This study combines classical QTL mapping with tissue-specific microarray analysis to identify novel genes potentially involved in the establishment of the cone photoreceptor population of the mouse retina.
Abstract
Purpose.
This investigation examines the genetic sources of marked variation in cone photoreceptor number among inbred lines of mice, identifying candidate genes that may control the proliferation, differentiation, or survival of this neuronal population.
Methods.
Cone photoreceptor populations were counted in C57BL/6J (B6/J) and A/J strains, and 26 recombinant inbred (RI) strains derived from them. Eyes from RI strains were also collected for microarray analysis. Quantitative trait locus (QTL) analysis was carried out by simple and composite interval mapping and validated using a consomic line. Candidate genes were evaluated based on genetic variance between the parental strains and analysis of gene expression. Expression data, deposited in GeneNetwork (www.GeneNetwork.org), were used to generate a coexpression network of established cone photoreceptor genes as a reference standard.
Results.
B6/J has 70% more cone photoreceptors than A/J. A significant QTL was mapped to chromosome 10 (Chr 10) and confirmed using B6.A<10> mice. Of 19 positional candidate genes, one—the myeloblastosis oncogene (Myb)—stood out. Myb has a potentially damaging missense mutation, high retinal expression, and a known role in cell proliferation. The ectonucleotide pyrophosphatase/phosphodiesterase 1 gene (Enpp1) was a second strong candidate, with an expression pattern that covaried with cone photoreceptors and that was differentially expressed between the parental strains. Enpp1 and several other candidate genes covaried with multiple genes within the cone photoreceptor gene network.
Conclusions.
The mouse retina shows marked variation in cone photoreceptor number, some of which must be controlled by polymorphisms in a gene or genes on Chr 10.
Neuronal populations frequently show a natural variation in size that is independent of the size of the structure within which they are situated. For example, the population of ganglion cells within the retina is highly variable between strains of mice and unrelated to variance in the retinal area,1 as it is between individual primates of the same species, including humans.2 Other retinal cell types in the mouse have also recently been shown to exhibit conspicuous variation in their sizes, including the populations of dopaminergic amacrine cells3 and cholinergic amacrine cells.4 Variation within the primate retina, including that of humans, has been most thoroughly documented with respect to the population of cone photoreceptors,5–7 in which this natural variation may underlie a functional difference in visual acuity. The cause (or causes) of such variation in primates is unknown but is presumed to reflect the action of allelic variants of genes that modulate cellular production or survival during early development.
We asked whether mice, like humans, show such a natural variation in their population of cone photoreceptors. Subsequently, we sought to identify potentially novel genes in determining the size of the cone photoreceptor population. We demonstrate a significant variation between two laboratory strains of mice, B6/J and A/J. Using 26 RI strains of mice derived from these two parental lines, we describe the mapping of a sizable portion of this variation to a QTL on Chr 10. We confirm the presence of a gene (or genes) on Chr 10 that modulates cone photoreceptor number using consomic mice of the chromosome substitution strain B6.A<10>.
Using two complementary approaches, we identified promising candidate genes that may underlie this natural variation in cone photoreceptor number. First, we identified genes with known coding or regulatory genetic variants, or both, between the parental strains and known to be expressed in the retina. Second, we generated genomewide eye mRNA expression data for 26 strains of the AXB/BXA RI strain set. With this resource, we were able to identify all genes within the QTL whose expression was highly correlated to the variation in cone photoreceptor number across this RI strain set. Additionally, the transcript abundance of each of these genes can be treated as a quantitative trait that can be mapped. We were, therefore, able to use this eye mRNA expression data set to identify candidate genes that both mapped an expression QTL (eQTL) to the physical location of the gene (cis-eQTL)8 and covaried with cone photoreceptor number.9 Finally, we used the eye mRNA expression data set for the AXB/BXA RI strain set to assess the degree of covariance between our most promising candidate genes with a group of known cone photoreceptor genes. Such coexpression networks have been used to predict the role of a gene10 and may provide further support for a candidate gene in determining cone photoreceptor number.
Materials and Methods
Animals
The total population of cone photoreceptors was estimated in B6/J mice, A/J mice, 26 strains of mice from the AXB/BXA RI strain set, and consomic B6.A<10> mice. An average of five retinas per strain were examined from animals ranging in age from 30 to 115 days old. All strains were obtained from The Jackson Laboratory (JAX) (Bar Harbor, ME). Mice from the RI strain set were housed at the University of Tennessee Health Science Center (UTHSC) before use. Consomic and parental strains were shipped to the University of California at Santa Barbara (UCSB); the former were euthanatized on delivery, and the latter were bred for several generations. All animals were reared and housed under standard lighting conditions of roughly 30 lumen per square foot (323 lux) and either 12:12 or 14:10 light/dark cycles at UCSB/UTHSC and JAX, respectively. The AXB/BXA RI strain set had been generated by inbreeding multiple pairs of progeny from an F1 intercross over 20 generations (either AB6F1 to make AXB strains or B6AF1 to make BXA strains), thereby yielding recombinant chromosomes that were homozygous at every location. A general description of this resource can be found at http://jaxmice.jax.org/type/recombinbred.html, and an expanded explanation of this particular strain set can be found at http://www.genenetwork.org/mouseCross.html#AXBXA. All RI strain mice were euthanatized by cervical dislocation; all others were euthanatized by intraperitoneal injection of sodium pentobarbital (120 mg/kg body weight). Eyes were dissected immediately from the orbit and immersed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2 at 20°C) for subsequent immunofluorescence labeling. These procedures were conducted under authorization by the Institutional Animal Care and Use Committees at both institutions and conformed to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Immunofluorescence and Cone Photoreceptor Quantification
Retinas were dissected and prepared as wholemounts, with care taken to ensure the entire retinal area was included in each dissection. A single retina from each mouse was labeled for cone photoreceptor outer segments using a cocktail of rabbit polyclonal antibodies to M and S cone opsins (1:1000; Millipore/Chemicon; Temecula, CA) followed by incubation with a donkey anti-rabbit IgG conjugated to Cy3 (1:200; Jackson ImmunoResearch, West Grove, PA). These retinas were mounted flat with the photoreceptor layer up on a glass slide and were coverslipped using 0.1 M sodium phosphate buffer as the mounting medium. Retinas were examined on an epifluorescence microscope (BH2; Olympus, Tokyo, Japan) equipped with X-Y stage encoders and a digital video camera (Sony, Tokyo, Japan) and linked to a computer running image analysis and measurement software (Bioquant Nova Prime; R&M Biometrics, Nashville, TN). We generated an outline of the entire retina and implemented an objective sampling protocol by which fields were counted in a 1-mm × 1-mm grid across the retinal surface, yielding on average 15 locations. The initial sample site was established in the uppermost left corner of the retina, positioning the sampling grid so that the greatest number of sample sites would overlie the retinal wholemount, allowing for adjustments, up to 100 μm, of fields obscured by retinal tears, relieving cuts, or the optic nerve head (Figs. 1e, 1f). Each sample site was examined using a ×40 objective, and every immunopositive outer segment within a field of known size (averaging 14,000 μm2) was counted, focusing up and down to ensure discrimination of the overlying outer segments. Sampled field densities were averaged to define mean cone photoreceptor density and then were multiplied by the retinal area to estimate the total number of cone photoreceptors per retina. A total of 145 retinas were sampled, counting on the order of 1500 to 2700 photoreceptors per retina, depending on the strain. All photoreceptor counts were conducted by the same observer.
Figure 1.
Cone photoreceptor density varies in different mouse strains. Sampled fields from the central and peripheral retina of A/J (a, b) and B6/J (c, d) mice show the greater density in B6/J. The sampling protocol, at 1-mm intervals across the retinal surface, is also shown for representative A/J (e) and B6/J (f) retinas.
QTL Mapping
The strains of an RI strain set are a mix of the parental genotypes. QTL mapping takes advantage of the nearly random recombination of parental genotypes to estimate the covariance between a given trait and the presence of the A or B haplotype throughout the genome.11 GeneNetwork implements standard methods of simple and composite interval mapping and estimates the genomewide P value of a type 1 error by random permutation. The marker regression tool plots the permutation tests used to assess the strength of the linkage for a trait. We used 1000 permutations to determine the suggestive and significant likelihood ratio statistic (LRS). The primary cone photoreceptor data derived from the parental strains and these RI strains have been permanently deposited in GeneNetwork as phenotype accession identifier number 10153 (cone photoreceptor number) in the mouse AXB/BXA Published Phenotypes database.
AXB/BXA Eye mRNA Expression Analysis
After cervical dislocation, whole eyes from 26 strains of the AXB/BXA RI strain set were removed and placed in tissue storage reagent (RNAlater; Qiagen, Valencia, CA). The extraction of RNA was carried out according to the manufacturer's recommendations (RNA STAT-60; Tel-Test Inc., Friendswood, TX). Total RNA was then concentrated (RNeasy MinElute Cleanup Kit; Qiagen). RNA purity was evaluated using the 260/280-nm absorbance ratio with a spectrophotometer (ND-1000; NanoDrop Technologies Inc., Wilmington, NC), and only samples with values greater than 1.8 were used. Most samples had values between 1.9 and 2.1. RNA integrity was assessed using a bioanalyzer (Agilent 2100; Agilent Technologies, Santa Clara, CA). The standard Eberwine T7 polymerase method was used to catalyze the synthesis of cDNA template from polyA-tailed RNA using an RNA amplification kit (Ambion/Illumina TotalPrep; Ambion, Austin, TX). Biotin-labeled cRNA was then evaluated using a spectrophotometer (ND-1000; NanoDrop Technologies Inc.). Samples that had 260/280-nm absorbance ratios between 2.0 and 2.3 were immediately used on oligomer bead chip slides (Sentrix Mouse WG-6 v2 oligomer BeadChip; Illumina Inc., San Diego, CA). The slides were hybridized and washed in accordance with standard Illumina protocols. A total of 54 pooled whole eye samples were processed using approximately 10 slides. This array consisted of 45,281 unique probe sequences, each 50 nucleotides in length. This particular data set was processed using the Illumina rank invariant method. Values were log2 transformed, and the variance of each array was stabilized to 4 U (SD, 2 U) and recentered to a mean of 8. Further details can be found at GeneNetwork under Eye AXBXA Illumina v6.2(Oct08) RankInv, accession number GN210.
Results
Cone photoreceptor density was observed to vary conspicuously between the parental strains, being greater in the B6/J strain in both the central (Figs. 1a, 1c) and the peripheral retina (Figs. 1b, 1d). The size of the total cone photoreceptor population was estimated at 116,158 ± 2737 cones (mean ± SEM, and hereafter) for the A/J strain and at 196,897 ± 5435 cones for the B6/J strain, an approximate 70% increase in the B6/J strain compared with the A/J strain. Although retinal area was slightly smaller in the A/J strain than in the B6/J strain (A/J, 14.8 ± 0.10 mm2; B6/J, 15.3 ± 0.30 mm2), this difference was not significant and could not account for the difference in estimated total cone photoreceptor number between the parental strains.
The estimated total number of cone photoreceptors in 26 strains of the AXB/BXA RI strain set largely spans the range defined by the parental strains (Fig. 2). One strain, AXB23, had numbers greater than the B6/J strain (average, 204,419 ± 5531 cone photoreceptors). The fact that this RI strain set exhibits such a graded distribution of phenotypes indicates that this trait—cone photoreceptor cell number—is likely influenced by multiple polymorphic genes. The variation within any given strain was low (with one exception, BXA14), the average coefficient of variation being 0.06. This relatively meager intrastrain variation validates our sampling method despite the small proportion of the retinal surface sampled (only 1.5%, on average). The proportion of the variance in total cone photoreceptor number across all mice that could be attributed to an effect of strain was 0.67. This high heritability estimate12 suggested this trait should be amenable to successful mapping of the allelic sources of variation in cone photoreceptor number.
Figure 2.
Total cone photoreceptor number varies between the two parental strains (white bar, black bar) and between the 26 RI strains of the AXB/BXA strain set (gray bars). Mean ± SE is shown for each. n = the number of retinas sampled per strain. Note the meager variation within most of the strains yet the conspicuous interstrain variation in total cone photoreceptor number.
We mapped a strain-associated phenotypic variance to a genomic locus at the proximal end of Chr 10 and determined, through permutation testing, a suggestive threshold for the LRS at 10.09 and a significant threshold at 15.67 (corresponding to a genomewide P < 0.05). The peak of the QTL surpassed both thresholds, with an LRS value of 16.5 at the D10Mit213 marker (20.09 Mb; Fig. 3a). For this locus, the trait increases with B alleles (Fig. 3a, red line), with an additive effect for two B alleles contributing 21,600 cells, which accounts for roughly 27% of the parental difference. There were no additional significant or suggestive QTLs for this trait, which we determined with the composite interval mapping tool of GeneNetwork. We have named this QTL cone photoreceptor number control Chr10 (Cpnc10).
Figure 3.
(a) QTL for cone photoreceptor number resides on Chr 10. The whole genome map plots the correlation between the presence of A alleles (green) and B alleles (red) with increases in trait values (right y-axis). The LRS (blue) plots the strength of this linkage between genotype and phenotype across the genome, revealing a single QTL on Chr 10 at 20.09 Mb, with an LRS score of 16.5. A random permutation test indicates a significant LRS threshold of 15.67 for P < 0.05. (b) Haplotypes of the RI strains that contribute to the significant mapping of Cpnc10 (AXB24, BXA26, BXA24, and BXA1), along with the haplotypes of the other RI strains, are displayed above the first 60 Mb of Chr 10. Red bars: B haplotype; green bars: A haplotype; blue bars: regions of unknown haplotype. The SNP density map (vertical orange lines) along the x-axis of the expanded map of Cpnc10 shows the SNP-rich region extending from 20 to 27 Mb. Candidate genes were identified from this interval. Other conventions are as described for (a).
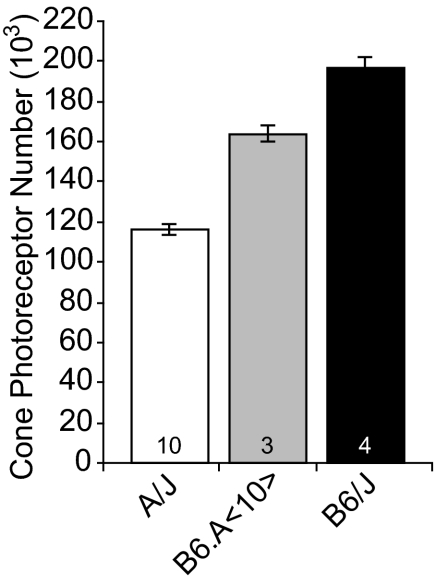
To confirm the presence of a gene or genes on Chr 10 modulating cone photoreceptor number, we analyzed retinas from the chromosome substitution strain B6.A<10>.13 These mice have an A haplotype for Chr 10 on an otherwise B6/J genetic background. We estimated the total size of their cone photoreceptor population at 163,906 ± 3813. Here the effect of substituting A for B alleles throughout Chr 10 was to reduce the size of the cone photoreceptor population by approximately 33,000 cones (Fig. 4). This reduction exceeded the size of the effect produced by Cpnc10 itself and confirmed the presence of the QTL on Chr 10 modulating cone photoreceptor number.
Figure 4.
Cone photoreceptor number is reduced in B6/J mice containing A alleles throughout Chr 10. Data for chromosome substitution strain B6.A<10> mice are shown (gray bar) along with the parental strains (white bar, black bar) for comparison. Conventions are as described for Figure 2.
Examination of the haplotypes of each AXB/BXA RI strain across the QTL revealed the strains that are critical to the significant mapping of Cpnc10: AXB24, BXA26, BXA24, and BXA1 (Fig. 3b). The robustness of this QTL was evidenced by the persistence of a highly suggestive peak even when the data from these particular strains were excluded, individually or altogether, from interval mapping (data not shown). The single nucleotide polymorphism (SNP) density map is also shown along the x-axis in Figure 3b, revealing the portion of Cpnc10 with high SNP density, where genes within this region have a greater probability of containing the functional variants responsible for variations in cone photoreceptor number. This reduces the number of potential candidate genes for Cpnc10 from roughly 100, underlying the suggestive portion of the QTL to approximately 70 genes within the SNP-dense region (20–27 Mb). From this reduced interval, we identified a subset of promising candidate genes based on two complementary approaches.
Using the SNP Browser of GeneNetwork and the Mouse Genome Browser of UCSC Genome Bioinformatics (http://genome.ucsc.edu/index.html), we examined the 8-Mb region of interest for Cpnc10. We sought genes with SNPs within their coding region, such as missense or nonsense mutations, which could potentially result in a functionally altered protein product between B6/J and A/J. We also looked for genes with SNPs within putative regulatory regions, including the predicted promoter, 5′UTR and 3′UTR (as defined on the GeneNetwork SNP Browser), which could potentially drive differences in transcript levels between the parental strains. Through this approach we identified four genes with at least one known coding mutation (missense or nonsense), 16 genes with at least one known SNP in regulatory regions, and eight more genes with both coding and regulatory SNPs. This reduced list of 28 genes was subsequently assessed for retinal expression using two mouse retinal expression data sets: the Serial Analysis of Gene Expression (SAGE) database, which provides data on gene expression in the embryonic and postnatal mouse retina (cepko.med.harvard.edu), and the Hamilton Eye Institute Retina Illumina v6.2 (Mar09) RankInv (www.GeneNetwork.org), which includes genomewide expression analysis of adult retinal mRNA for B6/J, among other strains. Fourteen genes—Bclaf1, 2610016C23Rik, Ahi1, Myb, Hbs1l, Aldh8a1, Vnn1, Stx7, Ctgf, Enpp3, Med23, Akap7, Epb4.1l2, and 2010003K15Rik—met both these criteria (coding and/or regulatory SNPs and retinal expression). Another four genes within the high-density SNP region of Cpnc10 (20–27 Mb) were also of interest: Mtap7, Sgk1, L3mbtl3, and Ptprk. Although these latter four genes have no known coding or regulatory SNPs, they may be worthy of further investigation because they are expressed in the embryonic retina and in the outer nuclear layer (ONL) of the adult retina, evidenced in the SAGE database. These 18 candidate genes identified using this approach are summarized in Table 1.
Table 1.
Candidate Genes
| Gene | Start (Mb) | GenBank ID | Coding SNP ID | Regulatory SNP ID | |
|---|---|---|---|---|---|
| 1 | Bclaf1 | 20.03 | NM_001025392 | rs13460732, rs29337668 | |
| 2 | 2610016C23Rik | 20.07 | NM_027930 | rs48866598: P200L | rs29335130 |
| 3 | Ahi1 | 20.67 | NM_026203 | rs29367573, rs33849385 | |
| 4 | Myb | 20.84 | NM_010848 | rs29363766: Q500R | rs4228162 |
| 5 | Hbs1l | 21.02 | NM_001042593 | rs29323595, rs29369264, rs29323314, rs29336840 | |
| 6 | Aldh8a1 | 21.10 | NM_178713 | rs29318312, rs48185419 | |
| 7 | Vnn1 | 23.61 | NM_011704 | rs29380693: M3T | rs46507393, rs29336794, rs29343431, rs29346467, rs13470937, rs13470936 |
| 8 | Stx7 | 23.87 | NM_016797 | rs13460363 | |
| 9 | Ctgf | 24.32 | NM_010217 | rs8254419: M24T | rs29350272, mCV22788637, rs29365883, mCV22788630, rs3090586 |
| 10 | Enpp3 | 24.49 | NM_134005 | rs29346172: N476S | |
| 11 | Med23 | 24.59 | NM_027347 | rs29313977, rs29369897, rs29365798, rs29359873, rs29358040, mCV23515679, rs29338172, rs29358849 | |
| 12 | Akap7 | 24.89 | NM_018747 | rs29321461 | |
| 13 | Epb4.1l2 | 25.13 | NM_013511 | rs29380930: A640T | rs29321684, rs29313996, rs29332592, rs29361787, rs29360004, rs29331657, rs29319000, rs13470177, rs13470178 |
| 14 | 2010003K15Rik | 25.25 | NC_000076.5 | rs47049296: L32l | rs29382205, rs29321112 |
| 15 | Mtap7 | 19.87 | NM_008635 | ||
| 16 | Sgk1 | 23.87 | NM_011361 | ||
| 17 | L3mbtl3 | 26.00 | NM_172787 | ||
| 18 | Ptprk | 27.79 | NM_008983 | ||
| 19 | Enpp1 | 24.36 | NM_008813 | rs13470869: R650H rs13470868: S680R |
Candidate genes with known coding or regulatory SNPs and retinal expression are listed in rows 1–14. Rows 15–18 list four additional candidate genes that have no known coding or regulatory SNPs but are expressed in the developing retina. The candidate gene in row 19 is Enpp1, the expression of which covaries with Cpnc10 across the RI strain set and maps a cis-eQTL.
Of these 18 candidate genes, one—Myb (myeloblastosis oncogene), a well-studied transcriptional regulator that plays a role in proliferation and differentiation of hematopoietic cells—is especially noteworthy.14 Myb is highly expressed in progenitor cells within neurogenic regions of the adult murine brain, and it drives the proliferation of progenitor cells by suppressing terminal differentiation, maintaining cells in a proliferative state.15,16 The localization of Myb transcripts to the mouse retina on embryonic day (E) 12.5 and E14.5 has been shown with in situ hybridization.17 This expression is also maintained into maturity, including localization to the ONL, as evidenced by immunofluorescence.18 Between the B6/J and A/J strains, there is a missense mutation located in the c-terminus of Myb (Q500R). The prediction of the potential functional impact of this sequence difference was conducted using the online program Polymorphism Phenotyping v2 (http://genetics.bwh.harvard.edu/pph2/), or PolyPhen,19 revealing this amino acid change to possibly alter the function of Myb. No other missense mutations of the candidate genes from Table 1 were predicted to impact protein function. The known functional role of Myb in effecting proliferation, in conjunction with the potentially functional SNP between the parental strains, coupled with retinal expression during embryogenesis and in the ONL during maturity together support this gene as a highly promising candidate from genes 1 to 18 listed in Table 1.
Our second approach to identify promising candidate genes relied on the generation of genomewide eye mRNA expression data for 26 strains of the AXB/BXA RI strain set. This massive data set was uploaded to GeneNetwork and made publicly available for use with various tools of the Web site. Specifically, we used the trait correlations tool to identify genes within the high-density SNP portion of Cpnc10 whose mRNA expression covaried with cone photoreceptor number across the RI strain set. The mRNA expression of nine genes from this interval covaried with cone photoreceptor number (minimum Pearson's product moment correlation, r = |0.4|). Seven of these genes can be dismissed because of poor microarray probes that aligned to multiple transcripts, targeted sequences in the array that had SNPs between the parental strains, or aligned to the intronic region of a gene. For the remaining two genes, Bclaf1 and Enpp1, we sought to determine whether their variation in expression across the strains might be attributed to a regulatory SNP, potentially the variant driving Cpnc10. This was assessed by interval mapping of the expression data across the AXB/BXA RI strain set for both genes. The localization of a significant eQTL of a gene to its chromosomal position indicated the possibility of a cis-acting variant regulating the expression of the gene (cis-eQTL). This exercise eliminated Bclaf1, which mapped an eQTL at a locus disparate from the chromosomal position from the gene (trans-eQTL). However, Enpp1 did map a cis-eQTL (Fig. 5), its expression covarying with Cpnc10 (r = 0.49; P = 0.007); furthermore, there was a 1.26-fold increase in expression from A/J to B6/J. All three attributes of Enpp1 expression, elucidated through use of the eye mRNA expression data set for the AXB/BXA RI strain set, suggested Enpp1 as a promising candidate gene for Cpnc10.
Figure 5.
Expression QTL (eQTL) for Enpp1 centers at 23.46 Mb of Chr 10, the chromosomal locus of this gene indicated by the orange triangle. The potential for a regulatory variant modulating the expression of Enpp1, across the RI strain set, is supported by the presence of this cis-eQTL. Conventions as are as described for Figure 3a.
We also took advantage of the eye mRNA expression data set to generate a putative cone photoreceptor network. This network was generated using the network graph tool on GeneNetwork, and 15 genes were selected with well-established roles in cone photoreceptor function or fate, among them Arr3, Cnga3, Cngb3, Gnat2, Gngt2, Opn1mw, Opn1sw, Pde6c, Pde6h, and Thrb.20 The mRNA expression of 13 of the 15 genes selected for this network covaried with at least one other known cone photoreceptor gene of this group, resulting in a highly interconnected putative cone photoreceptor network. We used this network to determine the covariance, and presumably the biological relevance, of Enpp1 to known cone photoreceptor genes. Enpp1 covaries with more than one-third of these genes in the cone photoreceptor network, including Arr3 and Opn1mw, to which Cpnc10 also covaries (Fig. 6a), further supporting Enpp1 as a promising candidate gene. To the best of our knowledge, this is the first published use of the AXB/BXA whole eye mRNA expression data.
Figure 6.
(a) Covariations of Enpp1 mRNA expression and Cpnc10 (yellow boxes) to each other and to the expression of known cone photoreceptor genes (blue boxes) across the AXB/BXA RI strain set are depicted. Colors indicate the strength of covariance. Known cone photoreceptor genes form a highly interconnected network; Enpp1 covaries with multiple genes within this putative cone photoreceptor network. (b) Covariance of candidate genes from Table 1 (pink boxes), in addition to Enpp1 and Cpnc10, to the putative cone photoreceptor network genes is depicted. Note the lack of covariance of Myb with any other gene; other candidate genes from Table 1, including Epb4.1l2, Mtap7, and Stx7, covary with numerous members of the cone network. Other conventions are as described for (a).
We also determined the covariance of the candidate genes derived from the first strategy described here to this network (excluding entries 11 and 14 from Table 1 because of poor microarray probes). Note that some of these genes, such as Ebp4.1l2, Mtap7, and Stx7, covaried with several of the cone photoreceptor network genes, as can be seen with Enpp1, suggesting they may also be of interest for future studies. Interestingly, our promising candidate gene identified from the first approach, Myb, did not covary with any of the genes that make up this network (Fig. 6b).
Discussion
This study demonstrates that cone photoreceptor number varies conspicuously between two inbred laboratory mouse strains, B6/J and A/J. The high heritability of this trait was made apparent by quantifying cone photoreceptor number across 26 strains of the AXB/BXA RI strain set derived from these same parental strains. Conspicuous interstrain variation was present, spanning a 61% increase from the lowest RI strain (AXB24) to the highest RI strain (AXB23). In fact, two-thirds of the variation in cone photoreceptor number, observed across this population of 128 mice from these RI strains, can be ascribed to an effect of strain. With this large and highly heritable variation in cone photoreceptor number across the AXB/BXA RI strain set, we mapped a single significant QTL on Chr 10, Cpnc10, at 20.09 Mb. The magnitude of the effect of B alleles at this genomic locus accounted for 27% of the difference between the parental strains. We confirmed the presence of this QTL in the B6.A<10> chromosome substitution strain, wherein an anticipated reduction in cone photoreceptor number was found by substituting A alleles throughout Chr 10.
Several inbred strains of albino mice, including A/J, show sensitivity to light-induced damage.21 Comparison of cone photoreceptor number with pigmentation status (GeneNetwork trait ID 10099) across the RI strain set shows a correlation coefficient of only 0.087 (P = 0.67). This lack of covariance between cone photoreceptor number and coat color suggests pigmentation has a negligible effect on this trait. Furthermore, though the A/J strain contains the RPE65 Leu450 variant, which confers susceptibility to light damage,22 composite interval mapping reveals no hint of a secondary QTL at the Rpe65 locus when factoring out the proportion of variance produced at Cpnc10.
Roughly 100 genes lie within the 20- to 32-Mb interval defined by the suggestive LRS threshold. Because the interval distal to 27 Mb is SNP poor, the gene or genes underlying Cpnc10 likely reside proximal to this, between 20 and 27 Mb, a region encompassing roughly 70 genes. From these, 18 promising candidate genes were identified by virtue of coding or regulatory SNPs between the parental strains and/or by their expression localized to the retina. Myb in particular stands out because of the predicted functional impact of the missense mutation between the parental strains; additionally, the known function of Myb in proliferation also supports the potential role of this gene in effecting cone photoreceptor number. Indeed, the lack of coexpression between Myb and any other member of the putative cone photoreceptor network (Fig. 6b) supports the possibility that a functionally altered protein product between A/J and B6/J, and not transcript abundance of Myb, may be driving the variation in cone photoreceptor number between these two strains.
Generation of the AXB/BXA eye mRNA expression data set permitted a complementary approach to identify potential candidate genes associated with phenotypic variation in cone photoreceptor number. This approach led to Enpp1, whose mRNA expression increases by 25% from the A/J to the B6/J strain, is correlated to Cpnc10, and maps a cis-eQTL. The latter suggests the presence of a variant modulating its expression across the AXB/BXA RI strain set, potentially driving Cpnc10. Our search for known variants between the parental strains, A/J and B6/J, revealed no known regulatory SNPs for Enpp1. Although two missense mutations are located within the c-terminal nuclease-like domain of Enpp1 (R650H and S679R), they do not appear to alter the enzymatic activity of this protein.23 Rather, one can assume, based on the differential abundance of Enpp1 transcript and the cis-eQTL it maps, that a variant unknown at this time is modulating the expression of this gene, and, in turn, perhaps cone photoreceptor number. The coexpression of Enpp1 mRNA with multiple genes within the putative cone photoreceptor network, including Arr3 and Opn1mw, to which Cpnc10 also covaries, supports the possibility of a functional role for Enpp1 in effecting cone photoreceptor number. One known function of Enpp1 (ectonucleotide pyrophosphatase/phosphodiesterase) is as a degradative enzyme, located at the apical membrane of the retinal pigment epithelium (RPE). It has been shown that the degradation of ATP by this enzyme alters the balance of purines in the subretinal space, which may result in a change in cellular signaling between the RPE and the photoreceptors.24
We have shown that both Myb and Enpp1 are supported on multiple counts as promising candidate genes. It is, however, important to consider other potential candidate genes from Table 1, including Bcl-2 associated factor 1 (Bclaf1), which has been shown to be required for proper lung development, lymphocyte homeostasis and limb development,25 and connective tissue growth factor (Ctgf), with known roles in proliferation and survival, including loss of eye formation in zebrafish knockdowns.26 Based on these documented functions of Bclaf1 and Ctgf in development, in addition to their expression in the embryonic retina and possible regulatory mutations (Table 1), further study of these candidate genes is warranted.
Interestingly, several genes shown in Table 1 may be argued to have greater potential for a role in retinal degeneration rather than in the initial establishment of the cone photoreceptor population. For example, Abelson-helper integration site-1 (Ahi1) has been implicated in ciliopathy Joubert syndrome, in which retinal dystrophy is one of the phenotypes.27 Indeed, loss of outer segments leading to photoreceptor cell death has recently been shown in the Ahi1 conditional knockout, occurring when the mice are 2 to 3 weeks of age, after the establishment of the photoreceptor population.28,29 Additionally, given that many studies have shown disruptions to the cytoskeletal structure of photoreceptors to be a contributing factor in retinal degeneration,30 the documented association of erythrocyte protein band 4.1-like 2 with the cytoskeletal filament, f-actin of cone photoreceptors,31 suggests a potential role of another of these candidate genes (Epb4.1l2) in retinal degeneration. However, there is a lack of covariance (r = 0.111; P = 0.576) between cone photoreceptor number and the age of all RI mice sampled (GeneNetwork trait ID 10176), which spans a range of roughly 2 to 4 months. This suggests that the variation in the cone photoreceptor population across strains is not due to differences in the progression of retinal degeneration. This is additionally supported by the uniform morphology of cone photoreceptor outer segments seen across the retinal surface; no signs of outer segment degeneration were apparent in the retinas of the parental strains or in the RI strains derived from them.
In summary, by quantifying the variation in cone photoreceptor number across a large recombinant inbred strain set, we have identified a significant QTL at proximal Chr 10, Cpnc10. We confirmed the modulation of cone photoreceptor number by a genetic variant (or variants) on Chr 10 with the chromosome substitution strain B6.A<10>. The SNP-dense region of this QTL was systematically assessed to identify top candidate genes; in particular, we identified Myb and Enpp1. Finally, the use of microarray expression data in conjunction with the phenotype QTL analysis proved to be a powerful resource in the identification and assessment of candidate genes. In the immediate future, analysis of available knockout or conditional knockout mice for these various gene candidates can be pursued. A longer term approach would be to refine the mapping of Cpnc10 to roughly a 1-Mb interval. The resolution of QTL mapping is limited by the number of recombinations across the genome. Increased mapping resolution of a previously identified QTL can be achieved by using heterogeneous stock mice, which are descended from inter-crossings of multiple founder strains and are maintained to maximize the number of recombination events.32 More specifically, the Hitzemann Northport Heterogeneous Stock mice, which are derived from an eight-way inbred strain cross including the parental strains of the AXB/BXA RI strain set, B6/J and A/J, would be ideal for fine-mapping Cpnc10 to generate a substantially reduced number of positional candidate genes.
Footnotes
Supported by the National Institutes of Health Grants EY-011087, EY-019968, and P30 EY-013080.
Disclosure: I.E. Whitney, None; M.A. Raven, None; L. Lu, None; R.W. Williams, None; B.E. Reese, None
References
- 1. Williams RW, Strom RC, Rice DS, Goldowitz D. Genetic and environmental control of variation in retinal ganglion cell number in mice. J Neurosci. 1996;16:7193–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Curcio CA, Allen KA. Topography of ganglion cells in human retina. J Comp Neurol. 1990;300:5–25 [DOI] [PubMed] [Google Scholar]
- 3. Whitney IE, Raven MA, Ciobanu DC, Williams RW, Reese BE. Multiple genes on chromosome 7 regulate dopaminergic amacrine cell number in the mouse retina. Invest Ophthalmol Vis Sci. 2009;50:1996–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whitney IE, Keeley PW, Raven MA, Reese BE. Spatial patterning of cholinergic amacrine cells in the mouse retina. J Comp Neurol. 2008;508:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. Human photoreceptor topography. J Comp Neurol. 1990;292:497–523 [DOI] [PubMed] [Google Scholar]
- 6. Curcio CA, Sloan KR, Jr, Packer O, Hendrickson AE, Kalina RE. Distribution of cones in human and monkey retina: individual variability and radial asymmetry. Science. 1987;236:579–582 [DOI] [PubMed] [Google Scholar]
- 7. Finlay BL, Franco EC, Yamada ES, et al. Number and topography of cones, rods and optic nerve axons in New and Old World primates. Vis Neurosci. 2008;25:289–299 [DOI] [PubMed] [Google Scholar]
- 8. Schadt EE, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geisert EE, Lu L, Freeman-Anderson NE, et al. Gene expression in the mouse eye: an online resource for genetics using 103 strains of mice. Mol Vis. 2009;15:1730–1763 [PMC free article] [PubMed] [Google Scholar]
- 10. Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577 [DOI] [PubMed] [Google Scholar]
- 11. Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Gen Biol. 2001;2:0046.1–0046.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behav Genet. 1981;11:103–114 [DOI] [PubMed] [Google Scholar]
- 13. Singer JB, Hill AE, Burrage LC, et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448 [DOI] [PubMed] [Google Scholar]
- 14. Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol. 2008;20:247–256 [DOI] [PubMed] [Google Scholar]
- 15. Ramsay RG. c-myb a stem-progenitor cell regulator in multiple tissue compartments. Growth Factors. 2005;23:253–261 [DOI] [PubMed] [Google Scholar]
- 16. Malaterre J, Mantamadiotis T, Dworkin S, et al. c-Myb is required for neural progenitor cell proliferation and maintenance of the neural stem cell niche in adult brain. Stem Cells. 2008;26:173–181 [DOI] [PubMed] [Google Scholar]
- 17. Sitzmann J, Noben-Trauth K, Klempnauer KH. Expression of mouse c-myb during embryonic development. Oncogene. 1995;11:2273–2279 [PubMed] [Google Scholar]
- 18. Lee E, Chung YH, Park JY, et al. The distribution of c-myb immunoreactivities in the adult mouse retina. Neurosci Lett. 2004;366:297–301 [DOI] [PubMed] [Google Scholar]
- 19. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corbo JC, Myers CA, Lawrence KA, Jadhav AP, Cepko CL. A typology of photoreceptor gene expression patterns in the mouse. Proc Natl Acad Sci U S A. 2007;104:12069–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. LaVail MM, Gorrin GM, Repaci MA. Strain differences in sensitivity to light-induced photoreceptor degeneration in albino mice. Curr Eye Res. 1987;6:825–834 [DOI] [PubMed] [Google Scholar]
- 22. Danciger M, Matthes MT, Yasamura D, et al. A QTL on distal chromosome 3 that influences the severity of light-induced damage to mouse photoreceptors. Mamm Genome. 2000;11:422–427 [DOI] [PubMed] [Google Scholar]
- 23. Banakh I, Sali A, Dubljevic V, Grobben B, Slegers H, Goding JW. Structural basis of allotypes of ecto-nucleotide pyrophosphatase/phosphodiesterase (plasma cell membrane glycoprotein PC-1) in the mouse and rat, and analysis of allele-specific xenogeneic antibodies. Eur J Immunogenet. 2002;29:307–313 [DOI] [PubMed] [Google Scholar]
- 24. Reigada D, Lu W, Zhang X, et al. Degradation of extracellular ATP by the retinal pigment epithelium. Am J Physiol Cell Physiol. 2005;289:C617–C624 [DOI] [PubMed] [Google Scholar]
- 25. McPherson JP, Sarras H, Lemmers B, et al. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ. 2009;16:331–339 [DOI] [PubMed] [Google Scholar]
- 26. Katsube K, Sakamoto K, Tamamura Y, Yamaguchi A. Role of CCN, a vertebrate specific gene family, in development. Dev Growth Differ. 2009;51:55–67 [DOI] [PubMed] [Google Scholar]
- 27. Parisi MA, Doherty D, Eckert ML, et al. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J Med Genet. 2006;43:334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Louie CM, Caridi G, Lopes VS, et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet. 2010;42:175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Westfall JE, Hoyt C, Liu Q, et al. Retinal degeneration and failure of photoreceptor outer segment formation in mice with targeted deletion of the Joubert syndrome gene, Ahi1. J Neurosci. 2010;30:8759–8768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eckmiller MS. Defective cone photoreceptor cytoskeleton, alignment, feedback, and energetics can lead to energy depletion in macular degeneration. Prog Retin Eye Res. 2004;23:495–522 [DOI] [PubMed] [Google Scholar]
- 31. Spencer M, Moon RT, Milam AH. Membrane skeleton protein 4.1 in inner segments of retinal cones. Invest Ophthalmol Vis Sci. 1991;32:1–7 [PubMed] [Google Scholar]
- 32. Flint J. Mapping quantitative traits and strategies to find quantitative trait genes. Methods. 2011;53:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]