This report provides the first detailed analysis of the role of Vps11 in retinal pathogenesis in a vertebrate model of syndromic albinism.
Abstract
Purpose.
To establish the zebrafish platinum mutant as a model for studying vision defects caused by syndromic albinism diseases such as Chediak-Higashi syndrome, Griscelli syndrome, and Hermansky-Pudlak syndrome (HPS).
Methods.
Bulked segregant analysis and candidate gene sequencing revealed that the zebrafish platinum mutation is a single-nucleotide insertion in the vps11 (vacuolar protein sorting 11) gene. Expression of vps11 was determined by RT-PCR and in situ hybridization. Mutants were analyzed for pigmentation defects and retinal disease by histology, immunohistochemistry, and transmission electron microscopy.
Results.
Phenocopy and rescue experiments determined that a loss of Vps11 results in the platinum phenotype. Expression of vps11 appeared ubiquitous during zebrafish development, with stronger expression in the developing retina and retinal pigmented epithelium (RPE). Zebrafish platinum mutants exhibited reduced pigmentation in the body and RPE; however, melanophore development, migration, and dispersion occurred normally. RPE, photoreceptors, and inner retinal neurons formed normally in zebrafish platinum mutants. However, a gradual loss of RPE, an absence of mature melanosomes, and the subsequent degradation of RPE/photoreceptor interdigitation was observed.
Conclusions.
These data show that Vps11 is not necessary for normal retinal development or initiation of melanin biosynthesis, but is essential for melanosome maturation and healthy maintenance of the RPE and photoreceptors.
Oculocutaneous albinism (OCA) is a group of genetically inherited conditions that result in pigmentation defects in the eyes, skin, and hair.1 The types of albinism have been divided into tryosinase-negative (no pigmentation) and those that produce various levels of melanin, termed tyrosinase-positive. In many cases, the data are contradictory in regard to how pigmentation affects photoreceptor viability in albinism. In certain model systems, albinism can exacerbate photoreceptor degeneration2 or cause fewer rod photoreceptors to develop.3 In contrast, other models of albinism exhibit no defects in either photoreceptor formation or viability.4–7
Clinically, the type of albinism is referenced based on the gene that causes the phenotype (if known) and classified as either “syndromic” or “nonsyndromic”.8 Cases of syndromic albinism are associated with other systemic diseases in addition to OCA. These include such syndromes as Chediak-Higashi syndrome (CHS), Griscelli syndrome (GS), Elejalde syndrome (ES), Cross-McKusick-Breen syndrome (CMBS), and Hermansky-Pudlak syndrome (HPS). In some cases, the genetic defects associated with these syndromes are known. For example, there are at least eight genes (HPS1–HPS8) that cause HPS.9 These syndromes all exhibit defects in protein sorting and trafficking in lysosomes and lysosome-related organelles, such as melanosomes.8
Recently, other genes known to play roles in protein trafficking have been linked to syndromic albinism. We describe the characterization of a zebrafish mutant, termed platinum, which contains a mutation in vps11, one of the HOPS/C-Vps class of genes that have been identified based on HPS-like phenotypes in mutant model organisms.10,11 Although the specific pathways that are defective in C-Vps-related HPS are not yet known in vertebrates, these genes have been highly studied in yeast. The C-Vps class encodes four proteins (Vps11, Vps16, Vps18, and Vps33) that physically interact to form a complex on the cytosolic side of the yeast vacuolar membrane.12 Originally, studies focused on the ability of this complex to bind to vacuolar t-SNARE proteins and facilitate tethering and membrane fusion of a transport vesicle from the late-Golgi to the yeast vacuole.12,13 For example, loss of Vps11 in yeast results in an absence of normal vacuoles and the accumulation of intermediate transport vesicles.12,14 However, more recent evidence indicates that Vps11 functions at multiple steps in vesicle-mediated transport, including Golgi-to-endosome anterograde and retrograde transport, and in the endocytic pathway.15 Because the yeast vacuole is analogous to the vertebrate lysosome and melanosome, Vps11 is hypothesized to function in endosome-to-lysosome and endosome-to-melanosome protein trafficking, resulting in both OCA and systemic defects related to lysosomal abnormalities.
We showed that, consistent with other cases of syndromic albinism, loss of Vps11 results in both OCA and systemic defects, such as pericardial edema, hepatomegaly, and premature death. Specifically, platinum mutant zebrafish exhibited reduced pigmentation in body melanophore and retinal pigmented epithelium (RPE) due to a disruption in melanosome maturation. Vps11 was not required for photoreceptors or the RPE to form normally; however, platinum zebrafish exhibited subsequent degeneration of the RPE and eventual photoreceptor loss.
Materials and Methods
Fish Maintenance
Wild-type (AB strain) and platinum strains of zebrafish (Danio rerio) were maintained at the University of Notre Dame and Wayne State University School of Medicine. The fish were fed a combination of brine shrimp and flake food three times daily and maintained under a daily light cycle of 14 hours light (250 lux):10 hours dark at 28.5°C.16 All protocols used in this study were approved by the animal use committee at both institutions and are in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Whole Mount Bright-Field Imaging
Wild-type and platinum embryos were anesthetized with 2-phenoxyethanol before microscopy. Images were taken with a digital camera (Spot; Diagnostic Instruments, Sterling Heights, MI) attached to a stereomicroscope (model MZFL III or M165 FC; Leica, Bannockburn, IL).
To induce melanosome aggregation or dispersion, embryos were placed in either an open-ended white or black box for 24 hours, respectively.17 At 5 days after fertilization (dpf), clutches from three separate platinum carrier in-crosses were separated into two groups: one exposed to a black background and one exposed to a white background. At 6 dpf, melanophores in the dorsal head region were analyzed and imaged as described above.
Positional Cloning of the platinum Mutation
Bulked segregant analysis18 localized the platinum mutation to chromosome 10, near Z6390. Linkage mapping, using a total of 378 platinum mutants, defined the interval containing the mutation, and a candidate gene approach was used to evaluate genes in very close proximity to Z25833. Two genes, dlg2 (accession no. NM_001012378) and zgc:112329 (vps11; accession no. NM_001037708) were identified as strong candidates. The open reading frames for both candidate genes were amplified and sequenced from platinum mutant and wild-type embryos, as described.19 vps11 was found to contain a 1-bp insertion of an A at nucleotide position 1894 followed directly by a G to A transition. PCR amplification followed by direct sequencing was used to confirm the presence of the mutation in genomic DNA.
Phenocopy and Rescue Experiments
To phenocopy the platinum phenotype, a morpholino (5′-GACAGCAGTCGTGAACCGTACCAGA-3′) complimentary to the splice site of the second exon and second intron of vps11 mRNA was obtained from Gene Tools (Philomath, OR). Microinjection of the morpholino solution into the first- to fourth-cell-stage wild-type embryos was performed as previously described.20,21 Injected and noninjected embryos were phenotyped at 24 and 48 hours post fertilization (hpf), at which point embryos from each group were pooled separately, and total RNA was harvested with extraction reagent (Trizol; Invitrogen). RNA was reverse-transcribed using Superscript II (Invitrogen) and an Oligo(dT) primer (Invitrogen), and either contained (see Fig. 4, lanes 2 and 4) or excluded (Fig. 4, lanes 3 and 5) reverse transcriptase. The following gene-specific primers were used for PCR: vps11 S1; 5′-CGAGGACACATTGTGCTCG-3′, vps11 AS3; 5′-GTGTCTTGTCTCAGCGTACATG-3′, β-actin S; 5′-TCTGGCATCACACCTTCTAC-3′, β-actin AS; 5′-ATACCGCAAGATTCCATACC-3′. After PCR, the products were separated by electrophoresis on a 0.8% agarose gel and directly sequenced to confirm that proper splice alteration of vps11 had occurred.
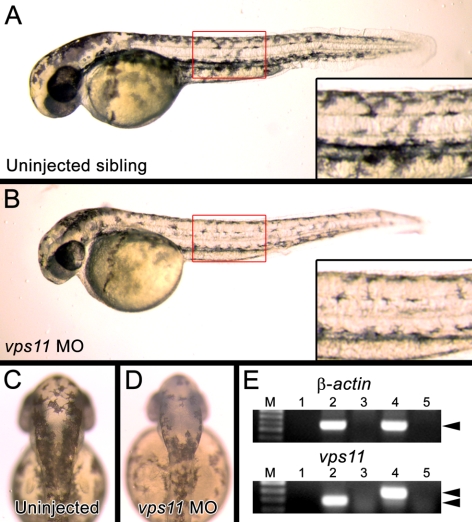
Figure 4.
(A) Normal pigmentation in an noninjected embryo at 48 hpf. Inset: darkly pigmented melanophores in the trunk of the embryo. (B) An 48 hpf embryo injected with the vps11 morpholino (vps11 MO). Inset: reduced pigmentation in the vps11 morphant melanophores. (C) The dorsal head region of an noninjected embryo at 48 hpf. (D) The dorsal head region of a vps11 morphant at 48 hpf containing melanophores with reduced pigmentation. (E) Alteration of vps11 mRNA splicing in vps11 morphants. Top: normal splicing of β-actin in both noninjected (lane 2) and vps11 morphants (lane 4). Bottom: inhibited splice-removal of the second intron of the vps11 transcript in vps11 morphants (lane 4, double arrowheads).
Rescue of the platinum phenotype was accomplished by microinjection of vps11 mRNA into first- to fourth-cell-stage embryos from a mutant carrier in-cross. A full-length vps11 cDNA was obtained from Open Biosystems (Thermo Scientific; Huntsville, AL) and subcloned into a vector (pBluescript; Stratagene, La Jolla, CA). The vps11 mRNA was obtained by in vitro transcription (mMessage Machine; Ambion; Austin, TX). Six known vps11 carrier pairs were in-crossed to obtain embryos for microinjection, which was performed in triplicate (n = 651). Quantification of the number of embryos with the platinum phenotype occurred at 48 hpf.
RT-PCR and Whole-Mount In Situ Hybridization
RNA was isolated (Trizol; Invitrogen) from wild-type embryos at multiple stages of development. Complementary DNAs were reverse transcribed using Oligo(dT) and reverse transcriptase (Superscript II; Invitrogen). The following primers were designed to amplify a 500- to 600-bp PCR product and spanned across an intron: β-actin (S: 5′-TCTGGCATCACACCTTCTAC-3′; AS: 5′-ATACCGCAAGATTCCATACC-3′); vps11 (S: 5′-CGAGGACACATTGTGCTCG-3′; AS: 5′-CCAATTCTACACGAGGATACTC-3′). PCR products were purified, cloned into the T-tailed vector pGEM-T Easy (Promega; Madison, WI), and sequenced to confirm that each product matched the expected cDNA.
Whole-mount in situ hybridization was performed exactly as described elsewhere.21 The following primers were designed to amplify a 600-bp product at the 3′ end of vps11 (S: 5′-AACGAGATCTGCACGACC-3′; AS: 5′-AGCTACAGTAATAAGTGGTTG-3′). PCR products were purified, cloned into pGEM-T Easy (Promega, Madison, WI), and sequenced to confirm 100% identity to the expected sequence. Digoxigenin-labeled sense and antisense probes were transcribed using SP6 and T7 polymerases (Roche; Indianapolis, IN) and hybridized with embryos as described,21 and color reaction was achieved in a chromogenic substrate (BM Purple; Roche). Embryos were post-fixed in 4% paraformaldehyde and either immediately imaged or processed for cryosectioning.
Histology and Transmission Electron Microscopy
Wild-type and platinum embryo siblings were collected at 2, 3, 5, and 7 dpf. They were then fixed, dehydrated, infiltrated, sectioned, and stained as described.22–26 For transmission electron microscopy, tissues were sectioned, collected onto grids, dried for 1 hour, and stained with 4.9% uranyl acetate (5 minutes) and 1% lead citrate (1 minute) before viewing.26
Immunohistochemistry
Immunohistochemistry was performed as described23 on frozen sections from wild-type and mutant siblings collected at 5 and 7 dpf. Primary antibodies included: mouse anti-PCNA monoclonal antibody (1:1000, clone PC10; Sigma-Aldrich, St. Louis, MO), rabbit anti-rhodopsin polyclonal antiserum27 (1:5000), rabbit anti-Pax6a polyclonal antiserum (1:200; AnaSpec, Fremont, CA), mouse anti-glutamine synthetase monoclonal antibody (1:500; Chemicon International, Temecula, CA) mouse anti-zpr-3 (1:200; University of Oregon Monoclonal Antibody Facility), mouse anti-zpr-1 (1:200; University of Oregon Monoclonal Antibody Facility), and goat anti-Vps11 polyclonal antiserum (1:500; Abcam, Cambridge, MA). AlexaFluor goat anti-primary IgG 488 or chicken anti-primary IgG 488 was used as a secondary antibody (Invitrogen-Molecular Probes, Eugene, OR). Nuclei were labeled (TO-PRO-3; Invitrogen-Molecular Probes) at a 1:750 dilution in 1× PBS/0.05% Tween-20. Confocal microscopy was performed with one of two microscopes (model 1024; BioRad, Hercules, CA, or model TCS SP5; Leica).
Results
The Zebrafish platinum Mutant Exhibits Reduced Pigmentation, Pericardial Edema, and Hepatomegaly
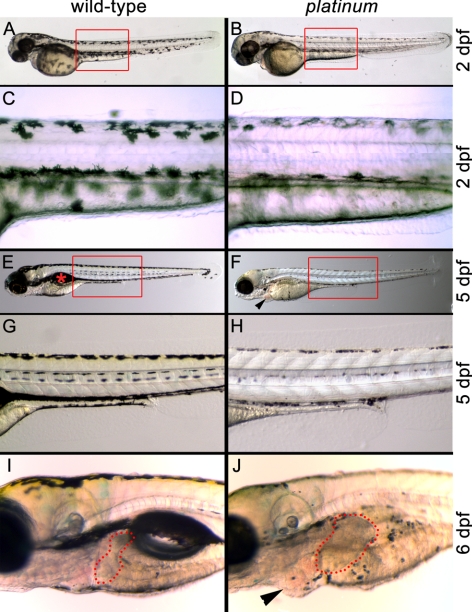
At 2 dpf, the platinum mutants exhibited reduced pigmentation in the eye and body (Figs. 1B, 1D). Quantification of the number of melanophores in the dorsal head of the wild-type and platinum embryos at 48 hpf revealed no significant difference in the number of melanophores in the platinum mutant relative to the wild-type sibling (43.3 ± 1.6 and 47.8 ± 1.9, respectively; n = 10; P = 0.15). However, pigment density in the melanosomes was clearly reduced in the platinum mutants, which resulted in an overall lighter appearance of the platinum mutants compared with the wild-type siblings at both 2 and 5 dpf (Fig. 1). In addition, the platinum mutants failed to form a swim bladder (Fig. 1F) and exhibited pericardial edema by 5 dpf (Fig. 1F) and significant liver hyperplasia by 6 dpf (Fig. 1J; 2.7-fold increase in liver area; n = 8; P < 0.001).
Figure 1.
Wild-type (left) and platinum siblings (right) at 2, 5, and 6 dpf. (A) A wild-type zebrafish embryo at 2 dpf with characteristic pigmentation. Red box: the region in (C). (B) A platinum mutant embryo at 2 dpf with reduced pigmentation. Red box: region shown in (D). (C) Darkly pigmented melanophores in the wild-type embryo at 2 dpf. (D) Reduced pigmentation within each melanophore in platinum mutants at 2 dpf. (E) A wild-type zebrafish larva at 5 dpf with characteristic pigmentation and swim bladder ( ). Red box: the region in (G). (F) A platinum mutant larva at 5 dpf showing reduced pigmentation and pericardial edema (arrowhead). Red box: the region in (H). (G). Normal pigmentation pattern of a wild-type larva at 5 dpf. (H) Small melanophores with reduced pigmentation in platinum mutants at 5 dpf. (I) Location and size of the wild-type liver (outlined in red) at 6 dpf. (J) A platinum mutant at 6 dpf showing reduced pigmentation, hepatomegaly (outlined in red), and pericardial edema (arrowhead).
). Red box: the region in (G). (F) A platinum mutant larva at 5 dpf showing reduced pigmentation and pericardial edema (arrowhead). Red box: the region in (H). (G). Normal pigmentation pattern of a wild-type larva at 5 dpf. (H) Small melanophores with reduced pigmentation in platinum mutants at 5 dpf. (I) Location and size of the wild-type liver (outlined in red) at 6 dpf. (J) A platinum mutant at 6 dpf showing reduced pigmentation, hepatomegaly (outlined in red), and pericardial edema (arrowhead).
The Zebrafish platinum Mutant Exhibits Normal Melanophore Formation, Migration, and Aggregation/Dispersion
Many zebrafish mutants described with pigmentation defects exhibit problems in melanophore formation and migration.28–32 At 24 and 28 hpf, when melanophores are first visualized in the anterior region of the developing organism,33 the platinum mutants could not be distinguished from the wild-type siblings (Fig. 2A) and 100% of the embryos (n = 68) contained melanophores migrating down the body axis (data not shown). At 2 dpf in both the wild-type siblings and platinum mutants, melanophores had migrated posteriorly to form the characteristic dorsal and ventral stripes (Figs. 1C, 1D, 2C, 2D) and left a gap in the ventral stripe for the developing caudal fin to appear (Figs. 2C, 2D).34
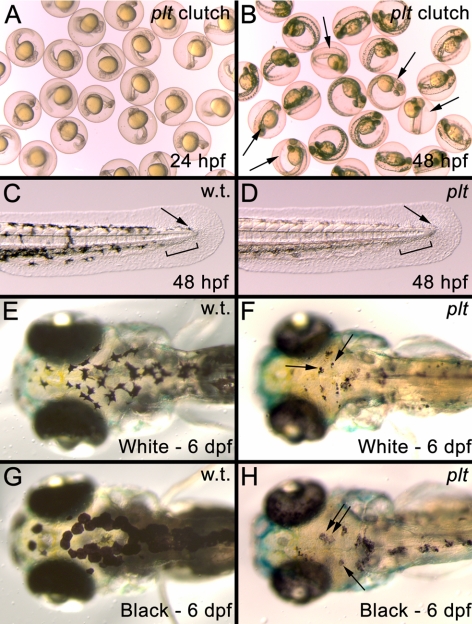
Figure 2.
(A) Image of embryos from a cross of two platinum carriers at 24 hpf. (B) The same clutch shown in A at 48 hpf. The platinum mutants (arrows) can now be distinguished from their wild-type siblings. (C) The developing tail bud of a wild-type embryo at 48 hpf showing that melanophores have migrated to the posterior of the animal (arrow) and left a gap in the ventral stripe for the developing caudal fin to appear (black bar). (D) The developing tail bud of a platinum embryo at 48 hpf showing normal melanophores migration to the tip of the tail bud (arrow) and the gap left in the ventral stripe for the developing caudal fin (black bar). (E, F) Melanophore aggregation in a wild-type larva (E) and platinum mutant (F, arrows), after 24 hours in a white environment. (G, H). Melanophore dispersion in a wild-type larva (G) and platinum mutant (H, arrows), after 24 hours in a black environment.
Zebrafish respond to visual stimuli, such as background color and light levels, by changing their color pattern to adapt to the background.17 Melanophore aggregation makes the fish appear lighter, while dispersion causes the fish to appear darker. To test whether platinum melanophores had the ability to aggregate and disperse, platinum and wild-type siblings were exposed to both black-and-white backgrounds at 5 dpf. At 6 dpf, wild-type embryos exposed to a white background exhibited melanosome aggregation (Fig. 2E), whereas wild-type embryos exposed to a black background exhibited dispersion of the melanosomes (Fig. 2G). All platinum zebrafish also exhibited melanophore aggregation and dispersion at 6 dpf (Figs. 2F, 2H).
The platinum Mutant Zebrafish Contain a Mutation in the Vacuolar Protein Sorting 11 (vps11) Gene
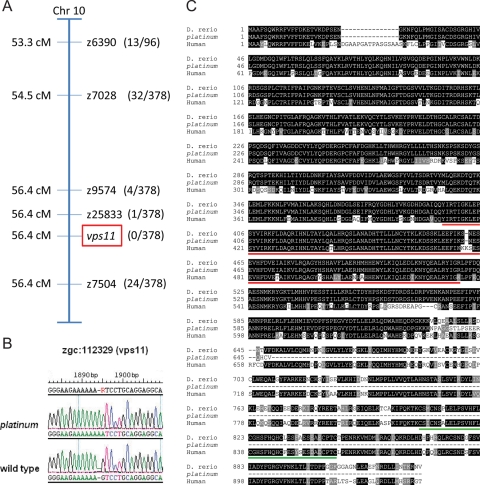
The platinum mutation was initially localized to chromosome 10 using bulked segregant analysis,18 and subsequent linkage mapping defined the interval containing the mutation (Fig. 3A). A candidate gene approach was used to evaluate genes in the identified interval. DNA sequence analysis revealed that the platinum mutants have a single-base change and a single-base insertion at nucleotide 1894 in the vps11 open reading frame, resulting in a frameshift mutation (Fig. 3B). The deduced amino acid sequence of the wild-type Vps11 protein is 927 residues. The platinum mutation results in 17 aberrant amino acids between the frameshift insertion and a premature translation termination codon, which results in a deduced protein of only 648 residues (Fig. 3C). Both the Clathrin repeat domain and the RING-H2 domain found in the human VPS11 protein are conserved in wild-type zebrafish (Fig. 3C). However, the truncation of the Vps11 protein in the platinum mutant zebrafish results in the absence of the RING-H2 domain (Fig. 3C).
Figure 3.
(A) The linkage map showing the localization of the platinum mutation to the region surrounding vps11 on chromosome 10. Recombination frequencies are shown in parentheses. (B) The platinum mutant zebrafish contained a single-base change and a single-base insertion at nucleotide 1894 in the vps11 open reading frame. (C) Protein sequence comparison of human VPS11, wild-type zebrafish (D. rerio) Vps11 and the deduced amino acid sequence of the Vps11 protein in platinum mutants, showing identical (black) and conserved (gray) residues. The Vps11 protein in platinum mutant zebrafish contains the conserved clathrin repeat domain (red underscore), but not the RING-H2 domain (green underscore).
Phenocopy and Rescue Experiments Suggest Loss of Vps11 Results in the platinum Phenotype
Knockdown of Vps11 in the wild-type (AB strain) embryos was performed using an antisense morpholino designed to inhibit proper splicing of vps11 mRNA. Embryos that were microinjected with the morpholino at the first to fourth cell stages (n = 85) could not be distinguished from noninjected embryos (n = 167) at 24 hpf (data not shown). However, at 2 dpf, compared with control embryos, 88% (75/85) of the embryos injected with the vps11 morpholino showed reduced pigmentation in the eye, body trunk (compare Figs. 4A and 4B), and dorsal head (compare Figs. 4C and 4D), phenocopying the platinum mutation. RT-PCR performed on RNA harvested from injected and noninjected embryos indicated that the vps11 morpholino inhibited removal of the second intron of the vps11 transcript (Fig 4E), which was confirmed by DNA sequencing.
Rescue of the platinum phenotype was accomplished by microinjection of vps11 mRNA into first- to fourth-cell-stage embryos from a heterozygous mutant in-cross. At 48 hpf, significantly fewer of the injected embryos (P = 0.002) exhibited the platinum phenotype (49/293), compared with the noninjected embryos (97/358) (Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5957/-/DCSupplemental). Although the rescued animals were not genotyped, taken together with the positional cloning and phenocopy results, these data suggest that loss of Vps11 results in the platinum phenotype.
Analysis of vps11 Expression
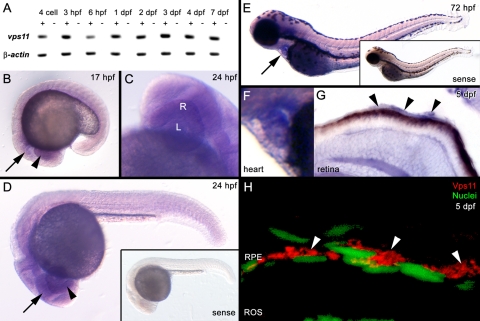
RT-PCR showed that vps11 is maternally expressed, as transcripts were detected before zygotic transcription at 3.5 hpf, and continues to be expressed through early development (Fig. 5A). Whole-mount in situ hybridization showed that vps11 was ubiquitously expressed, with stronger expression in the developing eye and central nervous system at 17 and 24 hpf (Figs. 5B-D), heart at 72 hpf (Figs. 5E, 5F), and RPE at 5 dpf (Fig. 5G). Immunolocalization of Vps11 confirmed strong cytosolic expression of Vps11 protein surrounding the RPE nuclei at 5 dpf (Fig. 5H).
Figure 5.
(A) Temporal expression pattern of vps11 and β-actin (control) at multiple time points. cDNA reactions contained (+) or excluded (−) reverse transcriptase. (B–G) Spatial expression of vps11 as detected by whole-mount in situ hybridization. (B) A 17 hpf embryo showing ubiquitous vps11 expression, with strong expression in the developing eye field (arrowhead) and brain (arrow). (C, D) A 24-hpf embryo, showing vps11 expression in the developing retina (R, arrowhead), lens (L), and brain (arrow). (D, inset) shows the sense strand control embryo. (E) A 72-hpf embryo. Arrow: strong expression in the heart. Inset: the sense strand control embryo. (F) Expression in the heart at 72 hpf. (G) A retinal section from a 5 dpf embryo showing vps11 expression in the retina and RPE (arrowheads). (H) A retinal section showing immunolocalization of Vps11 (red, arrowheads) in the RPE at 5 dpf. RPE nuclei are stained with TO-PRO-3 (green, nuclei).
Major Retinal Cell Types Form Normally in Zebrafish platinum Mutants
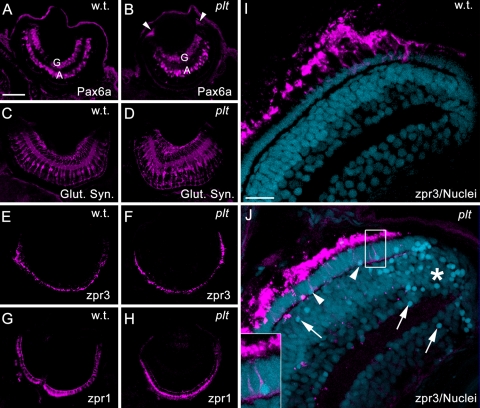
Previous reports of a loss of two other Vps proteins in zebrafish, Vps18 and Vps39, showed deleterious effects on RPE and photoreceptor viability.35,36 To determine whether loss of Vps11 resulted in retinal pathology, we examined retinas from wild-type and platinum mutants at 5 dpf for the presence of proteins associated with specific retinal cell types, the total number of retinal nuclei per nuclear layer and overall eye size. Immunolocalization of Pax6a revealed that both the wild-type (Fig. 6A) and platinum mutant retinas (Fig. 6B) contained amacrine and ganglion cells at 5 dpf. The platinum mutant retinas also exhibited persistent Pax6a expression in the retinal progenitors located in the ciliary marginal zone (Fig. 6B). Immunolocalization of glutamine synthetase revealed that the platinum mutant retinas contained a normal contingent of Müller glial cells (Fig. 6D). However, many of the Müller glial cell bodies were hypertrophied (Fig. 6D). Immunolocalization of zpr3 (Figs. 6E, 6F) and zpr1 (Figs. 6G, 6H) revealed the presence of both rod and double cone photoreceptors, respectively, in the platinum mutant retinas. Although the platinum mutants were slightly microphthalmic at 5 dpf (94% of wild-type; n = 5; Supplementary Fig. S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5957/-/DCSupplemental), the reduction in eye size was not significant (P = 0.3), nor was there a significant difference in the total number of nuclei present in each nuclear layer (Supplementary Fig. S2D). However, pyknotic nuclei were observed in each retinal layer in the platinum mutants, concentrated primarily near the margins (Fig. 6J; Supplementary Fig. S2). Co-labeling with zpr3 revealed an average of 17 pyknotic rod nuclei per retinal section in the platinum mutant retinas (Fig. 6J; Supplementary Fig. S2). Thus, even though retinal cell types formed normally in the platinum mutants, retinal disease was present at 5 dpf.
Figure 6.
(A, B) Immunolocalization of Pax6a in ganglion (G) and amacrine cells (A). The platinum mutant retinas also exhibited Pax6a expression in the retinal margins (B, arrowheads). (C, D) Immunolocalization of glutamine synthetase (Glut. Syn.) in Müller glial cells, showing hypertrophied Müller glia in the platinum mutant retinas (D). (E, F) Immunolocalization of zpr3 in rod photoreceptor outer segments. (G, H) Immunolocalization of zpr1 in double cone photoreceptors. (I, J) Immunolocalization of zpr3 (magenta) with TO-PRO-3 nuclear stain (blue). (I) A wild-type retina showing healthy rod outer segments (magenta) and uniform staining of nuclei. (J) A platinum mutant retina. Note the collapsed rod outer segments and pyknotic nuclei (arrows and arrowheads), concentrated primarily near the margins (asterisk) and in the photoreceptor layer (arrowheads). Inset: a pyknotic rod photoreceptor nucleus. Scale bar, 50 μm (A–H); 15 μm (I, J).
The Zebrafish platinum Mutants Show Progressive Degeneration of the Retinal Pigmented Epithelium
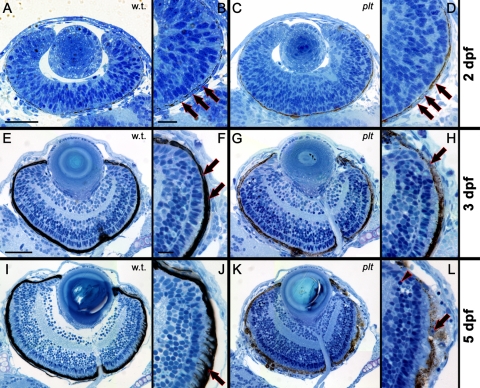
To more closely examine the retinal disease observed in the platinum mutants, wild-type and the platinum siblings were collected at 2, 3, and 5 dpf and processed for histologic staining.23 Histologic sections from the wild-type and platinum mutants were indistinguishable at 2 dpf (Figs. 7A–D). At 2 and 3 dpf, RPE nuclei and pigmentation were visible in both the wild-type and platinum mutants (Figs. 7B, 7D, 7F, 7H), and by 3 dpf, lamination of the retina into three distinct nuclear layers had occurred (Figs. 7E, 7G). At 3 and 5 dpf, the platinum mutant retinas could be distinguished from wild-type siblings based on reduced pigmentation of the RPE (compare Figs. 7G, 7K with 7E, 7I, respectively). In addition, the platinum mutant retinas contained pyknotic nuclei in the RPE and near the margins at 5 dpf (Fig. 7L). Finally, rod photoreceptor outer segments in the platinum mutants did not interdigitate with the RPE at 5 dpf (Fig. 7L), as they did in the wild-type retina (Fig. 7J).
Figure 7.
Histologic sections at 2, 3, and 5 dpf in wild-type (left) and platinum mutants (right). (A, B) A wild-type retina at 2 dpf. RPE nuclei (B, arrows) are surrounded by melanin pigment. (C, D) A platinum mutant retina at 2 dpf. RPE nuclei (D, arrows) and melanin pigment are visible. (E, F) A wild-type retina at 3 dpf, showing three distinct nuclear layers in the neural retina. The RPE is darkly pigmented (F, arrows). (G, H) A platinum mutant retina at 3 dpf. RPE nuclei are visible (H, arrow). RPE pigmentation is visible less than observed in wild-type retinas. (I–K) A wild-type retina at 5 dpf. Rod photoreceptor outer segments interdigitate with the darkly pigmented RPE (J, arrow). (K, L) A platinum mutant retina at 5 dpf. Note reduced RPE pigmentation and pyknotic nuclei in the RPE (L, arrow) and neural retina (L, arrowhead). Scale bar, (A, C, E, G, I, K) 50 μm; (B, D, F, H, J, L) 15 μm.
Loss of Vps11 Results in Multiple Retinal Abnormalities
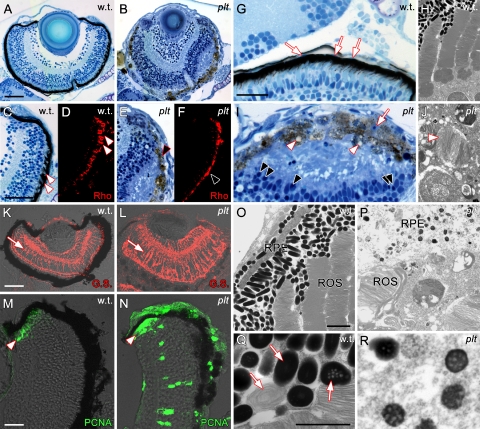
Histologic sections from the platinum mutants at 7 dpf showed a progressive loss of pigmentation of the RPE compared with the wild-type sections (Figs. 8B, 8A, respectively), which, in many cases, had pulled away from the neural retina (Figs. 8E, 8I). In addition, rhodopsin immunolocalization of rod photoreceptor outer segments revealed that the platinum mutants contained truncated rod outer segments (Fig. 8F). Large vacuoles were visible by light microscopy in the RPE of the platinum mutants (Fig. 8I). Ultrastructural analysis of wild-type retinas by transmission electron microscopy (TEM) showed healthy rod outer segments that interdigitated with RPE, which contained an abundance of mature melanosomes (Figs. 8H, 8O). In contrast, TEM of the platinum mutants showed complete degeneration of the rod outer segments (Figs. 8J, 8P). Many large vacuoles full of discs from rod outer segments were also observed in the platinum mutants (Fig. 8J).
Figure 8.
(A) Histologic section from a wild-type retina and a (B) platinum mutant retina at 7 dpf. (C) The margin of the wild-type retina shown in (A), showing the interdigitation of the photoreceptors and RPE (arrowheads). (D) Immunolocalization of rhodopsin expression (Rho) in the rod outer segments of wild-type rod photoreceptors at 7 dpf. (E) The margin of the platinum retina shown in (B), showing RPE degeneration and loss of the photoreceptor/RPE boundary. (F) Immunolocalization of Rho in a platinum mutant retina at 7 dpf, showing truncated and collapsed ROS (arrowhead). (G) Histologic section of a wild-type retina showing the elongated RPE nuclei (arrows) and interdigitation of the RPE with the underlying photoreceptors. (H) TEM of wild-type retina at 7 dpf, showing healthy rod outer segments and RPE with melanosomes. (I) Histologic section of a platinum mutant retina at 7 dpf. The neural retina has detached from the RPE. Note the presence of vacuoles (white arrowheads) in the RPE and pyknotic nuclei in both the RPE (white arrow) and photoreceptor layer (black arrowheads). (J) TEM of a platinum mutant retina at 7 dpf. Note the degeneration of the rod outer segments and large vacuoles full of discs from rod outer segments (arrowhead). (K) Glutamine synthetase (GS) localization in wild-type retina at 7 dpf showing Müller glial cells (arrow). (L) Immunolocalization of GS in a platinum mutant retina at 7 dpf, showing Müller glial cell hypertrophy. (M) Immunolocalization of PCNA-positive retinal progenitors in the CMZ of a wild-type retina at 7 dpf. (N) PCNA immunolocalization reveals abnormal proliferation in a platinum mutant retina at 7 dpf. PCNA-positive nuclei are present in all three nuclear layers and in the CMZ (arrowhead). (O) TEM of a wild-type retina, showing elongated and fully pigmented melanosomes in the RPE. (P) TEM of a platinum retina, showing reduced pigmentation in melanosomes and a degeneration of the RPE. (Q) Close-up of a region in (O) showing wild-type melanosomes in three (of the four) stages of maturation, including fully mature melanosomes completely full of pigment. (R) Close-up of the platinum mutant RPE shown in (P). The melanosomes observed in platinum mutants are in an immature phase of melanosome development (melanosomes in stage 3 are shown). Scale bar: (A–F, K, L) 50 μm; (G, I) 20 μm; (M, N) 15 μm; (O, P) 2 μm; (Q, R) 1 μm.
The platinum mutants also exhibited Müller glial cell hypertrophy and abnormal proliferation at 7 dpf. In the wild-type retinas, glutamine synthetase localization revealed the characteristic Müller glial cell morphology, with the soma located in the inner nuclear layer and processes that extend from the nucleus to the outer and inner limiting membranes (Fig. 8K). In contrast, both the Müller glial soma and processes were hypertrophied in the platinum mutants at 7 dpf (Fig. 8L). Immunolocalization of PCNA (proliferating cell nuclear antigen), a known marker for cell proliferation in zebrafish,22–24,26 was used. In wild-type retinas at 7 dpf, PCNA-positive nuclei were restricted to the ciliary marginal zone (CMZ, Fig. 8M), a location of continued neurogenesis in the zebrafish retina. In contrast, a significant number of PCNA-positive nuclei were identified in the INL and ONL (outside the CMZ) in platinum mutant retinas at 7 dpf (n = 5; P < 0.01 for both layers; Supplementary Fig. S3F, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5957/-/DCSupplemental). In addition, the percent area of PCNA-positive immunolocalization in the CMZ was significantly expanded in platinum mutants at 7 dpf (Supplementary Fig. S2G, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-5957/-/DCSupplemental; n = 5; P = 0.0004).
Finally, TEM was performed on the wild-type and platinum retinas to observe the extent of maturation of melanosomes in the RPE. Melanosomes go through four phases of maturation from early endosome to a fully pigmented and mature melanosome.9 Melanosomes at each of these stages were indentified in wild-type RPE (Fig. 8Q; stages II–IV shown). The vast majority of the melanosomes observed in wild-type RPE were fully mature (Fig. 8O), whereas, most of the melanosomes observed in the platinum mutants were immature (Figs. 8P, 8R).
Discussion
Although there are currently no mouse or fly mutants of vps11, mutations in three other C-Vps genes, vps16, vps18, and vps33, cause reduced pigmentation in Drosophila.9 A medaka (Oryzias latipes) mutant for vps11 was described with reduced pigmentation,37 and mutations in vps18 in zebrafish cause reduced RPE and body pigmentation similar to platinum mutants.35,36,38 Compared with the vps18 and vps39 mutants, the platinum (vps11) mutant looks morphologically identical with the vps18hi2499A retroviral insertion mutant and the leberknodel (vps39) mutant, as both exhibit reduced pigmentation and hepatomegaly.35,36 In vps11 mutants, the OCA phenotype was typical for tyrosinase-positive albinism. Body melanophores were observed at the appropriate stage in development, migrated properly, and maintained the ability to aggregate and disperse. Although some melanin was present, progressive loss of pigmentation in both the body and the RPE was observed in vps11 mutants.
We found that vps11 was maternally expressed in zebrafish, as transcripts were present before the midblastula transition (∼3.5 hpf), which represents the onset of zygotic transcription in zebrafish.39 Whole-mount in situ hybridization showed that vps11 is expressed ubiquitously, with higher levels of expression in the retina at 24 hpf, heart at 72 hpf, and RPE at 5 dpf. These data are consistent with the retinal defects and pericardial edema observed in vps11 mutants, and with previously reported data on human VPS11, which is ubiquitously expressed with relatively higher expression in heart, liver, and pancreas.40
Consistent with other cases of syndromic albinism, loss of Vps11 results in both OCA and systemic defects, both related to problems with endosome-to-lysosome and endosome-to-melanosome protein trafficking. Melanosomes undergo four stages of maturation, each with a characteristic increase in pigmentation.9 Changes in each stage of melanosome maturation require vacuole protein trafficking from the Golgi or endosome. Mutations affecting each of the HPS genes result in defects in each stage of melanosome maturation.9 Loss of C-Vps proteins have also been reported to affect the maturation of melanosomes. Vps33a mutant mice show very few fully mature melanosomes, and those that are present are abnormally small and rounded.41 We observed the identical phenotype in loss of Vps11 in platinum melanosomes (Figs. 8P, 8R). Loss of Vps18 and Vps39 in zebrafish also causes an accumulation of immature melanosomes with a reduction in the number of mature melanosomes.35,36 Since Vps11, Vps16, Vps18, and Vps33 bind as a complex, loss of one of these proteins very likely affects the function of the entire cluster in melanosome maturation. Notably, in each case reported, loss of any of these proteins resulted in the absence of stage IV melanosomes in the RPE, indicating that this complex functions in the final stages of melanosome maturation.
The truncation of the Vps11 protein in platinum mutant zebrafish resulted in the absence of the RING-H2 domain (Fig. 3C), which is a zinc finger binding domain that contains an octet of cysteines and histidines and represents the largest class of ubiquitin ligases. Although no report has linked a specific ubiquitination reaction in vivo to functions of the C-Vps complex, in vitro studies and the occurrence of this domain in C-Vps proteins are suggestive of ubiquitin-mediated regulation. Both Vps11 and Vps18 contain a RING-H2 domain. Consistent with our data, previous reports indicate that loss of the RING domains in Vps11 and Vps18 are required for C-Vps function.12,42 In addition, in vitro data indicate that the RING-H2 domain of both Vps11 and Vps18 exhibit E3 ubiquitin ligase activity.43,44 However, hVPS11 does not ubiquitinate the same protein domain on the Golgi-localizing adapter protein GGA3, as does hVPS18,44 and the presence of hVPS11 strongly increases the ubiquitination of GGA3 by hVPS18,44 indicating a potentially complex regulation of C-Vps complexes by ubiquitination.
Photoreceptor cell survival depends on a healthy RPE, which is involved in many important functions including uptake and turnover of photoreceptor outer segments, retinol biosynthesis, and maintenance of the blood–retina barrier.45 However, it is unclear how the level of pigment in the RPE affects these functions. In chick, frog, and zebrafish albino strains, photoreceptors form normally and are maintained in normal number.6,7 In contrast, multiple albino mouse strains exhibit reduced numbers of rod photoreceptors, in which the number of rod photoreceptors corresponds to the level of ocular pigmentation.4 This defect can be rescued by introducing an enzyme that promotes melanin deposits, Tyrosinase, into transgenic albino mice, providing strong evidence that tyrosinase is required in the mammalian RPE.3 Similar to HPS patients, platinum mutant zebrafish have a functional tyrosinase gene and, therefore have the ability to synthesize some melanin pigment. We showed that loss of Vps11 did not affect the development of RPE or photoreceptors, as both were present and not pyknotic at 2 and 3 dpf. However, by 5 and 7 dpf, the gradual loss of pigment in the RPE correlated with the developing retinal disease, including photoreceptor loss, truncation of rod outer segments, and the presence of pyknotic nuclei in multiple retinal layers. In addition, Müller glial soma and processes were hypertrophied in the platinum mutants at 7 dpf, a characteristic of reactive gliosis. It is difficult to determine whether RPE disease precedes photoreceptor loss. However, the loss of Vps11 most likely causes a toxic buildup of protein products in the RPE, caused by problems in protein trafficking and, possibly, by ubiquitination. RPE death would lead to photoreceptor loss and subsequent reactive gliosis. These data strongly suggest that individuals with syndromic albinism, like HPS, may exhibit progressive retinal disease in addition to congenital ocular defects, such as nystagmus and strabismus.
Supplementary Material
Acknowledgments
The authors thank Deborah Bang and the staff of the Freimann Life Science Center at the University of Notre Dame and Xixia Luo at Wayne State University School of Medicine for providing zebrafish husbandry and care.
Footnotes
Supported by National Institutes of Health Grants R21EY019401 (RT), R21EY018919 (DRH), and R01EY018417 (DRH, RT), the Center for Zebrafish Research at the University of Notre Dame, and Wayne State University School of Medicine start-up funds (RT), as well as an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology, Wayne State University.
Disclosure: J.L. Thomas, None; T.S. Vihtelic, None; A.D. denDekker, None; G. Willer, None; X. Luo, None; T.R. Murphy, None; R.G. Gregg, None; D.R. Hyde, None; R. Thummel, None
References
- 1. King RA, Summers CG. Albinism. Dermatol Clin. 1988;6:217–228 [PubMed] [Google Scholar]
- 2. Naash MI, Ripps H, Li S, Goto Y, Peachey NS. Polygenic disease and retinitis pigmentosa: albinism exacerbates photoreceptor degeneration induced by the expression of a mutant opsin in transgenic mice. J Neurosci. 1996;16:7853–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jeffery G. The albino retina: an abnormality that provides insight into normal retinal development. Trends Neurosci. 1997;20:165–169 [DOI] [PubMed] [Google Scholar]
- 4. Donatien P, Jeffery G. Correlation between rod photoreceptor numbers and levels of ocular pigmentation. Invest Ophthalmol Vis Sci. 2002;43:1198–1203 [PubMed] [Google Scholar]
- 5. Gorgels TG, Van Norren D. Two spectral types of retinal light damage occur in albino as well as in pigmented rat: no essential role for melanin. Exp Eye Res. 1998;66:155–162 [DOI] [PubMed] [Google Scholar]
- 6. Grant S, Patel NN, Philp AR, et al. Rod photopigment deficits in albinos are specific to mammals and arise during retinal development. Vis Neurosci. 2001;18:245–251 [DOI] [PubMed] [Google Scholar]
- 7. Jeffery G, Williams A. Is abnormal retinal development in albinism only a mammalian problem?—normality of a hypopigmented avian retina. Exp Brain Res. 1994;100:47–57 [DOI] [PubMed] [Google Scholar]
- 8. Scheinfeld NS. Syndromic albinism: a review of genetics and phenotypes. Dermatol Online J. 2003;9:5. [PubMed] [Google Scholar]
- 9. Raposo G, Marks MS. Melanosomes: dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Pietro SM, Dell'Angelica EC. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 2005;6:525–533 [DOI] [PubMed] [Google Scholar]
- 11. Wei ML. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 2006;19:19–42 [DOI] [PubMed] [Google Scholar]
- 12. Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell. 2000;6:661–671 [DOI] [PubMed] [Google Scholar]
- 14. Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Srivastava A, Woolford CA, Jones EW. Pep3p/Pep5p complex: a putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics. 2000;156:105–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press; 1995 [Google Scholar]
- 17. Logan DW, Burn SF, Jackson IJ. Regulation of pigmentation in zebrafish melanophores. Pigment Cell Res. 2006;19:206–213 [DOI] [PubMed] [Google Scholar]
- 18. Willer GB, Lee VM, Gregg RG, Link BA. Analysis of the zebrafish perplexed mutation reveals tissue-specific roles for de novo pyrimidine synthesis during development. Genetics. 2005;170:1827–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song Y, Selak MA, Watson CT, et al. Mechanisms underlying metabolic and neural defects in zebrafish and human multiple acyl-CoA dehydrogenase deficiency (MADD). PLoS One. 2009;4:e8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220 [DOI] [PubMed] [Google Scholar]
- 21. Thummel R, Li L, Tanase C, Sarras MP, Jr, Godwin AR. Differences in expression pattern and function between zebrafish hoxc13 orthologs: recruitment of Hoxc13b into an early embryonic role. Dev Biol. 2004;274:318–333 [DOI] [PubMed] [Google Scholar]
- 22. Kassen SC, Ramanan V, Montgomery JE, et al. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–1031 [DOI] [PubMed] [Google Scholar]
- 23. Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR. Inhibition of Muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev Neurobiol. 2008;68:392–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vihtelic TS, Soverly JE, Kassen SC, Hyde DR. Retinal regional differences in photoreceptor cell death and regeneration in light-lesioned albino zebrafish. Exp Eye Res. 2006;82:558–575 [DOI] [PubMed] [Google Scholar]
- 25. Vihtelic TS, Yamamoto Y, Sweeney MT, Jeffery WR, Hyde DR. Arrested differentiation and epithelial cell degeneration in zebrafish lens mutants. Dev Dyn. 2001;222:625–636 [DOI] [PubMed] [Google Scholar]
- 26. Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44:289–307 [DOI] [PubMed] [Google Scholar]
- 27. Vihtelic TS, Doro CJ, Hyde DR. Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis Neurosci. 1999;16:571–585 [DOI] [PubMed] [Google Scholar]
- 28. Arduini BL, Henion PD. Melanophore sublineage-specific requirement for zebrafish touchtone during neural crest development. Mech Dev. 2004;121:1353–1364 [DOI] [PubMed] [Google Scholar]
- 29. Cornell RA, Yemm E, Bonde G, et al. Touchtone promotes survival of embryonic melanophores in zebrafish. Mech Dev. 2004;121:1365–1376 [DOI] [PubMed] [Google Scholar]
- 30. Dutton KA, Pauliny A, Lopes SS, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125 [DOI] [PubMed] [Google Scholar]
- 31. Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. Nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767 [DOI] [PubMed] [Google Scholar]
- 32. Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–3436 [DOI] [PubMed] [Google Scholar]
- 33. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310 [DOI] [PubMed] [Google Scholar]
- 34. Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238:2975–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maldonado E, Hernandez F, Lozano C, Castro ME, Navarro RE. The zebrafish mutant vps18 as a model for vesicle-traffic related hypopigmentation diseases. Pigment Cell Res. 2006;19:315–326 [DOI] [PubMed] [Google Scholar]
- 36. Schonthaler HB, Fleisch VC, Biehlmaier O, et al. The zebrafish mutant lbk/vam6 resembles human multisystemic disorders caused by aberrant trafficking of endosomal vesicles. Development. 2008;135:387–399 [DOI] [PubMed] [Google Scholar]
- 37. Yu JF, Fukamachi S, Mitani H, Hori H, Kanamori A. Reduced expression of vps11 causes less pigmentation in medaka Oryzias latipes. Pigment Cell Res. 2006;19:628–634 [DOI] [PubMed] [Google Scholar]
- 38. Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572 [DOI] [PubMed] [Google Scholar]
- 39. Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456 [DOI] [PubMed] [Google Scholar]
- 40. Huizing M, Didier A, Walenta J, et al. Molecular cloning and characterization of human VPS18, VPS 11, VPS16, and VPS33. Gene. 2001;264:241–247 [DOI] [PubMed] [Google Scholar]
- 41. Nguyen T, Wei ML. Characterization of melanosomes in murine Hermansky-Pudlak syndrome: mechanisms of hypopigmentation. J Invest Dermatol. 2004;122:452–460 [DOI] [PubMed] [Google Scholar]
- 42. Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21:543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yogosawa S, Hatakeyama S, Nakayama KI, et al. Ubiquitylation and degradation of serum-inducible kinase by hVPS18, a RING-H2 type ubiquitin ligase. J Biol Chem. 2005;280:41619–41627 [DOI] [PubMed] [Google Scholar]
- 44. Yogosawa S, Kawasaki M, Wakatsuki S, et al. Monoubiquitylation of GGA3 by hVPS18 regulates its ubiquitin-binding ability. Biochem Biophys Res Commun. 2006;350:82–90 [DOI] [PubMed] [Google Scholar]
- 45. Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.