Abstract
The molecular mechanisms underlying major phenotypic changes that have evolved repeatedly in nature are generally unknown. Pelvic loss in different natural populations of threespine stickleback fish has occurred by regulatory mutations deleting a tissue-specific enhancer of the Pituitary homeobox transcription factor 1 (Pitx1) gene. The high prevalence of deletion mutations at Pitx1 may be influenced by inherent structural features of the locus. Although Pitx1 null mutations are lethal in laboratory animals, Pitx1 regulatory mutations show molecular signatures of positive selection in pelvic-reduced populations. These studies illustrate how major expression and morphological changes can arise by single mutational leaps in natural populations, producing new adaptive alleles via recurrent regulatory alterations in a key developmental control gene.
Evolutionary biology has been animated by long-standing debates about the number and type of genetic alterations that underlie evolutionary change. Questions about the roles of genetic changes of infinitesimally small versus large effects; the origin of traits by either natural selection or genetic drift; and the relative importance of coding and regulatory changes in evolution are currently being actively investigated (1-4). One of the classic examples of major evolutionary change in vertebrates is the extensive modification of paired appendages seen in different species (5). Although essential for many forms of locomotion, paired appendages have also been repeatedly lost in some fish, amphibian, reptile, and mammalian lineages, likely via selection for streamlined body forms (6).
Threespine stickleback fish (Gasterosteus aculeatus) make it possible to analyze the evolution, genetics, and development of major skeletal changes in natural populations (7). The pelvic apparatus of marine sticklebacks consists of prominent serrated spines that articulate with an underlying pelvic girdle that extends along the ventral and lateral sides of the fish (inspiring the scientific name Gasterosteus aculeatus, or bony stomach with spines). Although most sticklebacks develop a robust pelvic apparatus, over two dozen widely-distributed and likely independent freshwater stickleback populations show partial or complete loss of pelvic structures (8). Several factors may contribute to repeated evolution of pelvic reduction, including the absence of gape-limited predatory fish, limited calcium availability, and predation by grasping insects (9-12).
Genome-wide linkage mapping has identified a single chromosome region that explains more than two thirds of the variance in pelvic size in crosses with pelvic-reduced sticklebacks (13-15). This region contains Pituitary homeobox 1 (Pitx1), a gene expressed in hindlimbs but not forelimbs of many different vertebrates, and required for normal hindlimb development (13). Although the Pitx1 gene of pelvic-reduced sticklebacks shows no protein-coding changes compared to ancestral marine fish, its expression in the developing pelvic region is almost completely lost (13, 16). Based on the map location, changes in expression, and directional asymmetry shared in both Pitx1-null mice and pelvic-reduced sticklebacks, cis-regulatory mutations at the Pitx1 locus have been proposed as the basis of stickleback pelvic reduction (13). However, regulatory mutations are difficult to identify, and the actual sequences controlling pelvic reduction have remained hypothetical (2).
cis-regulatory changes at Pitx1 locus
Although Pitx1 represents a strong candidate gene for pelvic reduction, other genes in the larger chromosome region could be the real cause of pelvic loss, leading to secondary or trans-acting reduction of Pitx1 expression (2). To test this possibility, we generated F1 hybrids between pelvic-complete (Friant Low (FRIL) and pelvic-reduced (Paxton Lake Benthic (PAXB)) sticklebacks (see table S1 for geographic location of all populations used in this study, 17). F1 hybrid fish develop pelvic structures and contain both Pitx1 alleles in an identical trans-acting environment. Strikingly, the PAXB allele was expressed at significantly lower levels than the FRIL allele in the restored pelvic tissue of F1 hybrids (n=19, two-tailed t-test, P < 0.001, Fig. 1). Reduced expression of the PAXB allele was tissue-specific, since both Pitx1 alleles were expressed at similar levels in F1 hybrid head tissue. As a control we generated F1 hybrids between two pelvic-complete populations (FRIL and Little Campbell River (LITC), Fig. 1). In this cross, both Pitx1 alleles were expressed at comparable levels in both heads and pelves. Allele-specific down-regulation of Pitx1 in the FRIL × PAXB cross shows that pelvic-specific loss of Pitx1 expression is due to cis-regulatory change(s) at Pitx1 itself, and not to overall failure of pelvic development, or changes in unknown trans-acting factors.
Figure 1.
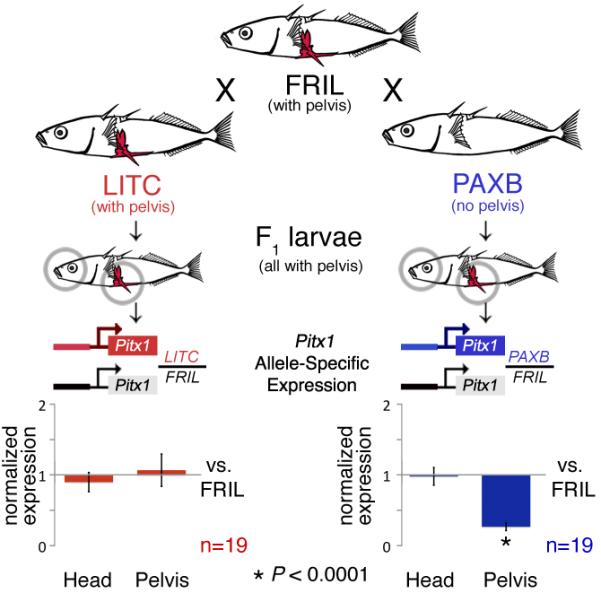
Alleles of Pitx1 from pelvic-complete (FRIL, LITC) and pelvic-reduced populations (PAXB) were combined in F1 hybrids, and brain and pelvic tissues were isolated to compare the expression of either the LITC or PAXB allele normalized to the level of expression of the FRIL allele in the same trans-acting environment. Expression of the PAXB Pitx1 allele is greatly reduced in the pelvis but not the head of F1 hybrids (two-tailed t-test, P < 0.0001), indicating a tissue-specific, cis-regulatory change in the Pitx1 locus.
Fine mapping of pelvic regulatory region
To further localize the position of the cis-acting changes, we looked for the smallest chromosome region co-segregating with bilateral absence of pelvic structures in a cross between pelvic-complete (Japanese marine (JAMA) and pelvic-reduced (PAXB) fish, 13). High resolution mapping identified a 124kb minimal interval, containing only the Pitx1 and Histone 2A (H2AFY), genes, which showed perfect concordance between PAXB alleles and absence of the pelvis (Fig. S1A).
Recombination in natural populations can also be used to narrow the size of regions controlling polymorphic traits in sticklebacks (18). We therefore tested whether markers in the Pitx1 region were associated with presence or absence of pelvic structures in lakes with dimorphic stickleback forms: benthic and limnetic sticklebacks from Paxton Lake, British Columbia (PAXB/PAXL), and pelvic-complete and pelvic-reduced sticklebacks from Wallace Lake, AK (WALR/WALC, Fig. S2) (13, 14). Microsatellite markers located in an intergenic region approximately 30 kb upstream of Pitx1 showed highly significant allele frequency differences in fish with contrasting pelvic phenoytpes (Fig. S1B, table S2; P < 10−35). In contrast, markers around the Pitx1 and H2AFY coding regions showed little or no differentiation above background levels. These results suggest that an approximately 23 kb intergenic region upstream of Pitx1 controls pelvic development. This region is conserved among zebrafish and other teleosts (Fig. 2A), suggesting it may contain ancestrally conserved regulatory enhancers.
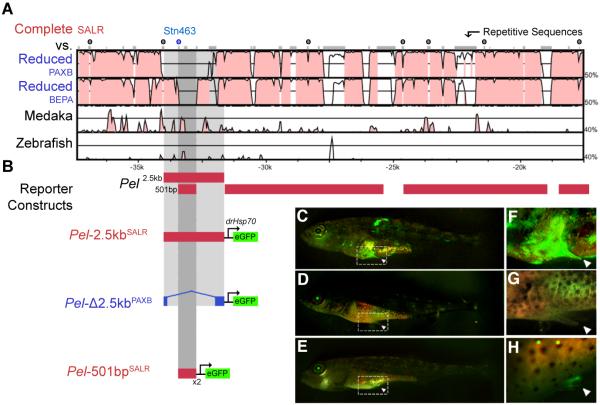
Figure 2.
(A) VISTA/mLAGAN alignment of Pitx1 candidate region from pelvic-complete stickleback (SALR), medaka and zebrafish. Red peaks, >40% sequence identity in 20bp sliding windows; grey bars at top, repetitive sequence;  symbols, microsatellite markers used in association mapping in Fig. S1). (B) Reporter gene expression in transgenic animals. (C) Pel-2.5kbSALR from a marine population drives tissue-specific EGFP (green) expression in the developing pelvic bud of Swarup stage 32 larvae (36); (F) detail. (D and G) Altered Pel-Δ2.5kbPAXB sequence from pelvic-reduced PAXB stickleback fails to drive pelvic EGFP expression. (E and H) A smaller fragment from marine fish, Pel-501bpSALR, also drives EGFP expression in the developing pelvic bud of multiple St. 30 larvae. This region is completely missing in PAXB.
symbols, microsatellite markers used in association mapping in Fig. S1). (B) Reporter gene expression in transgenic animals. (C) Pel-2.5kbSALR from a marine population drives tissue-specific EGFP (green) expression in the developing pelvic bud of Swarup stage 32 larvae (36); (F) detail. (D and G) Altered Pel-Δ2.5kbPAXB sequence from pelvic-reduced PAXB stickleback fails to drive pelvic EGFP expression. (E and H) A smaller fragment from marine fish, Pel-501bpSALR, also drives EGFP expression in the developing pelvic bud of multiple St. 30 larvae. This region is completely missing in PAXB.
A small enhancer drives pelvic expression of Pitx1
To test for regulatory functions in the Pitx1 intergenic region, we cloned different subfragments upstream of a basal promoter and enhanced green fluorescent protein (EGFP) reporter gene (Fig. 2B) (19). The hsp70 promoter drives modest or no EGFP expression except in the eye (19). A construct containing a 2.5kb fragment from a marine, pelvic-complete fish (Salmon River (SALR)) drove consistent EGFP expression in the developing pelvic region of transgenic sticklebacks (4 of 5 independent transgenics, Fig. 2C, F). A smaller 501bp subfragment also drove highly specific pelvic expression (7 of 9 transgenics; Fig. 2E, H). No consistent expression was seen in pectoral fins, or other sites of normal Pitx1 expression, including mouth, jaw, and pituitary (13, 16). Thus, the non-coding region upstream of Pitx1 contains a tissue-specific enhancer for hindfin expression, which we term “Pel”. Pel shows sequence conservation across distantly related teleost fish (Figs. 2A, S3), and contains multiple predicted transcription factor binding sites that might contribute to spatially restricted expression in the developing pelvic region (Fig. S4).
Transgenic rescue of pelvic reduction
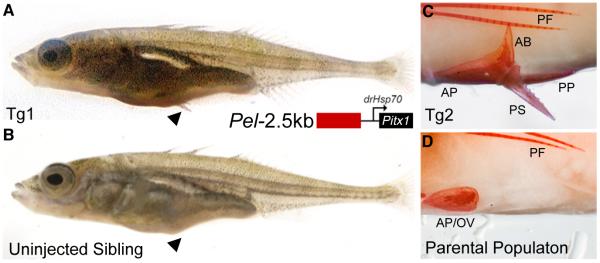
If regulatory changes in Pitx1 underlie pelvic reduction in sticklebacks, restoring pelvic expression of Pitx1 should rescue pelvic structures. We cloned the 2.5 kb Pel region from a pelvic-complete population (SALR) upstream of a Pitx1 minigene prepared from coding exons of a pelvic-reduced fish (Bear Paw Lake (BEPA)) (14). The rescuing construct was injected into fertilized eggs of BEPA fish, which normally fail to develop any pelvic spine, and show no more than a small vestigial remnant of the underlying pelvic girdle (Fig. 3B, D, pelvic score ≤ 3 Fig. S5, 12). Transgenic fry showed variable but enhanced development of external pelvic spines compared to control uninjected siblings (clutch 1, n=16 injected and 11 uninjected fish, Wilcoxon rank-sum test, W=1073.5, P < 0.01; clutch 2, n=4 injected and 18 uninjected fish, W=513, P < 2.3×10−9; Fig. 3A). Alizarin red skeletal preparations of two adult transgenic fish revealed prominent serrated spines articulating with an enlarged, complex pelvic girdle containing anterior, posterior and ascending branch structures (Fig. 3C; pelvic score summary, Fig. S5). These data provide functional evidence that Pel-Pitx1 is a major determinant of pelvic formation in sticklebacks.
Fig. 3.
(A) Juvenile pelvic-reduced BEPA stickleback expressing a Pitx1 transgene driven by the Pel-2.5kbSALR enhancer, compared to (B) uninjected sibling. External spines form only in transgenic fish (black arrowhead) (C and D) Alizarin red stained pelvic structures of adult transgenic fish compared to BEPA parental phenotype. BEPA fish normally develop only a small ovoid vestige (OV) of the anterior pelvic process (AP). Transgenic fish show clear development of the anterior process (AP), ascending branch (AB), and posterior process (PP) of the pelvis, and a prominent serrated pelvic spine. Pectoral fin (PF) rays develop in both fish.
Nature of mutations in pelvic-reduced fish
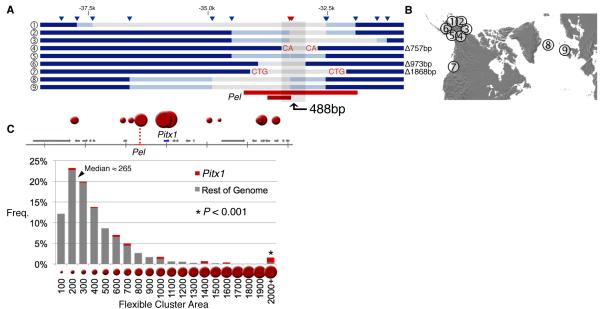
Bacterial Artificial Chromosome sequencing from the PAXB population identified a 1,868bp deletion present in the Pel-2.5kb region (Fig. S7). We cloned the PAXB deleted variant and found that it no longer drove expression in the developing pelvis (0 out of 8 transgenic animals; Fig. 2D, G), confirming that the molecular deletion in PAXB fish disrupts Pel enhancer function. We also identified a second 757bp deletion present in the pelvic-reduced BEPA population from Alaska and a third deletion of 973bp present in the Hump Lake, AK pelvic-reduced population (HUMP). The three different deletions in PAXB, BEPA and HUMP overlap in a 488bp region, each partially or completely removing the sequences found in the Pel-501bp enhancer (Figs. 4A, S4, S7, S8).
Fig. 4.
(A) SNP genotyping in additional pelvic-reduced populations identifies nine different deletions that overlap in a 488bp region. Triangles, SNP markers; grey bars, putative deleted regions flanked by two failed SNP genotypes; dark blue bars, markers flanked by two successful SNP genotypes; light blue bars, markers with successful genotypes only on one side; red bars, positions of Pel-2.5kb and Pel-501bp enhancers. Apparent deletions were confirmed by sequencing in Populations 4, 6 and 7, with the size of deletions indicated on the right, and micro-homologies of two to three base pairs at deletion junctions shown in red. (B) Location of populations surveyed. (C) TwistFlex prediction of highly flexible DNA regions (red circles) in Pitx1 locus (Pel region score: 3263) compared to frequency distribution of flexibility scores in rest of stickleback genome (median score: 265). Area of red circles is proportional to flexibility score.
To investigate whether a general mechanism and/or shared variants underlie repeated pelvic reduction in sticklebacks, we genotyped PAXB, BEPA, HUMP and ten additional pelvic-reduced populations from disparate geographic locations, as well as 21 pelvic-complete populations using 149 SNPs spanning 321kb around the Pitx1 locus (approximately 2kb spacing, Fig. S8, tables S1, S3). Nine of the 13 pelvic-reduced stickleback populations—but zero out of 21 pelvic-complete populations—showed consistent missing genotypes for multiple consecutive SNP markers located in and around the Pel enhancer (Figs. 4A, S8, two-tailed t-test, P < 0.001, df=12.279, tables S4, S5). For the PAXB, BEPA, and HUMP populations, the SNPs corresponding to the missing genotypes fall within the known deletion endpoints from DNA sequencing. The larger genotyping survey identified a total of nine different haplotypes with different staggered deletions, each consistently seen within a pelvic-reduced population, and each overlapping or completely removing the Pel enhancer region (Figs. 4 and S8).
Fragile sites
Several features suggest that Pitx1 may be located within a fragile region of the genome: the gene is located at the telomeric end of linkage group 7; the region contains many repeats and failed to assemble in the stickleback genome; the enhancer region is difficult to amplify and sequence; and close inspection of the deletion boundaries in PAXB and BEPA revealed short two or three base pair sequence identities present on both sides, one of which is retained following deletion (Figs. 4A, S7A). Similar nested deletions and small sequence identities may occur by re-ligation of chromosome ends following breakage and repair by non-homologous end joining (NHEJ) (Fig. S7B) (20, 21). In humans, NHEJ is associated with stalled replication forks at fragile chromosomal sites, which also are frequent in sub-telomeric regions (21). Fragile sites are also enriched in sequences with high DNA flexibility, a physical property that can be calculated from known twist angles between different stacked DNA base-pairs (20). DNA flexibility analysis of Pitx1 and the entire assembled stickleback genome showed a median flexibility score of 265 with a tail of extreme values. Strikingly, four of the top ten flexibility scores in the genome occur in the Pitx1 region, suggesting that this region is exceptionally flexible, and may be prone to deletion (Fig. 4C, Wilcoxon rank sum = 59624, P < 2×10−6).
Signatures of selection
Recurring deletions could explain how pelvic-reduction alleles arise repeatedly in widespread isolated populations. To test whether pelvic-reduction alleles have also been subject to positive selection, we looked for molecular signatures that commonly accompany selective sweeps, including reduced heterozygosity, and an over-representation of derived alleles (22). Patterns of allelic variation showed an excess of derived alleles near the Pel enhancer region of pelvic-reduced populations, as indicated by negative values of Fay & Wu’s H statistic (Fig. 5A, S9A, 23). We also observed a significant reduction in heterozygosity at, or near, the Pel enhancer in pelvic-reduced populations compared to marine populations (Fig. 5B, C; two-tailed t-test, P < 0.01). This reduction cannot be solely explained by population bottlenecks that occurred during freshwater colonization, since heterozygosity reduction near Pel is specific to pelvic-reduced, but not pelvic complete, freshwater populations (Figs. 5B, C; two-tailed t-test, P < 0.002). In flanking regions of Pitx1, and in unlinked control loci, we observed no significant difference in heterozygosity between freshwater fish with a complete or missing pelvis (Fig. 5C). Pelvic-reduced populations were significantly more likely to exhibit minimum heterozygosity close to the Pel enhancer region than either marine or freshwater populations with a robust pelvis (Fig. S9F, two-tailed t-test, P < 0.002). The local heterozygosity and H-statistic minima around the Pel enhancer region suggest that changes in this region have been selected in pelvic-reduced stickleback populations.
Fig. 5.
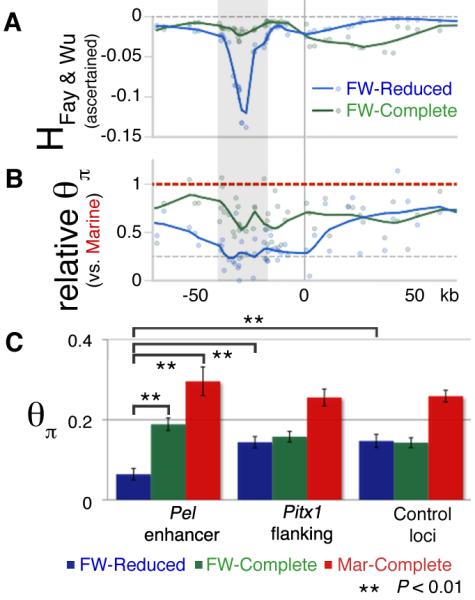
(A and B) Fay and Wu’s H and relative heterozygosity (θπ) statistics across the Pitx1 region. Blue (freshwater pelvic-reduced) and green (freshwater pelvic-complete) data points and LOESS smoothed (α=0.2) line indicates the behavior in each group. The Pel-containing regulatory region of Pitx1 (grey candidate region from Fig. S1B) shows both negative H values, indicating an excess of derived alleles; and reduced heterozygosity in pelvic-reduced fish, consistent with positive selection (see text). θπ values are plotted relative to the grouped marine mean (per SNP) to control for variation in ascertainment between SNPs. (C) Heterozygosity (θπ) from different genomic regions, grouped by population type. Freshwater fish show a general decrease in heterozygosity across both Pitx1 and control loci compared to marine fish (red bars), as expected from founding of new freshwater populations from marine ancestors. In the Pel enhancer region, but not in Pitx1-flanking regions, or in control loci, pelvic-reduced freshwater populations (blue bars) show even lower heterozygosity than pelvic-complete freshwater populations (green bars) (**, P < 0.01).
Discussion
Traditional theories of evolution posit that adaptation occurs through many mutations of infinitesimally small effect. In contrast, recent work suggests that mutation effect sizes follow an exponential distribution, with mutations of large effect contributing to adaptive change in nature (1). We narrowed the candidate interval for a pelvic QTL with large effects in sticklebacks to the non-coding region upstream of Pitx1, and identified a tissue-specific enhancer for pelvic expression that has been functionally inactivated in pelvic-reduced fish. Strikingly, re-introduction of the enhancer and Pitx1 coding region can restore formation of pelvic structures in derived populations that appear to be monomorphic for pelvic reduction. The combined data from mapping, expression, molecular, transgenic, and population genetic studies illustrate how major morphological evolution can proceed through a regulatory change in a key developmental control gene.
Large evolutionary differences which map to a particular locus can still be caused by many linked small-effect mutations that have accumulated in that gene (24, 25). However, we find that pelvic-reduction in sticklebacks maps to a type of DNA lesion that may produce a large regulatory change in a single mutational leap: deletions that completely remove a regulatory enhancer. Smaller functional lesions might be found in some pelvic-reduced populations, including four populations without obvious deletions. However, three of these populations show unusual morphological features, suggesting that their pelvic loss may have occurred through non-Pitx1-mediated mechanisms (8, 26).
The Pitx1 locus scores as one of the most flexible regions in the stickleback genome, which may reflect a susceptibility to double-stranded DNA breaks and repair by NHEJ (27-29). We hypothesize that sequence features in the Pitx1 locus may predispose the locus to structural changes, possibly explaining the high prevalence of independent deletion mutations fixed in different pelvic-reduced stickleback populations. A similar spectrum of independent small deletion mutations has been seen at the vernalization 1 locus of plants (30), suggesting that recurrent deletions in particular genes may also contribute to parallel evolution of other phenotypes in natural populations.
Mutations in developmental control genes are often deleterious in laboratory animals, leading to long-standing doubts about whether mutations in such genes could ever be advantageous in nature (31). Although Pitx1 coding regions are lethal in mice (32), we find clear signatures of positive selection in the Pitx1 gene of pelvic-reduced sticklebacks. Prior to this work, the primary evidence that pelvic reduction might be adaptive in sticklebacks came from repeated evolution of similar phenotypes in similar ecological environments and the temporal sequence of pelvic reduction in fossil sticklebacks (11, 12, 33). Interestingly, the molecular signatures of selection we have identified in the current study are centered on the tissue-specific Pel enhancer region, rather than the Pitx1 coding region. Regulatory changes in developmental control genes have often been proposed as a possible basis for morphological evolution (3, 34). However, many proposed examples of regulatory evolution in wild animals have not yet been traced to particular sequences (2), or do not show obvious molecular signatures of selection in natural populations (35). Identification of the Pel enhancer underlying pelvic reduction in sticklebacks connects a major change in vertebrate skeletal structures to specific DNA sequence alterations, and provides clear evidence for adaptive evolution surrounding the corresponding region in many different wild populations.
Supplementary Material
Acknowledgments
37. We thank M. McLaughlin for fish husbandry, M. Nonet for the gift of the pBH-mcs-YFP vector, Broad Institute for the public gasAcu1 genome assembly, and many individuals for valuable fish samples (table S1). Supported by: a Stanford Affymetrix Bio-X Graduate Fellowship (Y.F.C.); the HHMI EXROP program (G.V.); the Burroughs Wellcome Fund (M.D.S.); NSF grants DEB0211391 and DEB0322818 (M.A.B); a Canada Research Chair and grants from NSERC and the Guggenheim Foundation (D.S.); NIH grant P50 HG02568 (R.M.M., D.P., and D.M.K.); and an HHMI investigatorship (D.M.K.). Sequences generated for this study are available in Genbank (GU130433-7).
References
- 1.Orr HA. Nat. Rev. Genet. 2005;6:119. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 2.Hoekstra HE, Coyne JA. Evolution. 2007;61:995. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 3.Carroll SB. Cell. 2008;134:25. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Stern DL, Orgogozo V. Evolution. 2008;62:2155. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinchliffe JR, Johnson DR. The development of the vertebrate limb. Clarendon Press; Oxford: 1980. [Google Scholar]
- 6.Shapiro MD, Bell MA, Kingsley DM. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13753. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingsley DM, Peichel CL. In: Biology of the Three-Spined Stickleback. Ostlund-Nilsson S, Mayer I, Huntingford FA, editors. CRC Press; London: 2007. pp. 41–81. [Google Scholar]
- 8.Bell MA. Biological Journal of the Linnean Society. 1987;31:347. [Google Scholar]
- 9.Reist JD. Canadian Journal of Zoology-Revue canadienne de zoologie. 1980;58:1253. [Google Scholar]
- 10.Reimchen TE. Canadian Journal of Zoology-Revue canadienne de zoologie. 1980;58:1232. [Google Scholar]
- 11.Giles N. Journal of Zoology. 1983;199:535. [Google Scholar]
- 12.Bell MA, Ortí G, Walker JA, Koenings JP. Evolution. 1993;47:906. doi: 10.1111/j.1558-5646.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro MD, et al. Nature. 2004;428:717. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 14.Cresko WA, et al. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6050. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coyle SM, Huntingford FA, Peichel CL. J. Hered. 2007;98:581. doi: 10.1093/jhered/esm066. [DOI] [PubMed] [Google Scholar]
- 16.Cole NJ, Tanaka M, Prescott A, Tickle CA. Curr. Biol. 2003;13:R951. doi: 10.1016/j.cub.2003.11.039. [DOI] [PubMed] [Google Scholar]
- 17.Materials and methods are available as supporting material on Science Online
- 18.Colosimo PF, et al. Science. 2005;307:1928. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 19.Nagayoshi S, et al. Development. 2008;135:159. doi: 10.1242/dev.009050. [DOI] [PubMed] [Google Scholar]
- 20.Zlotorynski E, et al. Mol. Cell Biol. 2003;23:7143. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durkin SG, et al. Proc. Natl. Acad. Sci. U.S.A. 2008;105:246. [Google Scholar]
- 22.Nielsen R. Annu. Rev. Genet. 2005;39:197. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 23.Fay JC, Wu CI. Genetics. 2000;155:1405. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stam LF, Laurie CC. Genetics. 1996;144:1559. doi: 10.1093/genetics/144.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGregor AP, et al. Nature. 2007;448:587. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 26.Bell MA, Khalef V, Travis MP. J. Exp. Zoolog. B Mol. Dev. Evol. 2007;308:189. doi: 10.1002/jez.b.21132. [DOI] [PubMed] [Google Scholar]
- 27.Mishmar D, et al. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8141. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover TW, Arlt MF, Casper AM, Durkin SG. Hum. Mol. Genet. 2005;14:R197. doi: 10.1093/hmg/ddi265. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz M, et al. Genes Dev. 2005;19:2715. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cockram J, Mackay IJ, O’Sullivan DM. Genetics. 2007;177:2535. doi: 10.1534/genetics.107.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayr E. Populations, species and evolution. Harvard University Press; Cambridge, MA: 1970. [Google Scholar]
- 32.Lanctôt C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Development. 1999;126:1805. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 33.Hunt G, Bell MA, Travis MP. Evolution. 2008;62:700. doi: 10.1111/j.1558-5646.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 34.King MC, Wilson AC. Science. 1975;188:107. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 35.Jeong S, et al. Cell. 2008;132:783. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Swarup H. J. Embryol. Exp. Morphology. 1958;6:373. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.