.2 ± 0.07 | −2.26 ± 0.23 |
| MYOC | −3.05 ± 0.04 | −1.4 ± 0.04 |
| PLAT | −2.89 ± 0.38 | −4.95 ± 0.26 |
| PLAUR | −2.07 ± 0.12 | −2.93 ± 0.19 |
| RAB22A | −6.55 ± 0.17 | −6.42 ± 0.23 |
| RAB40B | −2.08 ± 0.07 | −2.5 ± 0.13 |
| SERP1 | −2.19 ± 0.29 | −1.7 ± 0.09 |
| SERINC3 | −3.28 ± 0.06 | −3.11 ± 0.24 |
| SERPINE1 (PAI-1) | −1.09 ± 0.09* | −2.01 ± 0.2 |
| TCF12 | −1.99 ± 0.17 | −1.88 ± 0.14 |
| TCF4 | −1.52 ± 0.1 | −1.27 ± 0.09 |
| TGFβR2 | −3.02 ± 0.2 | −3.91 ± 0.13 |
P > 0.05 compared with controls by Mann-Whitney U test.
Validation of Novel miR-204 Targets
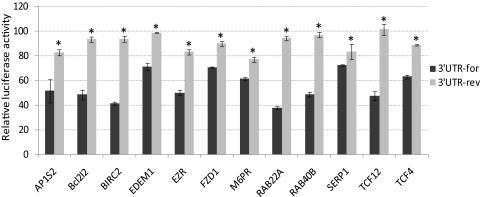
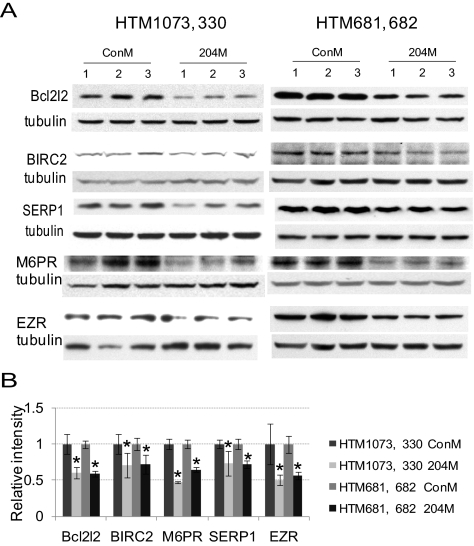
Twelve genes (AP1S2, Bcl2l2, BIRC2, EDEM1, EZR, FZD1, M6PR, RAB22A, RAB40B, SERP1, TCF12, and TCF4) with predicted target sequences for miR-204 that showed consistent and significant downregulation measured by gene array and real-time Q-PCR analysis in HTM cells were selected to verify the interaction of their predicted target sites with miR-204. Partial 3′UTRs containing the predicted target sites for miR-204 of these genes were cloned into a psiCheck2 dual-luciferase reporter vector using the specific primer pairs listed in Table 2. The vectors containing the 3′UTRs of these genes were cotransfected with 204M to HEK 293 cells. The results showed significantly lower expression of Renilla compared with the cells transfected with the same reporter vectors and ConM for all the selected genes. The effects of miR-204 on Renilla expression were eliminated when the 3′UTRs of these genes were cloned in reverse orientation (Fig. 1). The ability of miR-204 to downregulate the expression of the proteins encoded by five of these genes (Bcl2l2, BIRC2, SERP1, M6PR, and EZR) in HTM cells was confirmed by Western blot analysis (Fig. 2).
Figure 1.
Dual luciferase activity determined miR-204 potential targets. The 3′UTRs for the genes in the figure were amplified using primers listed in Table 2 and cloned into psiCheck2 dual-luciferase reporter vector and cotransfected with either 204M or ConM into HEK 293 cells. Negative control vectors (3′UTR-rev) were generated by cloning the same 3′UTRs of the genes in reverse orientation. The luciferase activities of Renilla/firefly were analyzed 24 hours after transfection. The data represent the percentage of changes in the ratio of Renilla/firefly activities compared with the controls ± SD (n = 3; *P < 0.05 compared with the 3′UTR by Mann-Whitney U test).
Figure 2.
Decreased protein expression of some targeted genes by miR-204. The expression levels of Bcl2l2 (HTM1073 and 681), BIRC2 (HTM330, 681), SERP1 (HTM1073, 681), M6PR (HTM330, 681), and Ezrin (EZR) (HTM1073, 682) were evaluated 3 days after transfection by Western blot analysis in two HTM cell lines transfected with either 204M or ConM. (A) Western blot analysis of the proteins' expression from three independent transfection experiments. (B) Densitometry analysis of the Western blot data normalized by β-tubulin (n = 3; *P < 0.05 compared with ConM-transfected cells by Mann-Whitney U test).
Overexpression of miR-204 Increased Susceptibility to Cell Death and Apoptosis in Response to Oxidative Stress
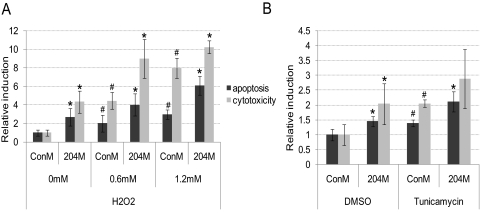
To evaluate whether the observed changes in gene expression induced by miR-204 could result in increased susceptibility to apoptosis, HTM cells were transfected with 204M or ConM and treated with either H2O2 for 4 hours or tunicamycin 1 μg/mL overnight. As shown in Figure 3A, miR-204 significantly increased both apoptosis and cytotoxicity in HTM cells in the presence and absence of H2O2 treatment. MiR-204 induced a similar increase in apoptosis in HTM cells treated with tunicamycin. However, the increase in cytotoxicity observed after tunicamycin treatment was not statistically significant (Fig. 3B).
Figure 3.
204M transfection increased HTM cell apoptosis and cell death. HTM681/p4 cells were transfected with ConM or 204M (120 pmol/5 × 105 cells) and plated in a 96-well plate (7 × 103/well). Forty-eight hours after transfection, cells were treated with H2O2 0, 0.6, or 1.2 mM in serum-free medium for 4 hours, tunicamycin 1 μg/mL, or DMSO overnight. Cells were then loaded with iodide in PBS and incubated for 30 minutes at 37°C, 5% CO2. Fluorescence was measured using an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Wells containing same amount of serum-free medium and PBS were used as blank control wells. Cytotoxicity was measured by LDH release into cell culture supernatant. (A) Apoptosis and cytotoxicity in 204M- or ConM-transfected HTM cells treated with H2O2. (B) Apoptosis and cytotoxicity in 204M- or ConM-transfected HTM cells treated with tunicamycin. The data represent fold changes of ConM-transfected cells without H2O2 or with DMSO treatment ± SD (n = 4–6; *P < 0.05 compared with their corresponding ConM-transfected controls; #P < 0.05 compared with ConM-transfected cells without H2O2 or tunicamycin treatment by Mann-Whitney U test).
MiR-204 Increased the Accumulation of Carbonylated Proteins after Chronic Oxidative Stress
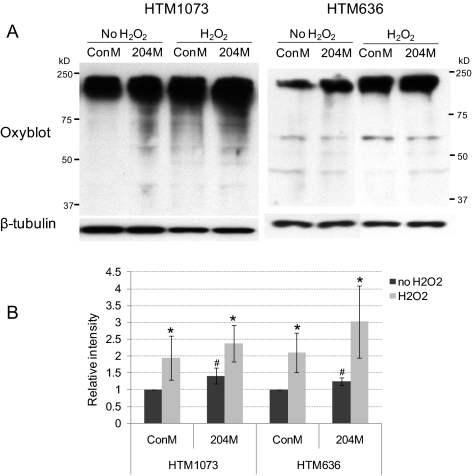
To test whether miR-204 could affect the accumulation of carbonylated proteins that result from chronic exposure to oxidative stress, HTM cells were transfected with 204M or ConM and treated with 250 μM H2O2 daily for 3 days. Transfection with miR-204 mimic resulted in a statistically significant increase in the accumulation of carbonylated proteins in cells not treated with H2O2. MiR-204 also led to an increase in the presence of oxidized protein induced by chronic H2O2 treatment, but this increase was not statistically significant with n = 3 (Fig. 4).
Figure 4.
204M increased protein oxidation in HTM cells. HTM636/p2 and HTM1073/p3 cells were transfected with ConM or 204M (120 pmol/5 × 105 cells). Twenty-four hours after transfection, the cell culture medium was replaced with fresh DMEM containing 10% FBS, and cells were treated with H2O2 250 μM once a day for 3 days. Total proteins were then collected in RIPA buffer containing 1.5% β-ME. Five microliters of cell lysates were used for protein oxidation. Five microliters of cell lysates were used for β-tubulin staining. (A) Western blot analysis of oxidized proteins. (B) Densitometry analysis of the Western blot data normalized by β-tubulin. The data represent fold changes of ConM-transfected cells without H2O2 treatment ± SD (n = 3; *P < 0.05 compared with their corresponding non-H2O2 controls; #P < 0.05 compared with ConM-transfected controls by Mann-Whitney U test).
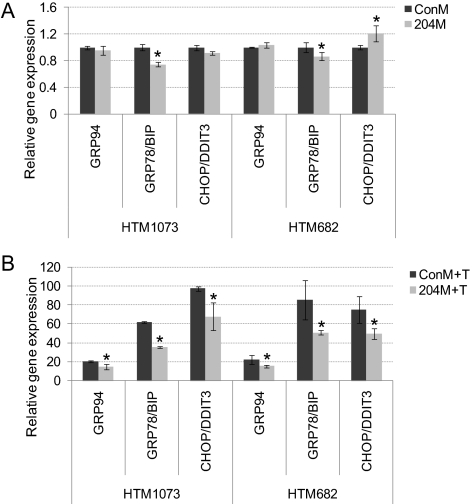
MiR-204 Inhibited the Induction of ER Stress Markers by Tunicamycin
To evaluate whether miR-204 could affect the cellular responses induced by the presence of unfolded proteins, ER stress markers (GRP94, GRP78/BIP, and CHOP/DDIT) were measured by real-time Q-PCR after tunicamycin treatment in two HTM cell lines. As shown in Figure 5A, miR-204 significantly decreased the expression of GRP78/BIP in both HTM cell lines and increased CHOP/DDIT3 in one HTM cell line even without tunicamycin treatment. After tunicamycin treatment, miR-204 significantly inhibited the induction of all three ER stress markers (GRP94, GRP78/BIP, and CHOP/DDIT) in both cell lines tested (Fig. 5B).
Figure 5.
204M decreased ER stress markers induced by tunicamycin. Two HTM cell lines (HTM1073/p5 or HTM682/p4) were transfected with ConM or 204M. Forty-eight hours after transfection, tunicamycin 1 μg/mL or the same concentration of DMSO in serum-free DMEM was added to the cells and incubated overnight. Total RNA was isolated, and real-time Q-PCR was performed using real-time PCR mix with specific primers, as described in Methods. Results were normalized with β-actin, and gene expression levels were expressed as the fold changes compared with those in ConM transfected without tunicamycin-treated cells. (A) ConM- or 204M-transfected cells treated with DMSO. (B) ConM- or 204M-transfected cells treated with tunicamycin. Data represent the mean of fold changes ± SD (n = 3–4; *P < 0.05 compared with ConM-transfected cells by Mann-Whitney U test.
Attenuation of Inflammatory Factors by miR-204
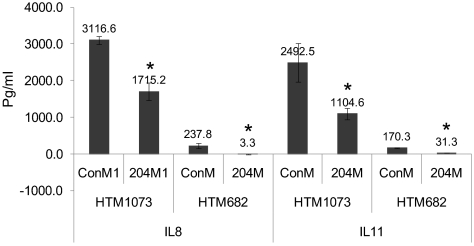
Affymetrix gene array data showed >1.5-fold significantly decreased expression of several inflammatory factors (IL-11, IL-1β, IL-1RAP, IL-8, SERPINE1, and CXCL3) by miR-204 (Table 3). Although the basal levels of expression of these inflammatory markers were different in the two cell lines analyzed, the inhibitory effects of miR-204 were highly consistent for both cell lines. Furthermore, the downregulation of IL-11, IL-1β, IL-8, and CXCL3 by miR-204 was confirmed by real-time Q-PCR in two additional HTM cell lines (Table 4). In addition, protein levels of IL-8 and IL-11 in the cell culture were also clearly reduced by miR-204 in two HTM cell lines (Fig. 6).
Figure 6.
204M reduced the production of IL-8 and IL-11. HTM1073/p4 or HTM682/p3 was transfected with ConM or 204M (120 pmol/5 × 105 cells). Twenty-four hours after transfection, the cell culture medium was replaced with fresh DMEM containing 10% FBS. Three days after transfection, the cell culture supernatant was collected, and the levels of IL-8 and IL-11 were measured using ELISA kits in accordance with the manufacturer's instructions. Data represent picogram per milliliter ± SD in three transfected samples. *P < 0.05 compared with ConM-transfected cells by Mann-Whitney U test.
Discussion
Our results showed that miR-204 induced extensive changes in gene expression in HTM cells, including the downregulation of the validated target TGFβR2.7 Array data showed significant downregulation of a large number of genes with target sequences for miR-204 predicted by at least 1 of the 3 databases for miRNAs: Microcosm, TargetScan, and PicTar-Vert. In most cases, such downregulation was confirmed by Q-PCR. The potential targets downregulated by miR-204 in HTM cells are known to be involved in a variety of biological processes, suggesting that this miRNA may play a role in the regulation of multiple cellular functions. Consistent with this concept, miR-204 overexpression resulted in significant alteration in HTM cells, including increased apoptosis and cell death in response to oxidative stress and ER stress induced by tunicamycin, increased accumulation of oxidized proteins after oxidative stress, decreased induction of ER stress response markers in response to tunicamycin treatment, and decreased expression of inflammatory mediators such as IL-8 and IL-11.
Specifically, our results confirmed direct targeting of miR-204 to the 3′UTRs of AP1S2, Bcl2l2, BIRC2, EDEM1, EZR, FZD1, M6PR, RAB22A, RAB40B, SERP1, TCF12, and TCF4 and demonstrated a significant decrease in the expression of Bcl2l2/Bcl-w, cIAP1/BIRC2, SERP1/RAMP4, M6PR, and EZR proteins induced by miR-204. Two of these genes, Bcl2l2/Bcl-w and cIAP1/BIRC2, are known to inhibit apoptosis. Bcl-w is a member of the Bcl-2 family, which inhibits apoptosis by interacting with proapoptotic members of the Bcl-2 family, such as Bad, Bax, and Bik, blocking the formation of the homodimers and, thus, the activation of the apoptotic cascade.15,16 cIAP1/BIRC2 belongs to the family of antiapoptotic regulators known as inhibitors of apoptosis (IAP) proteins. Expression of cIAP1/BIRC2 is induced in conditions of ER stress through the phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway and contributes to cellular adaptation to stress by inhibiting the ER stress–induced apoptotic program that is activated after prolonged ER stress.17–21 Cell cycle-dependent expression of cIAP2 at G2/M phase has also been shown to contribute to cell survival during mitotic arrest.22 Targeting of these antiapoptotic genes, together with the increased apoptosis and cell death induced by miR-204 in HTM cells after both oxidative stress and ER stress induced by tunicamycin, supports the concept that miR-204 might contribute to the regulation of cell survival under stress conditions. Targeting of Bcl2l2/Bcl-w and cIAP1/BIRC2 might also contribute to the proposed tumor suppressor effects of this miRNA23 because the overexpression of these two proteins observed in a diversity of cancer cells is believed to contribute to cell survival and cancer progression.24–27
In addition to cIAP1/BIRC2, our results showed direct targeting of two genes, SERP1/RAMP4 and M6PR, which are also relevant in the cellular response to the accumulation of damaged proteins and ER stress. SERP1/RAMP4 is a component of the ER translocation sites that have been shown to stabilize and suppress the aggregation of membrane proteins during ER stress and to facilitate subsequent glycosylation when the stress is removed.28 Animals lacking RAMP4 showed induction of the unfolded protein response in tissues with high secretory activity,29 suggesting that, at high levels of secretion, RAMP4 becomes critical for efficient folding of newly synthesized proteins. M6PR plays a key role in the transport of most of the soluble acid hydrolases from the Golgi complex to the endosomal/lysosomal system,30–32 which is known to contribute to the elimination of misfolded proteins and to prevent the accumulation of intracellular protein aggregates.33,34 Consistent with the inhibition of proteins involved in preventing the accumulation of misfolded proteins, miR-204 led to an increase in the presence of carbonylated proteins. However, cells transfected with miR-204 mimic also showed decreased induction of ER stress markers when challenged with tunicamycin. This observation suggests that miR-204 might have a general inhibitor effect on the induction of the UPR in conditions of ER stress that is likely to involve additional target genes yet to be identified.
Together with the observed effects on apoptosis and the accumulation of damaged proteins, miR-204 showed a significant inhibitory effect on the expression of several inflammatory mediators. Consistent with previous observations,6 the basal levels of expression of inflammatory markers were different in the two cell lines analyzed. These differences may reflect a strong level of individual variation in the human population or may result from differences in the presence of senescent cells in the culture given that chronic induction of inflammatory markers has been reported to be associated with the acquisition of a senescent phenotype in HTM cells.35 Although it is unclear what specific gene targets might mediate this effect, the observed inhibition of the UPR could potentially contribute to such a decrease in expression of inflammatory mediators since the activation of the UPR has been identified as one of the pathways leading to the inflammatory response.36,37 Specifically, targeting of cIAP1/BIRC2 could contribute to the downregulation of inflammatory mediators because this protein has been shown to play a role in the initiation of innate immunity signaling.38
In conclusion, the changes in gene expression induced by miR-204 in HTM cells suggest the involvement of this miRNA in the regulation of multiple cellular functions. Specifically, miR-204 appears to play an important role in the regulation of responses to ER stress, apoptosis, and production of inflammatory mediators. Identification of additional target genes will be necessary to fully understand the biological functions of miR-204.
Footnotes
Supported by National Eye Institute Grants EY01894, EY016228, and EY05722 and by Research to Prevent Blindness.
Disclosure: G. Li, None; C. Luna, None; J. Qiu, None; D.L. Epstein, None; P. Gonzalez, None
References
- 1. Aikata H, Takaishi H, Kawakami Y, et al. Telomere reduction in human liver tissues with age and chronic inflammation. Exp Cell Res. 2000;256:578–582 [DOI] [PubMed] [Google Scholar]
- 2. Kitada T, Seki S, Kawakita N, Kuroki T, Monna T. Telomere shortening in chronic liver diseases. Biochem Biophys Res Commun. 1995;211:33–39 [DOI] [PubMed] [Google Scholar]
- 3. Manestar-Blazic T, Volf M. The dynamic of senescent cells accumulation can explain the age-specific incidence of autoimmune diseases. Med Hypotheses. 2009;73:667–669 [DOI] [PubMed] [Google Scholar]
- 4. Liton PB, Challa P, Stinnett S, Luna C, Epstein DL, Gonzalez P. Cellular senescence in the glaucomatous outflow pathway. Exp Gerontol. 2005;40:745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130:731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Modulation of inflammatory markers by miR-146a during replicative senescence in trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2010;51:2976–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang FE, Zhang C, Maminishkis A, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55 [DOI] [PubMed] [Google Scholar]
- 11. Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen L, Yan HX, Yang W, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50:358–369 [DOI] [PubMed] [Google Scholar]
- 13. Jukic DM, Rao UN, Kelly L, et al. MicroRNA profiling analysis of differences between the melanoma of young adults and older adults. J Transl Med. 2010;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J Biol Chem. 2010;285:5461–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garofalo M, Quintavalle C, Zanca C, et al. Akt regulates drug-induced cell death through Bcl-w downregulation. PLoS ONE. 2008;3:e4070. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Gibson L, Holmgreen SP, Huang DC, et al. bcl-w, a novel member of the bcl-2 family, promotes cell survival. Oncogene. 1996;13:665–675 [PubMed] [Google Scholar]
- 17. Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcik M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem. 2004;279:17148–17157 [DOI] [PubMed] [Google Scholar]
- 18. Graber TE, Baird SD, Kao PN, Mathews MB, Holcik M. NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ. 2009;17:719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamanaka RB, Bobrovnikova-Marjon E, Ji X, Liebhaber SA, Diehl JA. PERK-dependent regulation of IAP translation during ER stress. Oncogene. 2009;28:910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varfolomeev E, Vucic D. (Un)expected roles of c-IAPs in apoptotic and NFκB signaling pathways. Cell Cycle. 2008;7:1511–1521 [DOI] [PubMed] [Google Scholar]
- 21. Jin HS, Lee DH, Kim DH, Chung JH, Lee SJ, Lee TH. cIAP1, cIAP2, and XIAP act cooperatively via nonredundant pathways to regulate genotoxic stress-induced nuclear factor-κB activation. Cancer Res. 2009;69:1782–1791 [DOI] [PubMed] [Google Scholar]
- 22. Effect of screening for hepatitis C virus antibody and hepatitis B virus core antibody on incidence of post-transfusion hepatitis: Japanese Red Cross Non-A, Non-B Hepatitis Research Group. Lancet. 1991;338:1040–1041 [PubMed] [Google Scholar]
- 23. Lee Y, Yang X, Huang Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568 [DOI] [PubMed] [Google Scholar]
- 25. Grzybowska-Izydorczyk O, Cebula B, Robak T, Smolewski P. Expression and prognostic significance of the inhibitor of apoptosis protein (IAP) family and its antagonists in chronic lymphocytic leukaemia. Eur J Cancer. 2010;46:800–810 [DOI] [PubMed] [Google Scholar]
- 26. Ma O, Cai WW, Zender L, et al. MMP13, Birc2 (cIAP1), and Birc3 (cIAP2), amplified on chromosome 9, collaborate with p53 deficiency in mouse osteosarcoma progression. Cancer Res. 2009;69:2559–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen N, Gong J, Chen X, et al. Caspases and inhibitor of apoptosis proteins in cutaneous and mucosal melanoma: expression profile and clinicopathologic significance. Hum Pathol. 2009;40:950–956 [DOI] [PubMed] [Google Scholar]
- 28. Yamaguchi A, Hori O, Stern DM, Hartmann E, Ogawa S, Tohyama M. Stress-associated endoplasmic reticulum protein 1 (SERP1)/ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J Cell Biol. 1999;147:1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hori O, Miyazaki M, Tamatani T, et al. Deletion of SERP1/RAMP4, a component of the endoplasmic reticulum (ER) translocation sites, leads to ER stress. Mol Cell Biol. 2006;26:4257–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olson LJ, Sun G, Bohnsack RN, Peterson FC, Dahms NM, Kim JJ. Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor. Biochemistry. 2010;49:236–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hille-Rehfeld A. Mannose 6-phosphate receptors in sorting and transport of lysosomal enzymes. Biochim Biophys Acta. 1995;1241:177–194 [DOI] [PubMed] [Google Scholar]
- 32. Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009;1793:605–614 [DOI] [PubMed] [Google Scholar]
- 33. Knaevelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584:2635–2645 [DOI] [PubMed] [Google Scholar]
- 34. Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells. 2010;15:923–933 [DOI] [PubMed] [Google Scholar]
- 35. Li G, Luna C, Liton PB, Navarro I, Epstein DL, Gonzalez P. Sustained stress response after oxidative stress in trabecular meshwork cells. Mol Vis. 2007;13:2282–2288 [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang K. Integration of ER stress, oxidative stress and the inflammatory response in health and disease. Int J Clin Exp Med. 2010;3:33–40 [PMC free article] [PubMed] [Google Scholar]
- 37. Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801 [DOI] [PubMed] [Google Scholar]