Abstract
Tetraodon nigroviridis is a freshwater pufferfish 20–30 million years distant from Fugu rubripes. The genome of both tetraodontiforms is compact, mostly because intergenic and intronic sequences are reduced in size compared to other vertebrate genomes. The previously uncharacterized Tetraodon genome is described here together with a detailed analysis of its repeat content and organization. We report the sequencing of 46 megabases of bacterial artificial chromosome (BAC) end sequences, which represents a random DNA sample equivalent to 13% of the genome. The sequence and location of rRNA gene clusters, centromeric and subtelocentric satellite sequences have been determined. Minisatellites and microsatellites have been cataloged and notable differences were observed in comparison with microsatellites from Fugu. The genome contains homologies to all known families of transposable elements, including Ty3-gypsy, Ty1-copia, Line retrotransposons, DNA transposons, and retroviruses, although their overall abundance is <1%. This structural analysis is an important prerequisite to sequencing the Tetraodon genome.
[The sequence data described in this paper have been submitted to the EMBL data library under accession nos. AJ245809, AJ270048, AJ245808, AJ270029–AJ270047, DS42722 and AL305790–AL352938.]
The human genome is in the process of being completely sequenced, and an attempt is being made in parallel to systematically identify functionally relevant sequences. Current gene identification methods are based on software predictions or comparisons with expressed sequence tags and still lack accuracy and completeness. Comparisons between the human genomic sequence and the complete sequence of another vertebrate should be a useful complement to rapidly and accurately reveal regions of functional interest. Indeed, two vertebrate genomes that are evolutionarily distant should only show strong conservation of sequences of functional importance (protein coding regions; tRNA; rRNA) while other segments submitted to random mutations will show much less similarity. It has been amply demonstrated that the genomic sequence of a tetraodontiform such as Fugu rubripes is a powerful yet efficient tool to reveal such coding regions (Elgar 1996; Elgar et al. 1999).
We have chosen Tetraodon nigroviridis to develop such comparative analyses on a genome scale (Roest Crollius et al. 2000) because of its widespread availability and trivial and inexpensive maintenance in the laboratory. It was also reasoned that studying a species related to Fugu but distant by 20–30 million years (Crnogorac-Jurcevic et al. 1997) would enable the identification of functionally important sequences that appeared after the human/teleostean divergence. We have initiated a random sequencing approach of the Tetraodon genome based on bacterial artificial chromosome (BAC) end templates and have generated 46 Mb of DNA or 13% of the genome. The average read length is 1 kb, which contributes to making this approach a very fast and cost-effective method of genome scanning. BAC end sequencing provides an added advantage by physically linking two sequences over a relatively short distance (75–200 kb), allowing direct comparisons between linked sequences in Tetraodon and other genomes. It also represents an ideal genomic resource for long-range physical mapping, as well as an STC resource (Mahairas et al. 1999) to assist shotgun sequencing in specific regions.
This Tetraodon genome sample was exploited in combination with fluorescence in situ hybridization experiments, to decipher the organization of repeat sequences. This study serves several purposes. First, repeat sequences occur naturally in multiple copies in the genome either in tandem or in dispersed distribution, and therefore can seriously hamper clustering studies or sequence assemblies. In any case such sequences must be identified and eliminated, generally by masking, during sequence comparison procedures to avoid the formation of unwanted repeat alignments. Second, major satellite and rRNA gene clusters form heterochromatic blocks in the genome that are easily recognizable cytogenetically. These blocks can serve as useful markers when the chromosome formula is difficult to establish, as is the case in pufferfishes (Barat and Khuda-Bukhsh 1984; Miyaki et al. 1995; Grützner et al. 2000; Fischer et al. 2000). Finally, repeat sequences are important elements of the genome from an evolutionary point of view (Charlesworth et al. 1994). They can contribute an important fraction of the DNA in a genome, between <10% for tetraodontiforms (Brenner et al. 1993 and this work) to >50% in some mammalian species. In addition, repeat sequences and in particular transposable elements, can influence chromosome evolution by promoting chromosome breakage, deletions, inversions and amplifications (Lim and Simmons 1994; Dimitri et al. 1997; O'Neill et al. 1998). Transposable elements and tandem repeats are closely associated in heterochromatic regions of the genomes of many distant eukaryotes such as Drosophila (Pimpinelli et al. 1995) and plants (Presting et al. 1998), a situation that further supports the structural role of such repeats in genome evolution (Dimitri and Junakovic 1999). It is therefore of particular interest to investigate repeat distribution in Tetraodon considering its unusual evolution which positions it today as the smallest known vertebrate genome.
We have identified the major satellite sequences, which are localized in the centromeres and acrocentric arms. The complete sequence of rRNA genes has been determined and their cluster localized on a small heteromorphic chromosome. The detection of minisatellite sequences essentially reveals their paucity in the genome. A comprehensive cataloguing of microsatellites compared with Fugu, shows that this genome is particularly rich in polyA stretches. We have found homologies to transposable elements (TEs) belonging to all major families, although their overall abundance is low compared to other eukaryotes. Globally, the genome contains 6.17% of repeated sequence. Taken together, these results represent a structural basis on which new studies focused on genome organization, evolution, and coding potential can be initiated.
RESULTS
Genomic Clone Library Construction, Characterization, and Sequencing
In order to limit possible cloning biases and redundancy in sequencing templates, two BAC libraries were constructed from the same fish specimen, using different vectors (pBAC3e.6 and pBeloBAC11) and two restriction enzymes to fractionate genomic DNA (EcoRI and HindIII). The resulting library A (pBAC3e.6/EcoRI) and library B (pBeloBAC11/HindIII) comprise 20,352 and 22,658 clones respectively. Based on field inversion gel electrophoresis separation of 1792 control clones, the average insert size is 126 kb and 153 kb for libraries A and B respectively. Taking into account that 7% of the clones in each library have no visible insert, both libraries together represent 14.5 genomic equivalents of the Tetraodon genome. A total of 52,619 BAC end sequences have been generated (60% library A, 40% library B). Control clones were also re-sequenced and therefore represent duplicate sequences spread evenly in the library, which serve as indicators of possible errors which may have occurred at any point along the production line. The average raw sequence length is 1075 bases, reduced to 969.2 bases after clipping off vector and low quality sequence at both ends of each read. The resulting sequences contain 3.2% of uncalled bases (N).
A database of 47599 reads was created after removal of redundant (same BAC end sequenced more than once) and contaminating (E. coli, vector) sequences. This set is available for similarity searches at http://www.genoscope.cns.fr/tetraodon and is the basis of the studies described here. The fraction of unique DNA in the database has been estimated by performing a BLAST search (Altschul et al. 1990) of the database against itself. This estimate is essential to evaluate the efficiency of the sequencing strategy as well as the probability to obtain a match when querying the database. In the present case, redundancy can be contributed either by cloning biases, supernumerary reads of the same BAC end or repeated sequences. The major families of repeated sequences are described in this report and include rRNA genes, tandem and interspersed repeats. It is however impossible to exclude at this stage that other types of repeated elements remain undetected, rendering attempts at formally distinguishing between the different types of redundancy unreliable. On the other hand it is possible to clearly separate the unique fraction, i.e. sequences that do not find any other match in the database than themselves, from the redundant fraction. Unique sequences represent 87% of the reads, equivalent to approximately 41 Mb of DNA.
Genome Size and Compositional Patterns
Measurement of haploid DNA content by a variety of methods initially suggested that Tetraodon has a haploid genome size around 380 Mb (Hinegardner 1968; Pizon et al. 1984) However more recent estimates based on flow cytometry indicate a genome size of 350 Mb (Lamatsch et al. 2000). Tetraodon possess 21 chromosome pairs (Grutzner et al. 1999; Fischer et al. 2000) which range in size between approximately 11 and 28 Mb, based on measurements of metaphase chromosomes and correlation with the haploïd genome size of 350 Mb. Thus the largest chromosome is still approximately twice smaller than the smallest human chromosome. The genome is 45.5% G + C rich, with BAC end sequences ranging from 15% to 70% G + C. The relative abundance of dinucleotides (ρXY= fXY/fXfY, where fX denotes the frequency of the nucleotide X and fXY the frequency of the dinucleotide XY) deviates significantly from expected values for CpG (0.60), TpA (0.62), TpT/ApA (1.20) and TpG/CpA (1.21).
Ribosomal RNA Genes
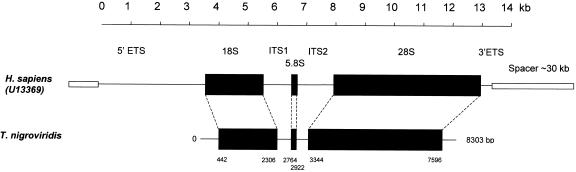
The typical eukaryotic rRNA gene array consists of a tandem repetition of a basic unit, separated from the next by an intergenic spacer (IGS). Each unit starts with a 5′ external transcribed spacer (ETS), followed by the 18S, 5.8S and 28S genes separated by two internal transcribed spacers (ITS1 and ITS2), and ending with a 3′ETS (Fig. 1). Gene sequences are extremely well conserved from mammals to bacteria, although the number and distribution of the genes and of the repeating units may vary between and within species.
Figure 1.
Schematic representation of the human (top) and Tetraodon (bottom) rRNA gene organization. In humans, a 30-kb nontranscribed spacer (open boxes, partially represented) separates a tandem repetition of the 18S, 5.8S, and 28S genes (black boxes) interspersed by intergenic transcribed spacers and external transcribed spacers (ITSs and ETSs, respectively; horizontal lines). In the 8303-bp Tetraodon sequence, the positions of the first and last base of each gene are indicated based on their homology with the human sequence. The sequence and position of the Tetraodon nontranscribed spacer is unknown. A multiple alignment of the Tetraodon sequence is available under EMBL accession number DS42722.
The high degree of sequence conservation of rRNA genes among vertebrates led us to select the complete and well annotated human repeated unit (U13369) to identify the Tetraodon homologous genes. The complete human transcribed unit was searched against the Tetraodon database and retrieved 606 reads (0.73% of the nucleotides in the database; Table 1). Assembly by Phred and Phrap of these sequences delineated one contig that covers the complete transcribed region. We have thus established the first consensus sequence of the transcribed rRNA repeated unit of a fish containing the 18S, 5.8S and 28S genes (Fig. 1). The sequence is 8303 bases long and includes a partial 5′ETS and 3′ETS. Compared to the homologous human sequence which measures 10502 bp, the Tetraodon sequence has smaller intergenic spacers and shows significant deletions in the 28S gene. Fluorescence in situ hybridization experiments with a 28S probe identify a small pair of chromosomes containing a characteristic heterochromatic region (Fig. 2B). This Nucleolar Organizer Region (NOR) is partly 4′,6-diamidino-2-phenylindole (DAPI)- and strongly propidium iodide (PI)-positive and entirely covered by the hybridization signal.
Table 1.
Summary of Tetraodon Sequence Resources, Genome Characteristics and Repeat Abundance, in Comparison with Fugua
| Sequence resource | T. nigroviridis | F. rubripes |
|---|---|---|
| Number of sequences | 47,599 | 52,668 |
| Sequencing protocol | Dye primer/LiCor | Dye terminator/ABI377 |
| Sequencing templates | BAC clone ends | Cosmid shotgun clones |
| Average raw sequence length | 1,075 bp | N.A.b |
| Average sequence length after clipping | 969 bp | 463 bp |
| Total DNA in database | 46,133 Mb | 24,385 Mb |
| Fraction of genome covered | ∼13% | ∼6% |
| Uncalled bases | 3.2% | 5.3%* |
| Genome characteristics | ||
| Genome size | ∼350 Mb | ∼400 Mb |
| Chromosome pairs | 21 | 22 |
| %GC | 45.5% | 47.67% |
| Repeat abundance | ||
| rRNA DNA | 0.77% | N.A. |
| Microsatellites | 3.21% | 2.12%* |
| Centromeric 118-bp satellite | 0.34% | 0.3% |
| Transposable elements | 0.90% | 1.89% |
| Minisatellites | 0.41% | N.A. |
| Subtelocentric 10-bp satellite | 0.54% | N.A. |
| Microsatellite distribution | ||
| Sequences with at least one motif | 79.5% | 40.0% |
| Motifs per sequence (average) | 1.67 | 0.56 |
| Fraction of all 501 motifs observed | 94.8% | 82.8% |
All figures for the Fugu genome are from Elgar et al. (1999), except for those indicated by (*) which are from this work.
N.A., not available.
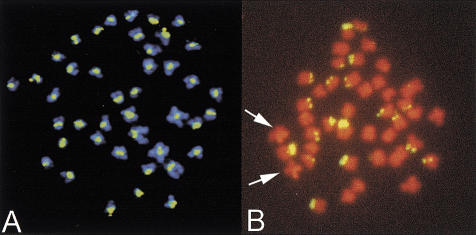
Figure 2.
Fluorescence in situ hybridization of repetitive probes on Tetraodon nigroviridis metaphase chromosome. (A) A cloned 180-bp fragment of the 118-bp satellite hybridizes uniformly to all centromeres. Fluorescein isothiocyanate signals are in green, and chromosomes are counterstained with DAPI. (B) A synthetic probe that includes 4 consecutive monomers of the 10 bp satellite labels specifically the short arms of 10 pairs of subtelocentric chromosomes. Fluorescein isothiocyanate signals are in yellow, and chromosomes are counterstained with DAPI and PI. Arrows indicate the 11th pair of subtelocentric chromosomes that carries the 18S-5.8S-28S rRNA gene clusters and which is strongly stained with propidium iodide.
The sequence of the complete 5S gene (120 bp) and its spacer (289 bp) has also been determined. In all vertebrates the 5S rRNA gene is organized in tandem repetitions and generally in separate cluster(s) from those formed by the 18S, 5.8S and 28S genes. A Tetraodon 5S rDNA PCR product was used as an in situ probe and gives a single signal on the short arms of one of the smallest chromosome pairs, but different from the pair bearing the other rRNA gene cluster. No real size polymorphism could be observed between the two arms. Localization of Tetraodon rRNA gene clusters (5S and 18S-5.8S-28S) on two different chromosome pairs will facilitate the unequivocal identification of the latter in a karyotype where the majority of chromosomes are of similar size (Grutzner et al. 1999; Fischer et al. 2000).
Centromeric Satellite Repeat
Centromeres of higher eukaryotes are often associated with tandem repetitions of a basic repeat unit that do not appear evolutionarily conserved between species, and no definite sequence-specific function has yet been determined for such repeats. However, it is clear that in most species, several—and sometimes all—chromosomes contain the same satellite sequence, indicating that a mechanism of concerted evolution is operating within populations (Elder and Turner 1995). The sequence of satellite repeats has been determined in several fish species, and some have been assigned to centromeres. For instance, tandemly repeated monomers of 355 bp and 168 bp are found in all centromeres of Hoplias malabaricus (Haaf et al. 1993) and of Sparus Aurata (Garrido-Ramos et al. 1994), respectively.
In Tetraodon, we have found a 118-bp repeated monomer in a large number of sequences (0.34% of nucleotides). Its organization in clusters is indicated by the observation that when a 118-bp tandem repeat is found at one end of a BAC, it is frequently found at the other end as well (27% of cases). A cloned monomer was hybridized to Tetraodon chromosomes and labels uniformly all centromeres (Fig. 2A), demonstrating its centromeric origin and pointing towards a concerted evolution of this satellite sequence. However, a more detailed comparison of the sequences of randomly chosen monomers reveals that this repeat is highly variable in a ∼60 bp region, while the remaining half is remarkably constant (Fig. 3A). This sequence variation is present within at least some centromeres, since examination of both end sequences belonging to the same BAC clones (the last eight sequences above the consensus in Fig. 3A) show that each end contains different variants. The monomer has a sequence composition of 57.6% A/T, close to the genome average (56.1% A/T).
Figure 3.
(A) Alignment of 22 different 118 bp monomers of the centromeric satellite sequence. The first 4 monomers with names starting with 118 are cloned PCR products; the first clone was used as in situ probe in Figure 1C. The last 8 monomers, with names identical two by two except for the last letter, are extracted from the two ends of the same BAC clones. (B) Smith-Waterman alignment between the Tetraodon and the Fugu 118 bp repeat unit. The optimal alignment was obtained by comparing in forward and reverse orientation the Tetraodon sequence to a database of 118 versions of the Fugu monomer obtained by shifting the starting position by one base. (C) Alignment of 25 consecutive subtelocentric satellite monomers, together with the resulting consensus sequence. The only nonvariable base is the thymidine in fifth position.
A Fugu tandem repeat sequence of identical monomer size has also been described (Brenner et al. 1993) with a probable centromeric origin (Elgar et al. 1999). A gapped alignment between the two monomer sequences shows 56.6% identity (Fig. 3B).
Subtelocentric Satellite Repeats
A second abundant tandem repeat of monomer size 10 bp was found in Tetraodon BAC end sequences. A prominent feature of this repeat is its high sequence variability, while the monomer size is strictly conserved. For instance, the alignment of 25 consecutive monomers found in a BAC end sequence (accession number AL315101; Fig. 3C) shows that this stretch is composed of 21 variant monomers. Interestingly, a thymidine is always found in the 5th position in the monomer in all sequences examined. Other bases show 4% to 48% variation on the sample described in Figure 3C.
The organization of this repeat in potentially very large arrays was suggested by the observation that out of all BAC clones that contain the repeat at at least one end, 30% of clones contain the repeat at both ends. We have investigated the genome distribution of this repeat. A 40-mer oligonucleotide probe, containing twice the consensus sequence interspersed by the two most abundant variants, was hybridized on Tetraodon metaphase chromosomes. The probe specifically hybridizes to the complete length of the short arms of 10 out of 11 pairs of subtelocentric chromosomes (Fig. 2B). The subtelocentric pair that does not hybridize is the pair bearing the 18S-5.8S-28S rRNA genes.
Similarity searches with the BAC end AL315101 in Fugu sequences identifies sequences that contain a 20-mer tandem repeat. The Tetraodon 10-mer consensus sequence (GGCGTCTGAG) is 80% identical to half of the Fugu 20-mer consensus sequence (GGCATCTGATCCTGGTAGCT), which may point toward a common origin for this satellite sequence in Tetraodontidae.
Minisatellite Repeats
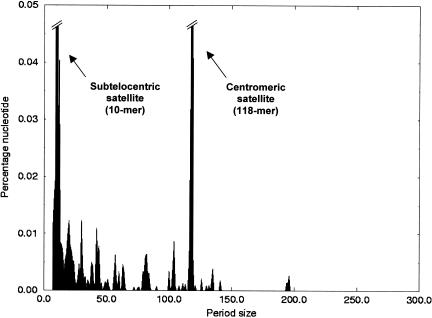
The definition of a minisatellite repeat is not well standardized in the literature and can vary in terms of repeat unit size (or period) and total array size (Franck et al. 1991; Charlesworth 1994). We chose to use this category loosely and include all tandem repeats that are neither microsatellite nor satellite sequences. Thus, our definition includes all sequences of repeat unit larger than 6 bases, tandemly repeated at least 3 times, and that are not satellite sequences. We used the software Tandem Repeat Finder (Benson 1999) with default parameters, except for the maximum period size that was set to 300 bases. Indeed, no motif of more than 300 bases repeated at least 3 times can be detected in sequences of average size 1 kb. Figure 4 shows the percentage of bases in the genome contributed by repeats of period sizes comprised between 7 and 300 bases. The two major peaks correspond to the subtelocentric (10-mer) and centromeric (118-mer) satellite sequences. Clearly no other tandem repeat contributes any substantial amount of DNA. The total fraction of nucleotides represented by minisatellites, excluding the 10-mer and 118-mer repeat, is 0.41%.
Figure 4.
Distribution of the percentage of DNA contributed by tandemly repeated sequences in the Tetraodon genome according to their period size (7 to 300 bases).
Microsatellite Repeats
Microsatellite repeats are defined as short tandem repetitions of monomer units of 1 to 6 bases that are present in most if not all eukaryotic genomes. Their widespread distribution and high heterozygosity have promoted their use as polymorphic markers in genetic mapping (Dib et al. 1996) and population genetics (Jarne and Lagoda 1996). Their identification and characterization is essential in whole genome studies based on sequence analysis because their high frequency and repetitive nature tends to hinder clustering analysis and homology studies. Early characterization of the Fugu genome (Brenner et al. 1993) has shown that microsatellites are the second most abundant class of repeats in this species, and a more exhaustive classification has since been performed (Edwards et al. 1998; Elgar et al. 1999). A direct comparison of microsatellite distribution in Fugu and Tetraodon genomes is possible because both species benefit from large, publicly available sequence samples that have been randomly generated from genomic clones (Elgar et al. 1999 and this work).
Our method, based on the Smith-Waterman algorithm, underestimates the total content of microsatellite sequences in the sample, because only one alignment is produced per motif per sequence. Thus, for instance, if two (CA)n are present in a sequence, only one will be reported. Despite this bias, we observe that 3.21% of the Tetraodon genome consists of microsatellites, versus 1.29% measured by Edwards et al. (1998) in Fugu. This disparity between two figures measured in closely related genomes is not negligible and is most probably due to the different strategies used in both studies. To resolve this, we repeated our study on Fugu genomic sequence (13.7 Mb, Fugu Landmark Mapping Project), a sample size similar to that used by Edwards et al. (1998), and found a total microsatellite content of 2.12%.
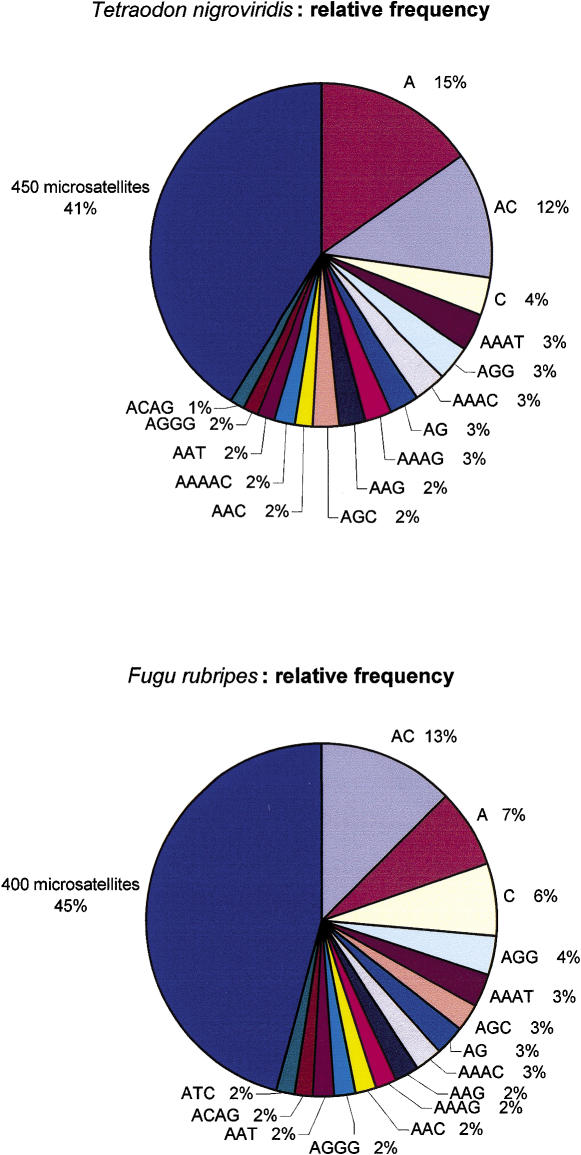
The motif frequency distribution is relatively similar between the Tetraodon and Fugu genomes when analyzed with our approach (Fig. 5), except for one noticeable difference: the polyA repeat is twice as frequent in Tetraodon (15%) than in Fugu (7%). Table 1 summarizes other features of microsatellite distribution in both genomes. There are twice as many reads containing at least one microsatellite in Tetraodon compared to Fugu, which correlates with the Tetraodon sequences being twice as long (969 bp and 473 bp in Tetraodon and Fugu respectively). Provided microsatellites are similarly distributed in both genomes, this constitutes good evidence that their identification is not dependent upon differences in sequence quality or sequencing chemistry between the two samples. A microsatellite occurs on average once every 588 bases in Tetraodon and once every 850 bases in Fugu. The longest microsatellite in Tetraodon is a 502-bp AGAT repeat, and the most abundant in nucleotides are AC (18%) and A (13%) which together constitute 31% of all microsatellites. In Fugu, the same repeats represent only 20% of all microsatellites.
Figure 5.
Distribution of microsatellite relative frequencies in Tetraodon (top) and Fugu (bottom).
Transposable Elements (TEs)
Considering the relative small size of the Tetraodon genome and the impact TEs may have on genome size, it is of interest to investigate their presence in pufferfishes, which have the smallest known vertebrate genome. We have performed a detailed cataloguing of TEs in Tetraodon and show that elements belonging to all known families have been integrated in the genome (Table 2). This observation is based on comparisons between translated Tetraodon genomic sequences and all known eukaryotic TEs annotated in nonredundant proteic and nucleic databases. The 732 BAC end sequences displaying such homologies were then subdivided into the following families based on database annotation: Ty3/gypsy, Ty1/copia, Line, Retrovirus, TC1/mariner and Hobo. The Tetraodon sequences belonging to each group show little or no sequence similarity between each other and thus form distinct families in the genome as suggested by the database matches. The total DNA content of TE-like regions in Tetraodon is only 0.9%, a large fraction of which is contributed by Line elements (0.4%). Out of the 27 TEs that are present in Tetraodon DNA, 10 are more similar to anonymous Fugu sequences than to any cognate TE in public databases (Table 2). From this, we deduce that these TEs are also present in the Fugu genome. TEs belonging to all families are present in both species, except for Hobo and Ty1/copia, which are present in the Tetraodon sequence sample only. However, these families are underrepresented in Tetraodon and their absence in Fugu may simply be a reflection of the smaller amount of DNA currently available for screening in this species (Table 1).
Table 2.
TE Elements Identified in the Tetraodon Genome
| Description | Method | Acc. Nr. | P. value | Fugu | Family | % DNA |
|---|---|---|---|---|---|---|
| F. rubripes SUSHI retrotransposon | NUCX | Gb:AF030881 | 8.5e-99 | Y | Class I Ty3/gypsy | 0.163% |
| B. mori MAG retrotransposon | SPTR | Pir:S08505 | 7.2e-52 | N | ||
| D. melanogaster retrotransposon 17.6 | SPTR | Sw:P04323 | 1.4e-31 | N | ||
| D. melanogaster ZAM retrotransposon | NUCX | gb:AJ000387 | 2.9e-47 | N | ||
| D. ananassae Tom retrotransposon | SPTR | pir:S34639 | 3.1e-49 | Y | ||
| C. elegans CER1 retrotransposon | SPTR | sptr:Q17329 | 6.1e-14 | N | ||
| Z. mays Gypsy-like retrotransposon Reina | NUCX | gb:U69258 | 2.3e-47 | N | ||
| A. comosus Dea1 retrotransposon | NUCX | gb:Y12432 | 1.3e-59 | N | ||
| C. elegans gypsy like retrotransposon | SPTR | sptr:O45092 | 3.3e-18 | Y | ||
| S. tuberosum reverse transcriptase | SPTR | sptr:O64387 | 3.2e-14 | N | Class I Ty1/copia | 0.067% |
| N. tabacum Ttol retrotransposon | SPTR | gp:TOBAA_1 | 1.5e-48 | N | ||
| D. melanogaster I factor | SPTR | sptr:O44317 | 3.3e-08 | N | Class I Line | 0.407% |
| S. mansoni SR2 reverse transcriptase | SPTR | gp:AF025672_1 | 1.0e-06 | Y | ||
| P. spixii CR1-like LINE | SPTR | sptr:O42109 | 5.1e-05 | Y | ||
| D. melanogaster reverse transcriptase | SPTR | pir:A32713 | 7.2e-13 | Y | ||
| A. maritima retrotransposon R2 | SPTR | sptr:O44319 | 2.8e-13 | N | ||
| D. rerio LINE like element | NUCX | gb:AB004653 | 2.2e-28 | Y | ||
| B. baikalensis retrotransposon | NUCX | gb:U18939 | 2.0e-45 | N | ||
| Feline leukemia retrovirus | SPTR | sptr:Q85521 | 2.2e-08 | Y | Retrovirus | 0.055% |
| Walleye epidermal hyperplasia virus type 2 | SPTR | sptr:O36977 | 6.6e-76 | N | ||
| S. salar Tc1-like transposon | NUCX | gb:L12206 | 6.7e-102 | N | Class II Tc1-mariner | 0.211% |
| X. laevis TX1 transposon | SPTR | sw:P14381 | 7.2e-59 | Y | ||
| C. elegans Tc1-like transposase | SPTR | sw:Q21679 | 1.5e-10 | Y | ||
| C. elegans Mariner element | SPTR | sptr:Q23373 | 1.5e-14 | N | ||
| Pacific hagfish TC1-like element | SPTR | pir:B46189 | 7.1e-09 | N | ||
| A. albimanus TC1-like sequence | SPTR | sptr:Q16925 | 2.0e-33 | N | ||
| D. melanogaster Hobo element | NUCX | gb:M69216 | 6.3e-07 | N | Class II Hobo | |
| Total | 0.903% |
The first column indicates the description of the best alignment in public protein (SPTR) or in translated nucleic acid (NUCX) databases. In each case the accession number of the best match, the P value of the alignment with the Tetraodon sequence, its presence (Y) or absence (N) in Fugu, its classification, and its contribution in nucleotides to the Tetraodon genome is indicated.
Of the 732 BAC end sequences that contain a TE, the frequency of this occurring at both ends of a given BAC clone is 10 times higher than expected from the average frequency of TE sequences in the database. This would suggest that TEs have a tendency to be organized in clusters in the Tetraodon genome.
DISCUSSION
A large sample of the Tetraodon nigroviridis genomic sequence has been analyzed to characterize repeat organization in this genome, in comparison with the Fugu genome. The sequence of the Tetraodon genome is 45.5% GC rich, which is within the vertebrates range, between 40% for Bos taurus and 48% for Sus scrofa (Karlin and Mrazek 1997). However, we observe a suppression of the CpG dinucleotide (ρCG = 0.6) as has previously been observed in Fugu (Elgar et al. 1999), although not as strong as in mammals where the odds ratio ρCG is comprised between 0.22 (Mus musculus) and 0.33 (S. scrofa) (Karlin and Mrazek 1997). We also observe a suppression of the TpA dinucleotide and a clear overrepresentation of the TpT/ApA and TpG/CpA dinucleotides. The mechanisms that drive these deviations from the expected values are not yet understood. It is, however, clear that tetraodontiforms and perhaps teleosts in general do not present extremes of suppression or overrepresentation for the same dinucleotides as mammals.
The two major satellite sequences reported here (centromeric and subtelocentric) are located in the main heterochromatic blocks of the chromosome complement. The subtelocentric repeat displays a highly variable monomer sequence within the genome, but its 10-bp length appears strictly conserved. The centromeric satellite, on the other hand, is less variable, but here the conservation of the monomer length has probably extended well beyond the Tetraodon species. Indeed, a similar satellite repeat of exact same monomer length (118 bp) but different sequence (56.6% similar) has been found in Fugu and is presumably also of centromeric origin (Brenner et al. 1993; Elgar 1996; Elgar et al. 1999). This would suggest that for both types of satellites evolutionary constraints have been much stronger on monomer length than on monomer sequence composition. The processes that affect satellite sequence evolution are not yet understood, although a number of models have been proposed (for review see Charlesworth et al. 1994) to explain variations in the number of consecutive monomers rather than the sequence of the monomer itself. We can envisage two possible explanations for the conservation of monomer length despite their sequence variation. It is possible that a still-unknown structural role for such satellite sequences requires a fixed monomer length but places few requirements on sequence composition per se. The alternative is that maintenance of the monomer length may only be the consequence of an amplification mechanism that would generate motifs of identical size, but without any strict requirement on sequence composition, except perhaps for a few critical bases. The poor sequence homogeneity of the 10-bp subtelocentric satellite is at odds with the generally accepted notion of concerted evolution that tend to maintain the sequence similarity of repeating units within a population or a species (Elder and Turner 1995).
Microsatellite sequence distributions have been investigated in a number of vertebrate species, although different software, sample size, and even microsatellite definition were often used (Beckmann and Weber 1992; Edwards et al. 1998; Jurka and Pethiyagoda 1995; Moran 1993; Van Lith and Van Zutphen 1996). Precise comparisons are therefore limited to studies performed in identical conditions. The most striking differences between Tetraodon and Fugu concern the overall microsatellite content (3.21% and 2.12% of the genome, respectively) and the overrepresentation of the mononucleotide A in Tetraodon (15% versus 7%). Poly(A) tails are also the most abundant microsatellite family in the human genome, where they are often introduced by retrotransposons, and in particular by Line and Alu sequences (Boeke 1997). In the Tetraodon genome such retrotransposons are rare (Line) or absent (Alu), and cannot be considered as a source of overrepresentation for poly(A) repeats.
TEs are DNA sequences that can move or copy themselves within a host genome, to which they can contribute a large fraction. For instance, approximately 50% of the maïze (SanMiguel et al. 1996), 35% of the human (Smit 1996), and 10% of the Drosophila melanogaster (Finnegan 1989) genomes are made of such elements. They can be classified according to their transposition mechanisms. Class I elements replicate via an RNA intermediate and may be flanked by long terminal repeats (LTR-retrotransposons, such as Ty3-gypsy and Ty1-copia families) or end with an A-rich tail in 3′ (non-LTR retrotransposons, such as the LINE and SINE families). Class II elements are essentially DNA-based transposons that code for a transposase and include Tc1-mariner and Hobo families. Early studies in Fugu on a small sequence sample concluded that this genome was devoid of interspersed repeats (Brenner et al. 1993). However, a Ty3/gypsy LTR-retrotransposon and a Line element have since been described in this genome (Poulter and Butler 1998; Poulter et al. 1999) and additional homologies to reverse transcriptase identified (Elgar et al. 1999). TEs have been documented in many teleosts (Britten et al. 1995; Duvernell and Turner 1998; Flavell and Smith 1992; Ivics et al. 1996; Izsvak et al. 1995; Koga et al. 1996; Tristem et al. 1995; ). In Tetraodon, the representation of these sequences is below 1%, similar to the 1.89% found in Fugu. It appears, therefore, that although a wide variety of TEs have repeatedly integrated the genome of pufferfishes, their amplification and spreading has been drastically limited compared to other eukaryotes. It is possible that this situation is related to the fact that these genomes are the smallest among vertebrates. The mechanisms that have limited TE amplification in the pufferfish genomes are not known, but investigating their distribution and local organization in the chromosome complement may shed light on this unusual phenomenon.
The characterization of the Tetraodon genome presented here lays the foundation for comparative genomic studies that may take several orientations. From an evolutionary point of view, results of rRNA genes and satellite sequences, when compared to those of other teleosts, particularly Fugu, may help us understand the complex processes involved in repeat dynamics over relatively short evolutionary distances in vertebrates. Comparative genomics with Tetraodon will, however, take its full dimension in the context of gene identification and analysis (Roest Crollius et al. 2000). Gene identification in human and other vertebrates sequence is one of the primary goals in sequencing Tetraodon. However, a large sample of teleost genomic sequence will also be invaluable to help us understand phenomenons such as genome duplication (Amores et al. 1998; Wittbrodt et al. 1998), or the importance and extent of conserved synteny over long evolutionary distances.
METHODS
Fluorescence In Situ Hybridization
All specimens were provided by the same supplier. We don't know their geographic origin, but they were positively identified as Tetraodon on the basis of morphological characters and genotyping using mitochondrial sequences. Fishes were injected with 2μl/g b.w. of 0.05% colchicine solution 1 hr. 15 min. before killing. Cephalic kidney and spleen were separated on a 350-μm mesh stainless steel sieve directly in a 0.075-m KCl hypotonic solution. After a 30-min hypotonic treatment at 29°C, suspension was centrifuged, and the pellet was fixed for 20 min in a 3:1 methanol–acetic acid solution that was changed only one time. The fixed cell suspension was immediately dropped on cleaned slides and stored deep-frozen at −20°C after 30 min drying. All probes were labeled with digoxigenin (Boehringer Mannheim) and hybridized according to standard protocols. The centromeric probe was a 180-bp PCR product cloned in the pAmp1 system (Gibco BRL) using primers (5′- ATGCAGCACACAGATTTCCA-3′) and (5′-TCCATCATTCTGCACCAAAC-3′). The subtelocentric probe was a 40-base oligomer (GGCGTCTGAGGGCGTCTGATGGTGTCTGATGGCGTCTGAT) consisting of two consensus monomers interspersed with the two most frequent variants. The probe was synthesized with a 5′ digoxigenin label (Genosys Biotechnologies Ltd.).
BAC Library Construction and Sequencing
Two BAC libraries were constructed from erythrocyte DNA from a single Tetraodon specimen identified as such by morphological characters and genotyping using mitochondrial sequences. DNA was partially digested with EcoRI (library A) and HindIII (library B) and separated on a 1% agarose gel by pulse field gel electrophoresis. For each digest, three size-selected samples (∼50 ng) ranging from approximately 100 kb to 175 kb were ligated to 10-ng vector DNA (pBACe3.6 for library A; pBeloBAC11 for library B). The BAC vectors pBeloBAC11 (Kim et al. 1996) and pBACe3.6 (Genbank accession number U80929) were gifts from H. Shizuya, Department of Biology, California Institute of Technology, Pasadena, CA and P. de Jong, Roswell Park Cancer Institute, Human Genetics Department, Buffalo, NY, respectively. Ligation reactions were electroporated into DH10B electrocompetent cells (Gibco-BRL) and plated on 2YT agar containing 12.5 μg/ml chloramphenicol and 5% saccharose. Recombinant clones were picked in microtiter plates, grown in 2YT media containing 12.5 μg/ml chloramphenicol and 5% glycerol, and subsequently frozen at −80°C. In total, 20,352 clones were picked from library A (EcoRI/pBAC3e.6) and 22,658 from library B (HindIII/pBeloBAC11). A sub-library, termed the control library, was arranged by selecting 16 clones in the central part of each microtiter plate (1792 clones) of libraries A and B. DNA from all control clones was isolated, digested by NotI to release the insert, and separated by field inversion gel electrophoresis in order to characterize a representative amount of clones covering the entire libraries. All clones in the control library were also resequenced. Templates for sequencing were prepared by alkaline lysis and purified on Qiagen columns. Sequences were obtained by sequencing the same template with two different dye primers in the same reaction. Four reactions were required in total, one for each base. One reaction contained 25 ng/μl DNA, 0.1 μm each primer, and 4.5 μl ThermoSequenase mix (Amersham) in a final volume of 11 μl. Primers were TET3 (TGACACTATAGAAGGATCCG) and T7 (TAATACGACTCACTATAGGG) for BACs from library A and BELO1 (CTATTTAGGTGACACTATAG) and T7 for BACs from library B. Reactions were loaded on 4.8% acrylamide gels on LiCor4200 machines, and images were collected and analyzed by BaseImagir V4.00. Graph files were then transferred to a UNIX environment, and sequences that showed at least a 300-base window containing <6 ambiguous bases were further processed by routine quality checks and vector clipping prior to analysis.
Sequence Comparison and Assembly
All sequence comparisons between large sets of sequences were performed using standard algorithms such as BLAST (Altschul et al. 1990) or Smith-Waterman (Smith and Waterman 1981) implemented in LASSAP version 1.1.3 (Large Scale Sequence Comparison Package; [Glemet and Codani 1997]). Most calculations were performed on one digital quadriprocessor (AXP 21164; each processor at 440 MHz), although when required, we used up to four quadriprocessors simultaneously. Sequence assembly was performed with Phrap and Phred (Ewing and Green 1998)
Tandem Repeat Analysis
The Tetraodon sequences consist of 47,599 single reads of average size 969.57 bases (45,742 Mb of DNA). For minisatellite detection, the software Tandem Repeat Finder (version 2.02, [Benson 1999]) was used with the following parameters: match: 2, mismatch: 7, delta: 7, PM: 80, PI: 10, minscore: 50, maxperiod: 300. The output was filtered to retain motifs of period size of at least 7 bases, repeated 3 times or more. When adding the percentage of bases contributed by each motif size, redundant motifs were eliminated by taking into account only the motifs with the smallest period size. For microsatellite analysis, our approach is very similar to that used for the identification of microsatellites in Fugu (Edwards et al. 1998), although some modifications were made. The repeat definition is the same, i.e., a motif of size 1 to 6 bases repeated at least three times, and of a total size of at least 12 bases. We also allowed up to 15% variation over the complete length of the sequence, between the microsatellite and the perfectly repeated motif of same length. However, here this definition is strictly observed regardless of the size of the repeat and implies that a 12-base microsatellite may also include up to one mismatch. This double constraint on size and identity is used when selecting microsatellites that respect the definition and eliminates the need for an arbitrary minimal score. The Fugu sequences were retrieved from the Human Genome Mapping Project web site (http://fugu.hgmp.mrc.ac.uk/fugu/fugu) and consist of 29,078 sequences (release 07/20/98) of average size of 473 bases (13,753 Mb of DNA). The reference microsatellite library consists of all 501 possible motifs from monomer to hexamer, repeated over 500 bases, in forward and in reverse complement (1002 sequences; (Jin et al. 1994). Comparisons between this library and pufferfish genomic DNA were performed exclusively with the Smith-Waterman algorithm (Smith and Waterman 1981) implemented in LASSAP version 1.1.3. The scoring matrix and gap costs were as follows: match +10, mismatch −30, ambiguity (N) −5, gap opening −40, gap extension −30. The results consist of the best local alignment per sequence and per motif (47.3 million alignments), to which two filters are applied. The first retains alignments that respect the definition of a microsatellite: a repetition of at least 3 motifs of at least 12 bases, with at least 85% identity over the complete length of the alignment. In cases where several similar motifs overlapped over the same region of a query sequence, a second filter was applied to retain only the motif with the highest percentage of identity.
Acknowledgments
We thank Patrick Lafaite and the Museum National d'Histoire Naturelle for assistance with photographic work and the sequencing teams of Genoscope, in particular Patrick Wincker and Philippe Brottier.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
After the submission of this article, an additional 100 Mb of Tetraodon DNA has been submitted to the EMBL data library under accession nos. AL163976–AL305789.
Footnotes
E-MAIL hrc@genoscope.cns.fr; FAX 33 1 608 72589.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Barat A, Khuda-Bukhsh AR. Karyomorphology of a sea-frog Tetraodon fluviatilis (Tetraodontidae, pisces) Current Science. 1984;53:1108–1109. [Google Scholar]
- Beckmann JS, Weber JL. Survey of human and rat microsatellites. Genomics. 1992;12:627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD. LINEs and Alus - the polyA connection. Nature Genetics. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–8. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Britten RJ, McCormack TJ, Mears TL, Davidson EH. Gypsy/Ty3-class retrotransposons integrated in the DNA of herring, tunicate, and echinoderms. J Mol Evol. 1995;40:13–24. doi: 10.1007/BF00166592. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–20. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T, Brown JR, Lehrach H, Schalkwyk LC. Tetraodon fluviatilis, a new puffer fish model for genome studies. Genomics. 1997;41:177–84. doi: 10.1006/geno.1997.4646. [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–4. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Dimitri P, Arca B, Berghella L, Mei E. High genetic instability of heterochromatin after transposition of the LINE-like I factor in Drosophila melanogaster. Proc Natl Acad Sci. 1997;94:8052–7. doi: 10.1073/pnas.94.15.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P, Junakovic N. revising the selfish DNA hypothesis: new evidence on accumulation of transposable elemnts in heterochromatin. Trends Genet. 1999;15:123–124. doi: 10.1016/s0168-9525(99)01711-4. [DOI] [PubMed] [Google Scholar]
- Duvernell DD, Turner BJ. Swimmer 1, a new low-copy-number LINE family in teleost genomes with sequence similarity to mammalian L1. Mol Biol Evol. 1998;15:1791–3. doi: 10.1093/oxfordjournals.molbev.a025906. [DOI] [PubMed] [Google Scholar]
- Edwards YJ, Elgar G, Clark MS, Bishop MJ. The identification and characterization of microsatellites in the compact genome of the Japanese pufferfish, Fugu rubripes: perspectives in functional and comparative genomic analyses. J Mol Biol. 1998;278:843–54. doi: 10.1006/jmbi.1998.1752. [DOI] [PubMed] [Google Scholar]
- Elder JF, Jr, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- Elgar G. Quality not quantity: the pufferfish genome. Hum Mol Genet. 1996;5:1437–42. doi: 10.1093/hmg/5.supplement_1.1437. [DOI] [PubMed] [Google Scholar]
- Elgar G, Clark MS, Meek S, Smith S, Warner S, Edwards YJ, Bouchireb N, Cottage A, Yeo GS, Umrania Y, et al. Generation and analysis of 25 Mb of genomic DNA from the pufferfish Fugu rubripes by sequence scanning. Genome Res. 1999;9:960–71. doi: 10.1101/gr.9.10.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–94. [PubMed] [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–7. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Fischer C, Ozouf-Costaz C, Roest Crollius H, Dasilva C, Jaillon O, Bouneau L, Bonillo C, Weissenbach J, Bernot A. Karyotype and chromosomal localization of characteristic tandem repeats in the pufferfish Tetraodon nigroviridis. Cytogenet Cell Genet. 2000;88:50–55. doi: 10.1159/000015484. [DOI] [PubMed] [Google Scholar]
- Flavell AJ, Smith DB. A Ty1-copia group retrotransposon sequence in a vertebrate. Mol Gen Genet. 1992;233:322–6. doi: 10.1007/BF00587596. [DOI] [PubMed] [Google Scholar]
- Franck JPC, Harris AS, Bentzen P, Denovan-Wright EM, Wright JM. Oxford Surveys on Eukaryotic Genes. Oxford, UK: Oxford University Press; 1991. Organization and evolution od satellite, minisatellite and microsatellite DNAs in teleost fishes; pp. 51–82. [PubMed] [Google Scholar]
- Garrido-Ramos MA, Jamilena M, Lozano R, Ruiz Rejon C, Ruiz Rejon M. Cloning and characterization of a fish centromeric satellite DNA. Cytogenet Cell Genet. 1994;65:233–237. doi: 10.1159/000133637. [DOI] [PubMed] [Google Scholar]
- Glemet E, Codani J. Lassap, a large scale sequence comparisons package. CABIOS. 1997;13:137–143. doi: 10.1093/bioinformatics/13.2.137. [DOI] [PubMed] [Google Scholar]
- Grützner F, Lutjens G, Rovira C, Barnes DW, Ropers HH, Haaf T. Classical and molecular cytogenetics of the pufferfish Tetraodon nigroviridis. Chromosome Res. 2000;7:655–62. doi: 10.1023/a:1009292220760. [DOI] [PubMed] [Google Scholar]
- Haaf T, Schmid M, Steinlein C, Galetti PM, Jr, Willard HF. Organization and molecular cytogenetics of a satellite DNA family from Hoplias malabaricus (Pisces, Erythrinidae) Chromosome Res. 1993;1:77–86. doi: 10.1007/BF00710610. [DOI] [PubMed] [Google Scholar]
- Hinegardner R. Evolution of Celullar DNA Content in Teleost fishes. The American Naturalist. 1968;102:517–523. [Google Scholar]
- Ivics Z, Izsvak Z, Minter A, Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci. 1996;93:5008–13. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvak Z, Ivics Z, Hackett PB. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio) Mol Gen Genet. 1995;247:312–22. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- Jarne P, Lagoda PJL. Microsatellites, from molecules to populations and back. Trends Ecol Evol. 1996;11:424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- Jin L, Zhong Y, Chakraborty R. The exact numbers of possible microsatellite motifs. Am J Hum Genet. 1994;55:582–583. [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: compilation and analysis. J Mol Evol. 1995;40:120–6. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrazek J. Compositional differences within and between eukaryotic genomes. Proc Natl Acad Sci. 1997;94:10227–32. doi: 10.1073/pnas.94.19.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UJ, Birren BW, Slepak T, Mancino V, Boysen C, Kang HL, Simon MI, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–8. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H. Transposable element in fish. Nature. 1996;383:30. doi: 10.1038/383030a0. [DOI] [PubMed] [Google Scholar]
- Lamatsch DK, Steinlein C, Schmid M, Schartl M. Noninvasive determination of genome size and ploidy level in fishes by flow cytometry: detection of triploid Poecilia formosa. Cytometry. 2000;39:91–5. doi: 10.1002/(sici)1097-0320(20000201)39:2<91::aid-cyto1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays. 1994;16:269–75. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- Mahairas GG, Wallace JC, Smith K, Swartzell S, Holzman T, Keller A, Shaker R, Furlong J, Young J, Zhao S, et al. Sequence-tagged connectors: a sequence approach to mapping and scanning the human genome. Proc Natl Acad Sci. 1999;96:9739–44. doi: 10.1073/pnas.96.17.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki K, Tabeta O, Kayano H. Karyotypes in siw species of pufferfishes Takifugu (Tetraodondontidae, Tetraodontiformes) Fisheries Science. 1995;61:594–598. [Google Scholar]
- Moran C. Microsatellite repeats in pig (Sus domestica) and chicken (Gallus domesticus) genomes. J Hered. 1993;84:274–80. doi: 10.1093/oxfordjournals.jhered.a111339. [DOI] [PubMed] [Google Scholar]
- O'Neill RJ, O'Neill MJ, Graves JA. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci. 1995;92:3804–8. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V, Cuny G, Bernardi G. Nucleotide sequence organization in the very small genome of a tetraodontid fish, Arothron diadematus. Eur J Biochem. 1984;140:25–30. doi: 10.1111/j.1432-1033.1984.tb08062.x. [DOI] [PubMed] [Google Scholar]
- Poulter R, Butler M. A retrotransposon family from the pufferfish (fugu) Fugu rubripes. Gene. 1998;215:241–9. doi: 10.1016/s0378-1119(98)00296-0. [DOI] [PubMed] [Google Scholar]
- Poulter R, Butler M, Ormandy J. A LINE element from the pufferfish (fugu) Fugu rubripes which shows similarity to the CR1 family of non-LTR retrotransposons. Gene. 1999;227:169–79. doi: 10.1016/s0378-1119(98)00600-3. [DOI] [PubMed] [Google Scholar]
- Presting GG, Malysheva L, Fuchs J, Schubert I. A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 1998;16:721–728. doi: 10.1046/j.1365-313x.1998.00341.x. [DOI] [PubMed] [Google Scholar]
- Roest Crollius H, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fizames C, Wincker P, Brottier P, Quetier F, Saurin W, et al. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nature Genet. 2000;25:235–238. doi: 10.1038/76118. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–8. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–8. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–7. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Tristem M, Kabat P, Herniou E, Karpas A, Hill F. Easel, a gypsy LTR-retrotransposon in the Salmonidae. Mol Gen Genet. 1995;249:229–36. doi: 10.1007/BF00290370. [DOI] [PubMed] [Google Scholar]
- Van Lith HA, Van Zutphen LF. Characterization of rabbit DNA microsatellites extracted from the EMBL nucleotide sequence database. Anim Genet. 1996;27:387–95. doi: 10.1111/j.1365-2052.1996.tb00505.x. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Meyer A, Schartl M. More genes in fish? Bioessays. 1998;20:511–515. [Google Scholar]