Abstract
We have performed a survey of the active genes in the important human pathogen Trypanosoma cruzi by analyzing 5013 expressed sequence tags (ESTs) generated from a normalized epimastigote cDNA library. Clustering of all sequences resulted in 771 clusters, comprising 54% of the ESTs. In total, the ESTs corresponded to 3054 transcripts that might represent one-fourth of the total gene repertoire in T. cruzi. About 33% of the T. cruzi transcripts showed similarity to sequences in the public databases, and a large number of hitherto undiscovered genes predicted to be involved in transcription, cell cycle control, cell division, signal transduction, secretion, and metabolism were identified. More than 140 full-length gene sequences were derived from the ESTs. Comparisons with all open reading frames in yeast and in Caenorhabditis elegans showed that only 12% of the T. cruzi transcripts were shared among diverse eukaryotic organisms. Comparison with other kinetoplastid sequences identified 237 orthologous genes that are shared between these evolutionarily divergent organisms. The generated data are a useful resource for further studies of the biology of the parasite and for development of new means to combat Chagas' disease.
[The sequence data described in this paper have been submitted to the dbEST database under nos. TENU0001–TENU5214 and the following: AA736292-AA736301, AA738502-AA738535, AA756982-AA756992, AA835598-AA835613, AA866501-AA866550, AA87464-AA874780, AA875669-AA875730, AA875809-AA875824, AA879318-AA897341, AA879376-AA879401, AA882494-AA882518, AA883036-AA883051, AI005678-AI005729, AI007342-AI007441, AI021797-AI021884, AI026370-AI026615, AI037797-AI037846, AI043247-AI043343, AI043427-AI043502, AI046026-AI046290, AI050095-AI050219, AI053146-AI053397, AI057644-AI057957, AI065169-AI065425, AI066117-AI066391, AI069556-AI069908, AI073286-AI073332, AI075466-AI075620, AI077051-AI077281, AI078888-AI079000, AI080790-AI080916, AI083097-AI083245, AI110290-AI110405, AI110412-AI110512, AW324789-AW325325, AW329885-AW330435, and AW621062-AW621094. The sequences are also available at www.genpat.uu.se/tryp/tryp.html.]
Some major health problems in the world are caused by eukaryotic parasites, and genomic studies of these pathogens are of utmost importance for finding new means of treatment. Identification of genes involved in unique metabolic pathways, in pathogenicity, and in mechanisms by which the parasites evade the immune defense is of particular interest. Genome projects for several medically important parasites have therefore been initiated and are in progress.
Trypanosoma cruzi, the causative agent of Chagas' disease affecting ∼18 million people in Latin America (WHO; www.who.int/ctd/chagas/burdens.htm), is a flagellated protozoan and an evolutionarily ancient organism that belongs to the order Kinetoplastida. Neither vaccines nor safe and efficient drug treatment are presently available against this debilitating disease. The genome project for T. cruzi involves both genomic sequencing (Andersson et al. 1998) and identification of functional genes through generation of expressed sequence tags (ESTs) (Brandão et al. 1997; Verdun et al. 1998), a cost-effective technique in gene discovery (Venter 1993; Okubo and Matsubara 1997). The ongoing parasite genome projects of the three kinetoplastids, Leishmania major (Blackwell 1997), Trypanosoma brucei (Melville 1997), and T. cruzi (Zingales et al. 1997), are critical in identifying orthologous genes involved in mechanisms among kinetoplastids. Gene orders are often conserved among Kinetoplastida (Bringaud et al. 1998), and orthologous genes may therefore also be used in physical and transcriptional mapping of any of the parasites, which would accelerate all three genome projects.
In this report we present the analysis of 5013 ESTs generated from a T. cruzi epimastigote library. This analysis provides a survey of genes transcribed during the insect stage of the parasite's life cycle. Clustering of all sequences resulted in >3000 different sequences, among which a large number of novel genes were identified.
RESULTS AND DISCUSSION
Generation and Assembly of ESTs
To accelerate gene discovery in T. cruzi, we generated 6059 cDNA sequences from a normalized epimastigote library constructed from the reference clone CL Brener (Urmenyi et al. 1999). Single-pass sequencing was performed from either the 5′- or the 3′-end of the cDNA clones with an average insert size of 650 bp. After assembly and subsequent removal of low-quality sequences, the resulting 5013 ESTs formed a total of 771 clusters (Table 1). Most of the clusters (74%) contained only 2 or 3 sequences and the largest cluster comprised 66 sequences. The consensus sequence generated for each cluster by the assembly program was later used together with all singleton sequences in the subsequent analyses. In total, 3054 transcripts were identified in this study. It is likely that most of these transcripts represent unique genes. However, some fraction of them could be nonoverlapping sequences derived from the same transcript.
Table 1.
Summary of the Assembly of All Expressed Sequence Tags (ESTs)a
| Clusters | 771 (2730) |
| Singletons | 2283 |
| Total no. of unique ESTs | 3054 |
The total number of ESTs included in the assembly is 5013, out of which 2620 are 5′-ESTs and 2393 are 3′-ESTs.
The redundancy of the cDNA library, estimated from the fraction of sequences that assembled into clusters was thus ∼ 54%, a relatively high value considering the normalization step. The most abundant cDNAs were from the mucin genes, which belong to a large gene family of highly divergent copies in the T. cruzi genome (Di Noia et al. 1998). A plausible explanation for a redundancy of genes such as these in the library might be that cross-hybridizing diverged sequences seem to escape the normalization procedure (Bonaldo et al. 1996). Moreover, several clusters showed sequence polymorphisms among different ESTs, suggesting that the sequences were derived from different gene copies and that the actual redundancy is lower. The divergence in the protein-coding regions of the mucin genes caused these cDNAs to resolve into several clusters. Several other multicopy gene families, previously identified in T. cruzi, could be found among the largest clusters (Table 2). Only 0.6% of the cDNA clones contained ribosomal RNAs and a few clones encoded known T. cruzi genomic repeats (Requena et al. 1996), indicating a low contamination of the cDNA library.
Table 2.
Summary of Most Abundant Trypanosoma cruzi ESTs
| No. of ESTs | Identitya |
|---|---|
| 107 (2.1%) | Mucin-like proteins |
| 71 (1.4%) | Histone H2A |
| 51 (1.0%) | Histone H3 |
| 31 (0.6%) | Mitochondrial HSP 70 |
| 20 (0.4%) | Unknown |
| 17 (0.3%) | RNA-binding protein |
| 17 (0.3%) | HSP 70 |
| 15 (0.3%) | Ribosomal protein S11A |
| 15 (0.3%) | Ribosomal protein L28 |
| 15 (0.3%) | Histone H1 |
The ten most frequent ESTs are shown. A more detailed result is available at http://www.genpat.tryp/tryp.html.
Other examples of separated clusters, besides the mucin genes, were the two clusters of ESTs encoding succinylCoA ligase, which differ by an in-frame deletion of 40 amino acids (http://www.genpat.uu.se/tryp/tryp.html).
A large number of full-length genes could also be obtained, because both 5′- and 3′-ESTs were generated. In total, 234 ESTs (4.7%) contained the spliced leader sequence or a part thereof. Complete protein-encoding regions of >140 short genes were obtained. Several clusters contained cDNAs, which differed in length of the 3′-UTRs. The 3′-UTRs frequently contained short nucleotide repeats of different lengths and composition as well as other repeat elements (Vazquez et al. 1994). Lists of ESTs showing alternative polyadenylation sites and ESTs containing the spliced leader or repeats are available (see http://www.genpat.uu.se/tryp/tryp.html).
Biological Survey of Identified Genes in T. cruzi
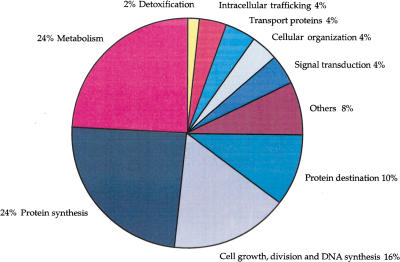
When searching for sequence similarities in public databases, ∼20% of the T. cruzi transcripts could be assigned a putative identity (Table 3). These identities were classified into different groups according to function (a list is available at http://www.genpat.se/tryp/tryp.html). A representation of the functional groups is shown in Figure 1.
Table 3.
Summary of BLAST Searches
| No. of unique ESTs | ||
|---|---|---|
| IDa | Putative identity | 610 (20%) |
| Unknown function | 398 (13%) | |
| No ID | 2046 (67%) |
A putative identity (ID) was assigned to T. cruzi ESTs when similarity values exceeded 50% (P ≤ 10−5 for BLASTN and P ≤ 10−4 for BLASTX), as described in Methods.
Figure 1.
Functional classification of Trypanosoma cruzi ESTs. The transcripts with putative identities to proteins other than highly abundant gene families in T. cruzi were divided into functional categories. The largest groups were genes involved in protein synthesis and metabolism. The group designated protein destination included genes for protein modification, folding, and proteolysis and the group named others mainly contained genes with protein kinase, DNA-binding, and Zn-finger motifs (www.genpat.uu.se/tryp/tryp.html).
A large fraction of genes with putative identity encoded proteins involved in translation (24%), including 61 ribosomal proteins, initiation and elongation factors, and proteins involved in tRNA synthesis.
An interesting group for gene regulatory processes in trypanosomes is the proteins involved in transcription and RNA processing, which amounted to 6% of transcripts and included several RNA polymerase subunits, RNA-binding proteins, and splicing factors.
About 4% of the functionally classified transcripts showed similarities to proteins involved in signal transduction, including multiple rab proteins and MAP kinases. A total of 17 novel genes with similarities to cyclophilins, kinesin-like proteins, and cell division checkpoint proteins were identified among proteins involved in cell cycle regulation and division.
A small group encoded enzymes involved in detoxification. Enzymes unique for trypanosomes participating in the trypanothione biosynthesis, such as glutathionyl spermidine synthetase and trypanothione synthetase, were found. Several thioredoxin-like proteins and different peroxidases were also identified.
Eleven percent of the transcripts were involved in energy metabolism. Multiple ATP synthase subunits, cytochrome C components, and cytochrome 450 were present.
New genes involved in the transport machinery, the secretory pathway, and degradation of proteins were identified and should facilitate studies of these less-defined processes in T. cruzi. Five ABC transporters previously not described were identified among the T. cruzi transcripts. Among genes involved in cellular organization, several subunits of dynein were identified.
Comparison of T. cruzi ESTs to the Complete Sets of Genes from a Unicellular and a Multicellular Organism
The T. cruzi transcripts were compared with the protein sequences of all predicted ORFs from the yeast genome project (Goffeau et al. 1997) and the genomic sequence of Caenorhabditis elegans (The C. elegans Sequencing Consortium 1998), amounting to 6217 and 19,099 ORFs, respectively. In total, 14.7% of the T. cruzi genes showed similarity to yeast and C. elegans ORFs (Table 4). Of these, 12% were shared by all three organisms and the matches were mainly to proteins with housekeeping functions.
Table 4.
Comparison of Trypanosoma cruzi ESTs to Saccharomyces cerevisiae, Caenorhabditis elegans, and Kinetoplastid Organisms
| Organism | No. of unique ESTs |
|---|---|
| S. cerevisiae | 448 (14.7%) |
| C. elegans | 450 (14.7%) |
| yeast + worma | 365 (12%) |
| yeast-restricted | 83 (2.7%) |
| worm-restricted | 85 (2.8%) |
| Kinetoplastid-restrictedb | 592 (19.4%) |
| T. cruzi | 355 (11.6%) |
| other than T. cruzi | 237 (7.8%) |
Number of unique EST sequences with matches to both yeast and worm. Cutoff value used for the comparison = P ≤ 10−4
Number of unique EST sequences with matches to kinetoplastid-restricted genes. Cutoff value used for the comparison = P ≤ 10−4 Detailed results are available at http://www.genpat.uu.se/tryp/tryp.html.
The comparisons of this fraction of ESTs to C. elegans and yeast ORFs are not conclusive, but might give an indication as to which T.cruzi genes may be shared with other eukaryotes. It is not possible to draw any conclusions from lack of homologs, which would also include genes acquired because of the adaptation to life as an intracellular parasite. The low percentage of similarity between yeast and the protozoan T. cruzi might reflect the evolutionary divergence of the trypanosomatids similar to what has been suggested for another protozoon, Toxoplasma gondii (Ajioka et al. 1998). The low percentage could also be due to a lower coding potential of 3′-ESTs compared with 5′-ESTs, estimated from singletons with putative identities to be ∼ 40% and 60%, respectively.
Identification of Genes Present in Kinetoplastids
Trypanosomes share a number of unique biological features with other flagellated protozoa of the order Kinetoplastida, such as trans-splicing, RNA editing, and the unusual organization of the mitochondrial DNA in the kinetoplast.
To identify genes that are shared among kinetoplastids, we compared all of the T. cruzi transcripts with a local database containing public DNA sequences from these organisms. After removal of all sequences with homologs in other organisms, 592 transcripts showed similarity to kinetoplastid sequences (Table 4). The search revealed 237 orthologous genes present in one or more kinetoplastids other than T. cruzi, >50% of which were of unknown identity. Among these genes were those encoding surface molecules such as the Gp63-homolog and ESAG from T. brucei. The rest of the hits to kinetoplastid sequences matched only T. cruzi sequences, a majority being genes or repeats of known identity and also previously known to be specific for T. cruzi.
T. cruzi can be estimated to have ∼12,000 genes from the gene density of about 1 gene per 3.5–4 kb, as revealed by genomic sequencing (Andersson et al. 1998) and the haploid genome size of ∼45 Mb in the T. cruzi reference clone CL Brener (J. Swindle, unpubl.). Because a considerable part of the T. cruzi genome comprises several large gene families as well as other repeat sequences (Requena et al. 1996), this number of genes should be an overestimate. The present study represents the hitherto largest sampling of generated T. cruzi ESTs and might correspond to almost one-fourth of the total gene repertoire in T. cruzi.
This study also reports the first clustering analysis of a large set of T. cruzi ESTs, allowing identification of, for example, alternative polyadenylation sites. Because a larger amount of ESTs than reported previously have been analyzed, a large number of new genes have been identified, giving a better representation of the T. cruzi gene content. The larger sampling revealed several new important genes involved in detoxification and a larger set of genes involved in metabolism than those presented in previous works by Brandão et al. (1997) and Verdun et al. (1998) by using the same cDNA library constructed by Urmenyi et al. (1999).
Taken together, the EST analyses performed in T. cruzi provide a valuable resource for future studies of parasite biology and for identifying functional genes in a complex genome containing a high number of large gene families.
METHODS
Template Preparation and DNA Sequencing
The cDNA library was constructed by using oligo(dT)-primed T. cruzi CL Brener epimastigote poly(A)+ RNA (Urmenyi et al. 1999) normalized to reduce the representation of abundant mRNA species (Bonaldo et al. 1996). The cDNA library was transformed into DH5-alpha strain of Escherichia coli, and > 23,000 individual colonies were randomly picked and ordered into 384-well microtiter plates. High-quality double-stranded plasmid DNAs were prepared by using the Wizard PLUS SV miniprep DNA purification system (Promega) or the PERFECTprep-96 plasmid DNA purification system (5 Prime-3 Prime Inc.).
Automated fluorescent cycle sequencing reactions were performed by using the ABI Prism-21M13 fluorescent dye-labeled primer kit (Perkin Elmer Cetus) and DYEnamic direct cycle sequencing (Amersham Life Science) with a T7 dye-labeled primer. The samples were analyzed on ABI Prism 377XL DNA sequencers.
Processing and Annotation of Sequences
The sequences were quality checked by using the software PHRED (Ewing and Green 1998). Vector sequence (GenBank accession no. U13869), including the modified polylinker, the spliced leader sequence (De Lange et al. 1984), and poly(A)-tail sequences, were removed before submission to the dbEST database. Sequences of a quality > 96% accuracy and longer than 100 bp were used for further analysis. The average length of the ESTs was 327 bp. In the similarity searches, low-entropy sequences were masked by using the program DUST (R. Tatusov and D. Lipman, unpubl.). The sequences were searched against the nonredundant Genbank (www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html) database at the National Center for Biotechnology Information by using BLASTN (Altschul et al. 1997) and gapped BLASTX (Altschul et al. 1997) (parameters
 |
and against Swissprot (Bairoch and Apweiler 1997) by using gapped BLASTX (parameters
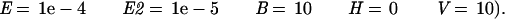 |
Results were parsed by using the program btab (Dubnick 1992). The matches in which similarity values exceeded 50% with the BLASTX program and P ⩽ 10−4 were listed as putative identities.
Assembly of EST Sequences
The T. cruzi EST sequences were assembled into overlapping sequences by using the fragment assembly program PHRAP (Green 1996). Initially, raw chromatogram data from 6059 cDNA sequences were used in the assembly. The vector sequences were masked by using cross_match (Green 1996), and the parameters used in the assembly for minmatch and minscore were set to 50 and 39, respectively, whereas the rest of the parameters were default parameters. The parameters for the assembly were tested to optimize the results and the contigs were manually checked for incorrectly assembled sequences.
Similarity Searches
Similarity searches were performed locally on an IBM SP parallel computer at the Parallel Computing Center, Stockholm, Sweden. Local databases included Genbank sequences in Flat File Release 110.0, SwissProt (Bairoch and Apweiler 1997) sequences in release 36, C. elegans (http://elegans.swmed.edu), and Saccharomyces cerevisiae (http://genome-www.stanford.edu/Saccharomyces) protein sequences. MT-BLAST (M. Tammi, unpubl.) wrapper by using gapped WU-BLASTX (W. Gish 1997, http://blast.wustl.edu) was run on 64 nodes, using default WU-BLAST parameters and matrix. The results were filtered afterward by using a value of P = 10−4.
The comparison to the kinetoplastid database was performed locally by using gapped WU-BLAST (W. Gish 1997, http://blast.wustl.edu). The database consisted of 9204 entries from the EMBL database and contained both cDNA and genomic sequences, including the complete sequence of chromosome 1 and parts of chromosome 3 from L. major Leishmania major. All T. cruzi ESTs already deposited into dbEST were excluded from this comparison.
Clones containing the spliced leader or part thereof were identified among the T. cruzi ESTs by BLASTN with search sequences containing the 5′-cloning site, including the tag sequence (GAATTCCAGCTCC) fused to the spliced leader sequence sequentially deleted from the 5′-end down to the four last base pairs.
Acknowledgments
We thank Daniel Nilsson for valuable help in programming. We are thankful to all colleagues within the T. cruzi network. Thanks are due to the Parallel Computing Center, Royal Institute of Technology, Stockholm. This work was supported by funds from the UNDP/WORLD BANK/WHO Special Programme for Research and Training in Tropical Diseases (T23/181/104), The Beijer Foundation, The Swedish Foundation for International Cooperation in Research and Higher Education (97/676), The Swedish Natural Science Research Council (B-AA/BU 06684–311), and The Swedish Medical Research Council (K99–31X-12633–02B). A-N Tran is supported by a PhD student fellowship from the Swedish Agency for Research and Cooperation with Developing Countries (SWE-1998–411A). B.M.P was supported by a grant from the Swedish Institute (210/51).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lena.aslund@genpat.uu.se; FAX 46-18-471 48 08.
REFERENCES
- Ajioka JW, Boothroyd JC, Brunk BP, Hehl A, Hillier L, Manger ID, Marra M, Overton GC, Roos DS, Wan K-L, et al. Gene discovery by EST sequencing in Toxoplasma gondii reveals sequences restricted to the Apicomplexa. Genome Res. 1998;8:18–28. doi: 10.1101/gr.8.1.18. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B, Åslund L, Tammi M, Tran A-H, Hoheisel J, Pettersson U. Complete sequence of a 93.4 kb region from chromosome III of Trypanosoma cruzi. Genome Res. 1998;8:809–816. doi: 10.1101/gr.8.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucleic Acids Res. 1997;25:31–36. doi: 10.1093/nar/25.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell JM. Parasite genome analysis: Progress in the Leishmania genome project. Trans R Soc Trop Med Hyg. 1997;91:107–110. doi: 10.1016/s0035-9203(97)90187-5. [DOI] [PubMed] [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Brandão A, Urmenyi T, Rondinelli E, Gonzalez A, de Miranda AB, Degrave W. Identification of transcribed sequences (ESTs) in the Trypanosoma cruzi genome project. Mem de Inst Osw Cruz. 1997;92:863–866. doi: 10.1590/s0074-02761997000600024. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Vedrenne C, Cuvillier A, Parzy D, Baltz D, Tetaud E, Pays E, Venegas J, Merlin G, Baltz T. Conserved organization of genes in trypanosomatids. Mol Biochem Parasitol. 1998;94:249–264. doi: 10.1016/s0166-6851(98)00080-2. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- De Lange T, Berkvens TM, Veerman HJ, Frasch AC, Barry JD, Borst P. Comparison of the genes coding for the common 5′ terminal sequence of messenger RNAs in the three trypanosome species. Nucleic Acids Res. 1984;12:4431–4443. doi: 10.1093/nar/12.11.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, D'Orso I, Åslund L, Sánchez DO, Frasch AC. The Trypanosoma cruzi mucin family is transcribed from hundreds of genes having hypervariable regions. J Biol Chem. 1998;273:10843–10850. doi: 10.1074/jbc.273.18.10843. [DOI] [PubMed] [Google Scholar]
- Dubnick M. Btab-a Blast output parser. Comput Appl Biosci. 1992;8:601–602. [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Goffeau A, Aert R, Agostini-Carbone ML, et al. The yeast genome directory. Nature. 1997;387 (Suppl 6632):5. [PubMed] [Google Scholar]
- Green, P. 1996. PHRAP documentation. http://bozeman.mbt.washington.edu/phrap.docs/phrap.html. University of Washington, Seattle.
- Melville SE. Parasite genome analysis. Genome research in Trypanosoma brucei: Chromosome size polymorphism and its relevance to genome mapping and analysis. Trans R Soc Trop Med Hyg. 1997;91:116–120. doi: 10.1016/s0035-9203(97)90189-9. [DOI] [PubMed] [Google Scholar]
- Okubo K, Matsubara K. Complementary DNA sequence (EST) collections and the expression information of the human genome. FEBS Lett. 1997;403:225–229. doi: 10.1016/s0014-5793(97)00042-2. [DOI] [PubMed] [Google Scholar]
- Requena JM, López MC, Alonso C. Genomic repetitive DNA elements of Trypanosoma cruzi. Parasitol Today. 1996;12:279–283. doi: 10.1016/0169-4758(96)10024-7. [DOI] [PubMed] [Google Scholar]
- Urmenyi TP, Bonaldo MF, Soares MB, Rondinelli E. Construction of a normalized cDNA library for the Trypanosoma cruzi genome project. J Eukaryot Microbiol. 1999;46:542–544. doi: 10.1111/j.1550-7408.1999.tb06072.x. [DOI] [PubMed] [Google Scholar]
- Vazquez MP, Schijma AG, Levin MJ. A short interspersed repetitive element provides a new 3′ acceptor site for trans-splicing in certain ribosomal P2 beta protein genes of Trypanosoma cruzi. Mol Biochem Parasitol. 1994;64:327–336. doi: 10.1016/0166-6851(94)00026-3. [DOI] [PubMed] [Google Scholar]
- Venter JC. Identification of new human receptor and transporter genes by high throughput cDNA (EST) sequencing. J Pharm Pharmacol. 1993;45 (Suppl. 1):355–360. [PubMed] [Google Scholar]
- Verdun RE, Di Paolo N, Urmenyi TP, Rondinelli E, Frasch AC, Sanchez DO. Gene discovery through expressed sequence Tag sequencing in Trypanosoma cruzi. Infect Immun. 1998;66:5393–5398. doi: 10.1128/iai.66.11.5393-5398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingales B, Rondinelli E, Degrave W, daSilveira JF, Levin M, Le Paslier D, Modabber F, Dobrokhotov B, Swindle J, Kelly JM, et al. The Trypanosoma cruzi genome initiative. Parasitol Today. 1997;13:16–22. [PubMed] [Google Scholar]