Abstract
A cosmid/bacterial artificial chromosome (BAC) contiguous (contig) map of human chromosome (HSA) 19p13.3 has been constructed, and over 50 genes have been localized to the contig. Genes and anonymous ESTs from ≈4000 kb of human 19p13.3 were placed on the central mouse chromosome 10 map by genetic mapping and pulsed-field gel electrophoresis (PFGE) analysis. A region of ∼2500 kb of HSA 19p13.3 is collinear to mouse chromosome (MMU) 10. In contrast, the adjacent ≈1200 kb are inverted. Two genes are located in a 50-kb region after the inversion on MMU 10, followed by a region of homology to mouse chromosome 17. The synteny breakpoint and one of the inversion breakpoints has been localized to sequenced regions in human <5 kb in size. Both breakpoints are rich in simple tandem repeats, including (TCTG)n, (CT)n, and (GTCTCT)n, suggesting that simple repeat sequences may be involved in chromosome breaks during evolution. The overall size of the region in mouse is smaller, although no large regions are missing. Comparing the physical maps to the genetic maps showed that in contrast to the higher-than-average rate of genetic recombination in gene-rich telomeric region on HSA 19p13.3, the average rate of recombination is lower than expected in the homologous mouse region. This might indicate that a hot spot of recombination may have been lost in mouse or gained in human during evolution, or that the position of sequences along the chromosome (telomeric compared to the middle of a chromosome) is important for recombination rates.
Comparative mapping, especially between the mouse and human genomes, gains importance as the Human Genome Project moves toward functional genomics, i.e., the functional characterization of large regions of sequenced DNA. A good comparative map allows identification of homologous mouse mutations to human disorders, which in turn aids gene identification (Probst et al. 1998; Wang et al. 1998) and the development of mouse models for identified human genes.
The most distal region of human 19p13.3 is abundant in G/C content, gene-rich, and is homologous to mouse chromosome 10 (Burmeister et al. 1998; Mohrenweiser et al. 1996). The gene for Peutz-Jehgers syndrome, which maps within this interval, has recently been identified as STK11 (Hemminki et al. 1998; Jenne et al. 1998). Five mouse mutations (jittery, hesitant, grizzled, mocha, and apathetic) have been located within this interval (Chung and Leibel, pers. comm.; Kantheti et al. 1998; Kapfhamer and Burmeister 1994; Kapfhamer et al. 1996). The mocha gene was recently identified as Ap3d (Kantheti et al. 1998). Jittery, hesitant, and apathetic are neurological mutations. Jittery and hesitant are allelic (Kapfhamer et al. 1996), whereas apathetic is not (Chung and Leibel et al.; W.K. Chung and M. Burmeister, pers. comm.). Interestingly, two human loci, Cayman ataxia, ATCAY (Nystuen et al. 1996) and a form of infantile febrile seizures, FEB2 (Johnson et al. 1998), are disorders whose symptoms overlap with the phenotypes of the jittery, hesitant, and apathetic mouse mutants. A careful comparative map will be useful in determining whether any of these mouse mutants are homologous to any of the human disorders.
Comparative mapping also is an important tool for understanding the evolutionary history of genomes. Here we show that within a conserved linkage group, an inversion of about 1 Mb has occurred. The small size of this rearrangement could have easily prevented discovery by traditional means such as linkage analysis and genetic mapping. The availability of human sequences and an abundance of mouse ESTs allowed the precise definition of the boundaries of two such breaks to a resolution of less than 5 kb. The break-point sequences contain a variety of small tandem repeats, including (TCTG)n, (CT)n, and (GTCTCT)n, some of which previously have been found near recombination hot spots. A determination of whether any of these repeats are typical for evolutionary breaks will require the identification of additional breakpoints at the sequence level.
RESULTS
A Complete Cosmid/Bacterial Artificial Chromosome Map of 19p13.3
An overlapping clone map was constructed spanning the entire p13.3 band of human chromosome 19. The map, consisting of cosmids and bacterial artificial chromosomes (BACs) plus a few P1-derived artificial chromosomes (PACs) and P1 clones, covers an estimated 7.5 Mbp. Complete clonal continuity was achieved, except for a single gap within the SHC2 gene ∼400 kb from the telomere. The gap is spanned by multiple cDNA clones, but no spanning genomic clones have been found in screening numerous libraries including chromosome 19-specific cosmid and fosmid libraries, as well as three genomic BAC or PAC libraries. Searching the BAC end database (see http://www.tigr.org/tdb/humgen/bac_end_search/bac_end_search.html) with the sequence of cosmid R34739 (Genbank accession AC006124) on the proximal side of the gap retrieved no BACs heading into the gap. Sequencing of cosmid F25549 on the distal side of the gap currently is in progress.
Selected genes and genetic markers were localized on the map by hybridization as described (Brandriff et al. 1994). Additional genes were identified by genomic sequencing. About 3.6 unique Mb of 19p13.3 have been submitted to Genbank as finished sequence. Another 3.9 Mb of sequence are in progress and are currently available in Genbank HTGS phase 1 or 2 preliminary sequence. Updated versions of the chromosome 19 metric physical map and underlying restriction maps, as well as current sequence data, are available from the Lawrence Livermore National Laboratory (LLNL) Human Genome Center web site (see http://bbrp.llnl.gov/bbrp/genome/genome.html) and the DOE Joint Genome Institute web site (see http://www.jgi.doe.gov/). Table 1 shows, for genes discussed here, the position from the telomere and the clone on which they reside. Most, but not the entire region, of 19p13.3 discussed here is available as finished sequence. Although the total number of genes in this very gene-rich region is still unknown, based on a few fully finished and annotated regions, we estimate that about 10% of genes from this region of 19p13.3 were mapped to mouse chromosome 10 as described here.
Table 1.
Gene and Marker Positions in 19p13.3 Region Homologous to Mouse Chromosome 10
| Gene | Genomic clone | |||||
|---|---|---|---|---|---|---|
| human gene/marker | gene/marker accession | mouse orthologue | location | LLNL clone ID | clone name | finished accession |
| SCK | AB001451 | Shc2 | 450 | R34739 | LLNL-R_310G3 | AC006124 |
| CDC34 | L22005 | Cdc34 | 540 | R27157 | LLNL-R_231G5 | |
| R31903 | LLNL-R_281B11 | |||||
| GZMM | L23134 | Lmet1 | 550 | R31903 | LLNL-R_281B11 | |
| BSG | D45131 | Bsg | 590 | F18382 | LLNL-F_140D2 | AC005559 |
| D19S814/D19S20 | GDB:591942 | 610 | F18382 | LLNL-F_140D2 | AC005577 | |
| ELA2 | M34379 | Ela2 | 850 | R33516 | LLNL-R_298A4 | AC004799 |
| DF | M84526 | Adn | 870 | R32285 | LLNL-R_285B9 | |
| D19S886 | Z52881 | 1020 | R32184 | LLNL-R_284B4 | AC004528 | |
| GPX4 | X71973 | Gpx4 | 1130 | R28738 | LLNL-R_248C2 | AC004151 |
| STK11 | AF035625 | 1250 | R30295 | LLNL-R_264D11 | ||
| R29144 | LLNL-R_252D12 | AC004221 | ||||
| CIRBP | D78134 | Cirbp | 1300 | R33114 | LLNL-R_293G10 | AC004258 |
| EFNA2 | AJ007292 | Efna2 | 1320 | R33114 | LLNL-R_293G10 | AC004258 |
| D19S883 | Z52832 | 1430 | BC38864 | CIT978SKB_25B13 | ||
| PCSK4 | AB001898-1914 | Pcsk4 | 1510 | BC38864 | CIT978SKB_25B13 | |
| TCF3 | M31523 | Tcfe2a | 1650 | R27377 | LLNL-R_234A9 | AC005321 |
| No human symbol | D10Bwg1364e | 1880 | BC41487 | CIT978SKB_31O20 | ||
| AP3D | AF002163 | Ap3d | 2120 | R26660 | LLNL-R_226E12 | AC005328 |
| R26634 | LLNL-R_226C10 | AC005545 | ||||
| F8682 | LLNL-F_39C10 | AC005257 | ||||
| AMH | K03474 | Amh | 2240 | F20489 | LLNL-F_162C9 | AC005263 |
| OAZ1 | U09202 | Oaz1 | 2260 | FOS37308 | LLNL-FOS_25A4 | AC004152 |
| D19S878 | Z52750 | 2350 | FOS39554 | LLNL-FOS_48D6 | AC004410 | |
| LMNB2 | M94362 | Lmnb2 | 2420 | F24881 | LLNL-F_208A9 | |
| GNG7 | AB010414 | Gng7 | 2510 | F23259 | LLNL-F_191B7 | AC005512 |
| D19S565 | Z51674 | 2550 | FOS39347 | LLNL-FOS_46C3 | AC005756 | |
| THOP1 | U29366 | Thop1 | 2780 | BC41195 | CIT978SKB_31C16 | AC006538 |
| TLE2 | M99436 | 3000 | R26610 | LLNL-R_226A10 | AC007766 | |
| AES | U04241 | Grg | 3040 | F23613 | LLNL-F_194H1 | AC005944 |
| D19S591 | GDB:365306 | 3060 | F23613 | LLNL-F_194H1 | AC005944 | |
| GNA11 | M69013 | Gna11 | 3090 | F23990 | LLNL-F_198G6 | AC005262 |
| GNA15 | M63904 | Gna15 | 3130 | F23990 | LLNL-F_198G6 | AC005262 |
| R31335 | LLNL-R_275C7 | AC005264 | ||||
| D19S120 | GDB:127658 | 3170 | R31045 | LLNL-R_272C5 | ||
| D19S424 | Z24649 | 3210 | R31341 | LLNL-R_275D1 | AC005331 | |
| D19S209 | Z16605 | 3290 | R34115 | LLNL-R_304C3 | ||
| HMG20B | AF072836 | Hmg20b | 3560 | R31109 | LLNL-R_272H9 | AC005786 |
| TBXA2R | D38081 | Tbxa2r | 3580 | R31449 | LLNL-R_276E1 | AC005175 |
| cDNA298622 (Sut1e) | N74289, W04390 | D10Sut1e | 3740 | F16403 | LLNL-F_119G3 | AC005777 |
| MATK | L18974 | Matk | 3760 | F16403 | LLNL-F_119G3 | AC005777 |
| DAPK3 | AB007144 | Dapk3 | 3900 | BC95216 | CIT978SKB_171N13 | |
| BC802058 | CITB-E1_2622I13 | |||||
| MAP2K2 | L11285 | Map2k2 | 4030 | BC802058 | CITB-E1_2622I13 | |
| R33590_1 (bZIP motif) | AC005620 | D10Bur1e | 4120 | R33590 | LLNL-R_298G6 | AC005620 |
| R33243_1 SIRT6 | AF233396 | Sirt6 | 4140 | R33590 | LLNL-R_298G6 | AC005620 |
| R33243 | LLNL-R_295B7 | AC006930 | ||||
| R33243_2 (Ank. motif) | AC006930 | D10Bur2e | 4170 | R33243 | LLNL-R_295B7 | AC006930 |
| F20887 | LLNL-F_166D11 | AC005578 | ||||
| EBI3 | L08187 | Ebi3 | 4200 | F20887 | LLNL-F_166D11 | AC005578 |
| SH3GL1 | X99656 | Sh3d2b | 4330 | R31167 | LLNL-R_273E7 | AC007292 |
| D19S894 | Z53052 | 4360 | BC48708 | CIT978SKB_50L17 | ||
Markers/genes are listed in order from p-telomere to centromere. Only those genes and markers shown in Figure 2 are listed. Accession numbers are Genbank unless otherwise indicated. Symbols for mouse orthologs are listed where available. Locations, in approximate kilobase from p-telomere, were determined from a combination of map and sequence data. Sequenced genomic clones containing each gene or marker are indicated. Multiple clones are listed for genes spanning more than one sequenced clone. Genbank accession numbers are given for clones with finished sequence.
A PFGE Map of Mouse Chromosome 10 with 19p13.3 Homology
In order to generate a pulsed-field gel electrophoresis (PFGE) map of mouse chromosome 10 in the region of homology to 19p13.3, probes were developed from published genes and the emerging sequence of 19p13.3 (see Methods for detail). Additionally, end points of cosmid walks (Wong et al. 1999, and D.E. Jenne, unpubl.) within the protease cluster around Lmet1, Ela2, and Bsg were used. When it was not clear whether a gene would localize to mouse chromosome 10, it was mapped on the “BSS-panel”, a public DNA resource of a backcross, and the data were deposited to the database maintained at the Jackson Laboratory (Rowe et al. 1994).
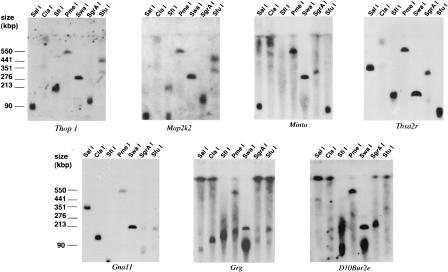
Probes for each gene that mapped genetically to the relevant region of mouse chromosome 10 were hybridized to PFGE filters prepared as described (Burmeister, 1992). Two unique probes recognizing three or more pulse-field fragments in common on a filter were considered sufficient evidence that the probes mapped to the same region. If only one or two bands were corecognized, genetic or polymorphism evidence also was considered for verification. Figure 1 shows PFGE analysis of several genes, demonstrating two of the breakpoints discovered here. The first two probes shown, Thop1 and Map2k2, clearly recognize several identical fragments on the mouse PFGE map, but are known to be about 1200 kb apart on human 19p13.3. Similarly, the human homologs of Grg and D10Bur2e are about 1100 kb apart in human, outlining the other end of the inversion. A summary of all PFGE data is shown in Table 2 and Figure 2. The human restriction map is shown on the LLNL Web site (http://www-bio.llnl.gov/rmap/).
Figure 1.
Pulsed-field gel electrophoresis (PFGE) mapping on mouse chromosome 10. DNA isolated in agarose plugs from mouse spleen was digested with a variety of restriction enzymes and separated in the 50–800 kb range on a BIORAD Chefmapper II using the conditions suggested by the autoalgorithm. Two filters were blotted from the same gel, and hybridized to radioactively labeled probes from mouse chromosome 10. Examples of overlapping hybridizing fragments are shown. Note that Thop1 and Map2k2 share many fragments, yet are >1000 kb apart on human 19p13.3, visualizing the inversion. Similarly, Grg and D10Bur2e share clearly many bands but are also ∼1000 kb apart in human.
Table 2.
Pulsed-Field Gel Electrophoresis (PFGE) Data for Markers on Mouse Chromosome 10 in the 19p13.3 Homology Region
| Probes | Genbank | Nru | Not | Mlu | Cla | Swa | Sal | Pme | SgrA | Sfu | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| *K17 | – | ≪70 | ≪70 | 450 | <60 | 320 | 190,250 | 210 | 110,90 | 190 | D. Jenne unpubl. |
| Shc2 | T05885 | ≪70 | ≪70 | 450 | <60 | 320 | 190,250 | 210 | 110,90 | 190 | ATCC #82569 |
| Bsg | D82019 | 70,90 | 60 | 450 | 220,190 | 270 | 190,250 | 190 | 270 | 190 | T. Miyauchi |
| Lmet1 | L76741 | 70,90 | 60 | 450 | 220,190 | 270 | 190,250 | 190 | 270 | 190 | D. Jenne |
| *K16 | – | 70,90 | 60 | <60 | 220,190 | 270 | 190,250 | 190 | 270 | 190 | D. Jenne unpubl. |
| *K14 | – | 100 | 150 | 250 | 280 | 310,180,150 | 140,80 | 440 | 320 | 450,270,150 | D. Jenne unpubl. |
| Ptb | X52101 | 100 | 150 | 250 | ND | 310,180,150 | 140,80 | 440 | 320 | 450,270,150 | K. Brady |
| Ela2 | X04576 | 100 | 150 | 250 | ≪70 | 100 | 300 | 440 | 320 | 450,270,150 | D. Jenne |
| Adn | X04673 | 100 | 150 | 250 | ≪70 | 100 | 300 | 440 | 320 | 450,270,150 | D. Jenne |
| *K79 | – | 90 | 90 | 250 | 80 | 90 | ND | 440 | 320 | 450,270,150 | D. Jenne |
| Gpx4 | AJ012104 | 90 | 70 | 250 | 250 | 220 | 280 | 440 | 180 | 450,270,150 | F. Chu |
| Cirbp | D78135 | 150 | 200 | 250 | 250 | 280 | ldr 440-440+ | 420,350,280 | 210 | 280 | IMAGE #329445 |
| Efna2 | U14941 | 150 | 200 | 250 | 250 | 280 | ldr 400-440+ | 420,350,280 | 210 | 280 | W. Chung |
| Pcsk4 | D01093 | ≪90 | 80 | ≪90 | 550,330 | 280 | ldr 440-100 | 420,350,280 | 210 | 280 | M. Mbikay |
| Tcfe2a | D16631 | 300 | 150 | 50,100 | 350 | 110 | ldr 440-100 | 420,350,120 | 70 | 190,140 | T. Kadesch |
| D10Bwg1364e | N28107 | 300 | 100 | 60 | 440,380,340,280 | 90 | 180 | 440,350 | 340 | 140 | K. Brady |
| Ap3d | AB004305 | 300 | 70 | 250 | ND | ND | ND | ND | 340 | ND | IMAGE #519977 |
| Amh/Sap62 | X83733 | <50 | 440 | 250 | 440,380,200,150 | 430 | 340 | 570,440 | 90 | 280,90 | R. Behringer |
| Oaz1 | U84291 | 100 | 440 | 80,100 | 440,200,150 | 420 | 340 | 90,440 | 200 | 110,390 | L. Ghoda |
| Lmnb2 | X54098 | 100 | 440 | 80,100 | 440,200,150 | 420 | 340 | 90,440 | 200 | 110,390 | G. Krohne |
| Gng7 | U38499 | 240 | 440 | 90 | 80,100,570 | 420 | 70,160,200 | 450 | 90,200 | 270,360 | G. Sutcliffe |
| Thop1 | M61142 | 240 | 440 | 120,250? | 570 | 220 | <70 | 450 | 100 | 270,360 | C. Abraham |
| D10Bur1e | AA530431 | 240 | 440 | 250 | 570 | 220 | <70 | 450 | 100 | 270,360 | IMAGE#931394 |
| D10Wad2 | – | <50 | 440 | 150,220 | ND | 220 | ND | ND | ND | ND | R.Baldocchi |
| Map2k2 | S68267 | <100 | 440 | 150,220 | 300,560,570 | 220 | <80 | 450 | 100 | 270,360 | B.Brott |
| Minta | – | 190 | 440 | 150,220 | 300,560,570 | 220 | 80 | 450 | 200,250 | 70 | M.Taketo |
| Dapk3 | AA023742 | 190 | <50 | 150,220 | 300,560,570 | 220 | 160 | 450 | 200,250 | 130 | IMAGE# 457325 |
| D10Sut1e | D21389 | 190 | 90 | 200 | 300,560,570 | 70 | 160 | 520 | <90 | 130 | IMAGE# 329416 |
| Matk | D45243 | 190 | 90 | 200 | 300,560,570 | 70 | 160 | 520 | <90 | 130 | R. White |
| Tbxa2r | D10849 | 80 | 90 | 200 | 300,560,570 | 120 | 320 | 520 | 90,270 | <70 | T. Namba |
| Hmg20b | AA106132 | 80 | 90 | 200 | 300,560,570 | 120 | 320 | 520 | 90,270 | <70 | IMAGE #522081 |
| Gna11 | M57617 | 210 | 100 | 260 | 120 | 200 | 320 | 520 | 70,170 | 180 | T. Wilkie |
| Gna15 | M80632 | 210 | 100 | 260 | 120 | 200 | 320 | 520 | 70,170 | 180 | T. Wilkie |
| Grg | L12140 | 210 | 1000 | 260 | 120 | 200 | 70 | 520 | 100,170 | 180 | PCR product |
| Sirt6 | AA839912 | 210 | ND | 260 | ≪90 | 200 | ND | 520 | ND | 180 | IMAGE #1259892 |
| D10Bur2e | W61992 | 210 | ND | 260 | 240 | 200 | 470-500 | 520 | 100,170 | 180 | IMAGE #374959 |
| Nfyb | X55316 | 210 | 1000 | 900,1200 | 250 | 80,100 | 600 | 550,360,340 | 450 | 370,280,260 | D. Mathis |
Fragment sizes detected by gene probes are shown in kilobases for several relevant enzymes, as determined by PFGE mapping. Markers/genes are shown in order from centromere to telomere. More than one gene on a line indicates that PFGE mapping could not resolve the order. Accession numbers are Genbank. All are mouse DNA sequences except for Thop1, where only rat sequence is available and given.
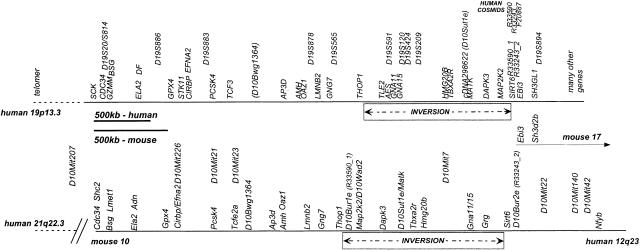
Figure 2.
Physical map of 19p13.3 and mouse chromosome 10 identifies inversion and synteny breaks. Overall physical map constructed from the cosmid contiguous sequence (contig) restriction maps (19p13.3) and schematic constructions of the mouse chromosome 10 pulsed-field gel electrophoresis (PFGE) map. The map shows overall conserved order and distances of many genes for ∼2500 kb — ∼1800 kb in mouse—followed by a region of ∼1200 kb — ∼900 kb in mouse — which is inverted in the two species relative to each other. In most cases, the mouse and the human gene names are identical or nearly identical (see also Table 1). The mouse genetic markers (D10Mit …) were placed onto the map using genetic breakpoints and yeast artificial chromosomes (YACs). The mouse map overall is shorter than the human map. To better compare the gene order of the two maps, they were scaled proportionately, such that the maps are easily compared, but note that the size bars are different in size.
Integration of Genetic Markers into the Physical Map
Several murine genetic markers were integrated fully into the physical map. First, several crosses with CAST/Ei and CASA/Rk of mouse mutants in this region have been described previously (Kantheti et al. 1996, 1998; Kapfhamer and Burmeister 1994) as well as one similar, unpublished cross with apathetic (Chung and Leibel, pers. comm.; W.K. Chung, unpubl.). These crosses have been continued and expanded to a total of over 6000 meioses. Yeast artificial chromosomes (YACs) were identified by two different means: first, we screened the pool of a commercially available mouse YAC library (Research Genetics, Huntsville, AL) with a number of probes and markers. Second, after Nusbaum et al. (1999) published a framework YAC map, these YACs (called WI-YACs, below) were ordered and tested not only with genetic markers but also with probes that we had previously placed on the physical map. YAC 59A4 was <300 kb in size and hybridized to both D10Mit226 and Efna2, thus placing D10Mit226 on the physical map near Efna2. D10Mit21 and D10Mit23 are colocalized on many of the WI-YACs (Nusbaum et al. 1999). In addition, YAC200A12 was positive for D10Mit23, Tcfe2a, and D10Bwg1364, but not D10Mit21, placing D10Mit21 proximal to D10Mit23. In fact, BLAST analysis of the published sequence for D10Mit23 shows that this marker is contained within intron 2 of Tcfe2a. D10Mit7 could only be located genetically (between Tbxa2r and Gna15) because that region is unstable or difficult to clone in YACs. Previously, we already had established the order D10Mit7 – D10Mit22 – D10Mit140 – D10Mit42 (Kapfhamer et al. 1996). These results were confirmed and expanded by YAC analysis: 390H10 was positive for D10Mit22, D10Mit140, and D10Mit42, as well as Nfyb, 298F6 and others were positive for D10Mit140, D10Mit42, and Nfyb, but negative for D10Mit22, whereas 411C1 was positive for D10Mit140 and D10Mit42, but not Nfyb. The existing primers for D10Mit22 appear to amplify two different loci that map close to each other, and thus the placement of D10Mit22 on YACs is somewhat ambiguous. None of these were positive for Grg, which together with the genetic data, places these markers distal to Grg as shown in Figure 2. D10Mit207 maps genetically distal to D10Mit175 and proximal to D10Mit226 (Dietrich et al. 1994, D. Kapfhamer and M. Burmeister, data not shown), and D10Mit175 maps to the region of homology to human 21q22 ∼100 kb from the breakpoint to 19p13.3 (Wiltshire et al. 1999). Additionally, genetic mapping has previously placed D10Mit207 proximal to Bsg (Kapfhamer et al. 1996), allowing placement of D10Mit207 between Bsg and the beginning synteny to 21q22 (Fig. 2).
An Inversion Within the Region of Homology Between HSA 19p13.3 and MMU10
The human and mouse maps were compared, considering both the order of markers and the distance between markers. For most of the region analyzed (CDC34 through THOP1), the order of markers was collinear between human 19p13.3 and mouse chromosome 10. However, as illustrated in Figure 2, there is an inversion of about 1200 kb, between the mouse and human sequences (size from the human sequence). Except for this inversion, we found the gene order between mouse and human to be conserved within this region, and gene order also is conserved within the inversion. Several landmarks of the inversion were confirmed by genetic mapping, using either previously published crosses (Chung and Leibel, pers. comm.; Kantheti et al. 1998; Kapfhamer and Burmeister 1994; Kapfhamer et al. 1996) or the publicly available BSS panel (Rowe et al. 1994). For example, several recombinant animals between Gna15 and Tbxa2r genetically confirmed the existence of an inversion and located D10Mit7 into that interval (data not shown). With over 30 genes tested, there was no evidence for genes from this region of 19p13.3 mapping elsewhere in the mouse genome.
While gene orders, except for the inversion, are conserved and no genes appear to be translocated, the apparent map distances are significantly shorter in mouse than determined from the human cosmid restriction map. While there is uncertainty in the size estimate from PFGE data (see Discussion), the observed reduction in size by about 30% (Fig. 2) is larger than would be expected for technical reasons alone. The only human gene for which we found no mouse orthologue in databases nor experimentally by Southern or Northern blot hybridization is azurocidin (AzuI; M96326, data not shown). However, this is a small (about 10 kb) human gene, and even if a few more mouse genes might be found missing, the overall shorter map distances over several Mbp in mouse are unlikely because of only a few missing genes.
The End of the Inversion is Not Identical with the Synteny Break
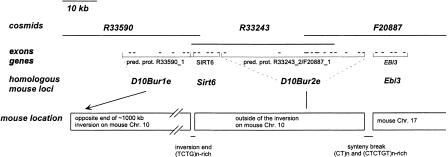
Following the end of the inversion, we have found that Nfyb (Table 2), as well as Tdg (data not shown), which map to human chromosome 12 (Li et al. 1991; Sard et al. 1997), are within less than 1000 kb of markers from the 19p13.3 synteny such as Grg (the mouse homolog of human AES). Because human AES is a gene within the inversion mapping close to THOP1 (which is outside of the inversion), this result suggested that the end of the inversion is very close to the synteny break on mouse chromosome 10 between regions of homology to human 19p13.3 and 12q23 or, on human 19p13 close to the break between homologies to mouse chromosomes 10 and 17. To locate the breakpoint precisely and to determine if the end of the inversion coincides with the end of the synteny between MMU10 and HSA 19p13.3, we analyzed sequences from HSA 19p13.3 cosmids near the synteny break. Figure 3 shows the three human cosmids spanning these breakpoints and the location of predicted genes. Mouse ESTs homologous to predicted genes were identified through BLAST searches, and the corresponding genes mapped either genetically (Ebi3) or by PFGE (all others). Ebi3 was mapped to mouse chromosome 17 (as do many genes proximal to EBI3), all other homologs to mouse chromosome 10. Two genes, Sirt6 and D10Bur2e, were found to map to mouse chromosome 10 outside the inversion based on PFGE (Fig. 1; Table 2) as well as genetic mapping (D10Bur2e, data not shown). These results identify the sequences between R33590_1 (D10Bur1e) and SIRT6 as the inversion breakpoint, and the sequences between R33243_2 (also F20887_1) (mouse 10) and EBI3 (mouse 17) as the synteny break.
Figure 3.
Identification of chromosomal breakpoints on human cosmid sequences. A schematic presentation of predicted and known genes on three cosmids near the synteny and inversion breakpoints is shown. Size and distance between exons is approximate and not to scale. The predicted gene sequences were blasted, and the inserts of IMAGE clones corresponding to identified mouse ESTs were PCR-amplified and mapped by pulsed-field gel electrophoresis (PFGE) and genetic mapping. The mouse loci names D10Bur1e and D10Bur2e are those assigned by the mouse nomenclature committee. The human predicted genes have not yet been named. The mouse Ebi3 gene was mapped genetically to mouse chromosome 17 on a publicly available panel (Rowe et al. 1994). Based on the boundary of the genes, the synteny break and the inversion break have been localized to small regions less than 5 kb in size, between the EBI3 and R33243_2, and between R33590_1 and SIRT6. The accession numbers for cosmids R33590, R33243, and F20887 are AC005620, AC006930, and AC005578.
The Sequences at the Evolutionary Breakpoints are Rich in Simple Repeats
Genetic and PFGE mapping of the IMAGE clones for the homologous mouse genes identified the breakpoints as a 5-kb region on human cosmid F20887 (between bases 17181 and 22227 of Genbank Accession No. AC005578) and a 3 kb region on cosmid R33590 (between 35974 and 38266 of Genbank Accession No. AC005620). Both breakpoints contain an abundance of simple sequence repeats, more than expected from a typical 5-kb human sequence: The breakpoint on F20887 contains a 1-kb region with several sets of (CT)n and (TCTCTG)n repeats. Similarly, the breakpoint on R33590 contains a 1-kb region full of simple repeats, mostly incomplete repeats, and interrupted (CAGA)n repeats.
The other boundary of the inversion is less precisely defined. It is located in a region of about 250 kb on 19p13.3 between THOP1 and AES (called Grg in mouse), between BC41195 (contains THOP1) and cosmid F23613 (contains AES). Because the last gene that is inverted in mouse (D10Bur1e) starts <100 kb distal to Thop1 (Table 2), the break is expected to be within 100 kb of Thop1. However, we currently can not exclude the possibility that some sequences were lost during the inversion.
Comparison of Physical and Genetic Maps: Hot Spots in Human are Cold Regions in Mouse
The availability of a physical map in both humans and mouse allows us to compare genetic and physical distances in both species. In human, the region between D19S886/D19S20 and D19S894 shows an increased recombination rate, especially in males. While <3000 kb in size, the genetic distance is 16 cM on average, and over 20 cM in males. Specifically, the small region between D19S886 and D19S883, <500 kb in size, has a genetic length of >5 cM (from Broman et al. 1998 and corresponding web site [see http://www.marshmed.org/genetics/]). Similarly, Mohrenweiser et al. (1998) identified the region between D19S565 and D19S120, slightly further centromeric, as a male hot spot. These results are not unexpected because telomeric, G/C-rich regions generally are recognized as hot spots of recombination in humans (Craig and Bickmore, 1993; Mohrenweiser et al. 1998).
The question therefore arises: is a recombination hot spot primarily a property of the position in the genome (in humans, most hot spots are near the telomeres) or mostly a result of the gene-rich and G/C-rich sequences (in humans, most telomeres are rich in G/C-content and in genes [Craig and Bickmore 1993] )? The region analyzed here is located in the middle of mouse chromosome 10 but contains homologous DNA to the hot spot region in human, and is also gene-rich. However, in contrast to the higher-than-average recombination rate on human 19p13.3, we found this region relatively lacking in recombination in mouse. In over 6000 meioses (F2, i.e., male and female) in five different crosses, we observed few recombinants in the region, especially between Gna15 and D10Mit21/23, resulting in a distance of <0.2 cM in 1500 kb instead of the expected 1 cM/2000 kb, or overall in <1 cM in a 5000 to 6000 kb region. Our data are confirmed by others. The Whitehead Institute MIT marker map lists all of the markers from the 3000 to 4000 kb human 19p13.3 homology region (D10207, 21, 23, 7, 140, and 22) as well as two markers from the human 21q homology region (D10Mit139, 175) at the same position, and only D10Mit42 (human 12q23 homology region) is one recombination event away. Because most of these data are based on crosses involving Mus musculus castaneus, we also analyzed over 1000 meioses in 2 previously described F2's involving C57BL/6J and C3H/HeJ (Kantheti et al. 1998; Kapfhamer et al. 1996). This confirmed that there is very little recombination in this region (only D10Mit42 and D10Mit175 were informative). Additionally, there also is a paucity of recombination in crosses involving Mus spretus. Rowe et al. (1994) have extensive data on their web page (http://www.jax.org/resources/documents/cmdata/bkmap/BSS10data.html) showing that a 1-cM “bin” contains all of the region of human 21q 22.3/MMU10 homology as well as the 19p13.3 homology region, estimated to be about 6000 kb (from data presented here in addition to the size of the mouse 10/human 21q22.3 region) (Burmeister et al. 1991; Cole et al. 1999). The number of recombinants thus is at least three times less than the expected 1600 to 2000 kb/cM in mouse in several different crosses involving many different mouse strains, identifying a “cold spot” or better, “cold region” of recombination.
DISCUSSION
Mouse Cdc34 Maps to Mouse Chromosome 10, Not 11
Several of the genes reported here were mapped to human 19p13.3 or mouse chromosome 10 for the first time, while others have never been mapped with this precision before. Cdc34 has been listed as located on mouse chromosome 11 for many years based on Plon et al. (1993) in which, however, the actual mouse mapping data are not shown. Sequence analysis of the mouse contig of this region (D.E. Jenne, unpubl.) unambiguously showed mouse Cdc34 on mouse chromosome 10 centromeric to the protease cluster (Lmet1/Bsg). Additionally, mapping a mouse Cdc34 probe on the “BSS” panel of the Jackson Laboratory (Rowe et al. 1994) identified a gene on mouse chromosome 10, as well as a Mus spretus-specific band that maps to mouse chromosome 11 (see exact position at http://www.jax.org/resources/documents/cmdata/bkmap/BSS10data.html). This latter band has been shown to be a Mus spretus-specific pseudogene (D.E. Jenne, D. Kapfhamer, and M. Burmeister, in prep.) and is likely the reason for the previous erroneous assignment of the Cdc34 gene to mouse chromosome 11.
Small Intrachromosomal Rearrangements–More Common Than Thought
We have presented here a comparative map showing a region of conserved marker order followed by an inversion of ∼1200 kb in human (shorter in mouse), followed by a very small region of ∼50 kb (human) that is not inverted. This in turn is followed by a break in the synteny. The size of the inversion and the subsequent not inverted DNA stretch are so small that they had not been detected previously, and are hardly detectable by genetic means. While the average conserved segment between mouse and human has been predicted to be ∼8 cM or ∼16 Mbp in size, corresponding to 5 to 6 different conserved segments per chromosome (Nadeau and Sankoff 1998a,b), here a region of ∼5000 kb contains 5 different segments — homology to 21q22.3, 19p13.3, 19p13 inversion, 19p13.3 not inverted segment, 12q23. Analysis at such a high resolution rarely has been achieved. However, whenever high resolution mapping was performed, the evidence suggests that small intrachromosomal rearrangements may be the rule rather than an exception, as summarized by Carver and Stubbs (1997). Similar examples are a 10-cM region of homology between human 5q and mouse 11, which is split into at least 4 different segments (Watkins-Chow et al. 1997), and a 2.5 Mbp region of MMU16 homologous to HSA 22q that is rearranged into three different blocks (Lund et al. 1999; Puech et al. 1997). Thus, intrachromosomal rearrangements of the type uncovered here seem to be quite common once they can be reliably detected.
The Homologous Region in Mouse is Shorter Than the 19p13.3 Map
The size of the region in mouse appears shorter than the homologous region on human 19p13.3 (2500 to 2900 kb compared to about 3800 kb. Figures are based on the more conservative estimates). For example, the inverted segment appears to be ∼900 to 1000 kb in mouse rather than 1200 kb in human. As evidenced in Figure 2, in which the maps were scaled proportionately in order to superimpose them, there is not a single interval that is more significantly affected than others, and with the exception of azurocidin, no single gene or segment is missing from the mouse chromosome 10 map. Based on the resolution of the PFGE map and the density of probes, we estimate that we would have been able to detect missing or translocated segments larger than ∼200 kb in mouse. There are two notes of caution for evaluating these results. First, a PFGE map is constructed by placing markers on fragments. Overlapping fragments result in a continuous map. While this usually gives unambiguous order of markers, the exact amount of overlap is not determined, resulting in some ambiguity of size at each point. Second, the conditions under which we used PFGE were to allow multiple rehybridization by loading 5 to 10 μg of DNA per lane. These conditions often result in apparently shorter fragment lengths (Doggett et al. 1992). Comparing the recently published chromosome 21 sequence (Hattori et al. 2000) with older PFGE maps of the same region prepared using the same PFGE techniques used here (Burmeister et al. 1991), the size error seems to be ∼10% (M.B., data not shown). However, precise restriction mapping of mouse BACs or sequencing of the mouse region certainly will be needed to confirm this observation and to determine why the mouse is shorter than the human map. Preliminary restriction map analysis in mouse (L.A. Gordon and Lisa Stubbs, unpubl.) confirms that our observation of a shorter map in mouse by ∼30% is not solely a PFGE artifact.
Sequences at Evolutionary Breakpoints
Here we identify an inversion and a synteny breakpoint at the sequence level at a resolution of a few kilobases. On chromosome 7, an evolutionary breakpoint recently has been narrowed to ∼300 kb (Thomas et al. 1999), and on mouse chromosome 10, the breakpoints between HSA 21 and 22 as well as between HSA 21 and 19 have been cloned in PACs but not yet sequenced (Wiltshire et al. 1999). In the recently completely sequenced HSA 22, breakpoints also can be identified. For example, Gnaz maps to mouse chromosome 10, and the next gene proximal on 22q is the Igl cluster on mouse chromosome 16. However, the region between them spans more than one cosmid or BAC, and thus is not as well defined. To our knowledge, evolutionary breakpoints rarely have been identified with the precision of a few kb achieved here. Very recently, comparative sequencing identified a 9-kb region containing a synteny breakpoint (Lund et al. 2000). Lund et al. (2000) did not notice any unusual sequences in that interval. The sequences at the two breakpoints identified here are rich in TCTG, CT, and GTCTCT repeats. TCTG tandem repeats previously have been identified in a murine recombination breakpoint hot spot region (Shiroishi et al. 1990). However, given that so far only these three evolutionary breakpoint sequences are available, further studies are needed to determine whether small tandem CT-rich repeats are involved in evolutionary breakpoints in general, and identification of more breakpoint sequences may well point to other elements.
The other end of the inversion breakpoint is less well defined, only within ∼100 kb (see results) but will be very interesting once the mouse sequence becomes available. Within this region near the inversion break interval is a zinc finger gene sequence, ZNF57, on BAC BC102889 (AC006130). Stubbs et al. (1996) have found that zinc finger clusters often are near evolutionary breakpoints on HSA 19, and Lund et al. (2000) have identified one synteny break within a zinc finger gene. How often zinc finger clusters coincide with evolutionary breakpoints will be revealed only once the sequencing of the homologous region in mouse will be completed. Mouse sequence homologous to human chromosome 19 will be available at http://www.jgi.doe.gov.
Relevance for Mapping Disease Genes
The comparative map presented here will also aid in the identification of genes and mouse models for human disorders. The human recessive disorder Cayman ataxia [ATCAY (Nystuen et al. 1996)] has been located proximal to D19S424. Similarly, a form of febrile seizures (FEB2) has been mapped to 19p13.3 proximal to D19S591 and distal to D19S395 (Johnson et al. 1998). Thus, the mouse homolog of ATCAY or FEB2 could be expected either on mouse chromosome 10 within the inversion, outside of the inversion on the proximal end, or on mouse chromosome 17. In order to determine whether jittery or apathetic are homologous to either of these disorders, it is important to consider the inversion, demonstrating the value of high-resolution comparative mapping also for disease gene identification.
METHODS
Construction of a Clone Map of Chromosome 19
Construction of the human chromosome 19 map has been previously described (Ashworth et al. 1995). Cosmid contigs originally were assembled by fingerprinting clones from a chromosome 19-specific cosmid library constructed at LLNL (de Jong et al. 1989). Contigs were ordered along the chromosome and the distance between them was determined by high resolution pronuclear fluorescence in situ hybridization (FISH) (Brandriff et al. 1994; Gordon et al. 1995). Gaps between contigs were closed by directed walking (Olsen et al. 1994, 1996) in cosmid and BAC (Osoegawa et al. 1998; Shizuya et al. 1992) or PAC (Ioannou et al. 1994) libraries. Approximate EcoRI restriction maps of all contigs were constructed as described in Lamerdin and Carrano (1993).
Genes were localized on the map by hybridization of one or more gene-specific probes, including cDNAs, polymerase chain reaction (PCR) products and oligos. Additional genes in 19p13.3 were identified by genomic sequencing of a tiling path of BAC and cosmid clones spanning the region. Updated versions of the metric physical map and underlying restriction maps, as well as current sequence data, are available from the LLNL Human Genome Center web site (http://www-bio.llnl.gov/bbrp/genome/genome.html).
To calculate a rough estimate of the proportion of genes on the map presented here, we chose three fully sequenced regions, each of 4 to 5 cosmids/fosmids spanning a total of ∼200 kb, counted the number of known and predicted genes in each segment and noted which of them were mapped here. This is a very rough estimate because not all predicted exons are indeed part of genes, and it is unknown how many predicted exons are part of any one gene. In the regions chosen, surrounding the genes for GPX4, TCF3 and TBXA2R/HMG20B, we had mapped 1/13, 1/9, and 2/12 genes — counting known and predicted genes — thus on average ∼10% of all genes.
Genetic Mapping
Crosses used have been described before (Kantheti et al. 1998; Kapfhamer and Burmeister 1994; Kapfhamer et al. 1996), except for a cross involving apathetic (W.K. Chung, in prep.). These crosses have been continued after their original description and now >6000 meioses have been analyzed. All animals first were screened by PCR as described (Kapfhamer et al. 1996) for flanking markers D10Mit42 or D10Mit140 and D10Mit175, and only recombinants within that interval were analyzed further. The small number of animals that showed recombination events were scored for markers D10Mit7, D10Mit21 and D10Mit23, and for restriction fragment length polymorphisms (RFLPs) in nearby genes.
When there was any doubt whether a gene mapped to mouse chromosome 10, DNA from the public BSS panel (Rowe et al. 1994) was used to demonstrate the mapping of a gene to mouse chromosome 10. In short, an RFLP testing blot with C57BL/6J and Mus spretus DNA (Jackson Laboratory) was prepared by cleaving the DNAs with nine different restriction enzymes. In this manner, Sh3d2b and Ebi3 were mapped to mouse chromosome 17, and Gpx4, Cdc34, and D10Bur1e were mapped to mouse chromosome 10. The detailed data are available at the cross' web site (http://www.jax.org/resources/documents/cmdata/bkmap/BSS10data.html).
Generation of Mouse Probes
Clones that were published as mapping to central mouse chromosome 10, human 19p13.3, or both were ordered from American Type Culture Collection (ATCC) or the authors of the publications (see Table 2 for each gene). As the sequence of human 19p13.3 began to emerge, sequences from the cosmids (some “excellent” Grail predicted exons or matches with human ESTs) also were used to screen the mouse EST database on NCBI's BLAST server (Altschul 1993; Altschul et al. 1994). When homologies >80% identity for >100 bp were found, they were considered likely mouse orthologs, for which IMAGE clones were obtained (Research Genetics). IMAGE clones were ordered from Research Genetics and the inserts were amplified with appropriate vector primers as indicated by the IMAGE consortium. Correct clone identity was confirmed by sequencing or by cross hybridization of two independent clones. After verification, all clones were used for PFGE mapping. Additionally, the public mouse mapping databases (called BSS and BSB panels) at the Jackson Laboratory (Rowe et al. 1994) were periodically screened for clones that map into the “bin” of interest. This latter tool was relatively imprecise because genes with homologies to human 21q22.3 are located in the same bin as all the genes on mouse chromosome 10 with homology to 19p13.3. In addition, partial cosmid walks, in particular in the protease cluster region (Wong et al. 1999; D.E Jenne, unpubl.), allowed identification and mapping of end clones of walks that helped in orienting genes in the protease region.
PFGE Mapping
Blocks for PFGE analysis from mouse spleen were prepared essentially as described (Herrmann et al. 1987). A mouse spleen was quickly dissected, the outer skin cut horizontally, the spleen cut into a few pieces, mixed with 2 ml of phosphate-buffered saline (PBS) and suspended with 6 strokes in a hand-held Teflon homogenizer. Patches of nonhomogenized skin or tissue were removed with a Pasteur pipette. The remaining cell suspension (about 2 ml) was moved to a new Falcon tube, warmed to 45°C on a water bath, and mixed with an equal volume of 1.6% low-melting-point agarose (Gibco-BRL) previously prepared in PBS and maintained at 45°C. The mix was pipetted into Plexiglas slot formers maintained on a flat surface (glass plate) over ice. Blocks are chilled for a minimum of 30 min. followed by transfer to 10 ml lysis buffer (1 mg/ml proteinase K in 1% N-laurylsarcosine, 10 mM Tris/HCl, 0.45 M EDTA, pH 8.0) and maintained at 50°C with slight shaking overnight. Blocks are then washed in TE in the presence of 40μg/ml PMSF to inactivate the proteinase K. Blocks are stored in 0.5 M EDTA at 4 °C and washed in TE prior to restriction digests. Each resulting block contains ∼10 μg of DNA in a volume of 50 μl. Restriction digests were performed for ∼15 hours, with a second enzyme addition after ∼5 hours, at 37°C (or 50°C in the case of SfiI or BssHII) in a final volume of 200 μl in the restriction enzyme buffers supplied by the manufacturer (New England Biolabs). After digestion, EDTA was added to the blocks that were loaded onto an agarose gel prepared according to manufacturer's instructions (Biorad). The gel was run on a Biorad Chefmapper II, programmed to separate 50 to 800 kb (or, in the case of the 1000 kb NotI band, 100 to 1200 kb). Agarose gels were blotted onto Hybond N + filters (Amersham) in 0.5 M NaOH/1.5 M NaCl. After ∼15 hours of blotting, one membrane was removed and a second membrane was added on the gel, and blotting continued for another 15 to 24 hours. Filters were neutralized in 2 x SSC.
For map construction, adjacent probes were analyzed on the same filter, stripped and rehybridized (indeed, some filters were stripped and rehybridized >50 times). If three or more fragments had identical migration rate, they were considered identical. If one or two fragments were identical in size, other evidence was needed to corroborate the overlap. If additional probes were available that might map in between, they were hybridized as well. If such probes were not available, polymorphisms also were considered. For example, in the case of Nfyb and Grg (Table 2), the two probes recognized only one fragment of identical size, a 1000-kb NotI fragment. Because there could be more than one NotI fragment of that size, additional evidence was needed. This was obtained by hybridizing a blot with NotI-digested DNA from CAST/Ei, JIGR-ji/ji and C57BL/6J. These 3 strains showed fragments of 3 different sizes (800 and 900 kb for CAST/Ei and JIGR-ji/ji) but not control probes hybridizing to NotI fragments in a similar size range. This result indicated the presence of polymorphisms. Thus, overall, three different fragments were recognized and the evidence that identical bands hybridized with Grg and Nfyb was considered sufficient. When drawing maps, there is always some ambiguity. PFGE data typically give a minimal and a maximal distance between markers. While the order of markers could be determined with high precision, there is considerable error in the distances. The results shown in Figure 1 are from DNA of JIGR-ji/ji mice but similar results were obtained with DNA from normal (C57BL/6J) mice.
The human and the mouse physical maps were drawn independently. For Figure 2, after scaling to equal scale, it became clear that the mouse map was shorter than the human map. After the final map was drawn, graphic programs were used to scale the map simultaneously with the size bar, until flanking markers were aligned.
Acknowledgments
We thank the many investigators around the world who contributed probes to our effort, some of whom are mentioned in Table 2, and Damaris Sufalko (Michigan) and Anca Georgescu (LLNL) for technical assistance. We appreciated the availability of clones from the IMAGE consortium, and useful discussions with Val Sheffield and Arne Nystuen (University of Iowa). We thank Heinz Himmelbauer (Berlin) and Michael Hortsch for useful comments on the manuscript. This work was supported in part by the March of Dimes, the National Institutes of Health grant NS32130, and the Alexander von Humboldt Foundation (M.B.). M.B. thanks Hans Lehrach, Heinz Himmelbauer, and Leo Schalkwyk (Max Planck Institute, Berlin) for hosting her for a sabbatical in Berlin. Work at LLNL was supported by the U.S. Department of Energy under contract No. W-7405-ENG-48.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Present address: DOE Joint Genome Institute, 2800 Mitchell Drive, B100, Walnut Creek, CA 94598
E–MAIL olsen2@llnl.gov; FAX (425) 296–5666.
E–MAIL margit@umich.edu; FAX (734) 647–4130.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.145200.
REFERENCES
- Altschul SF. A protein alignment scoring system sensitive at all evolutionary distances. J Mol Evol. 1993;36:290–300. doi: 10.1007/BF00160485. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Boguski MS, Gish W, Wootton JC. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Ashworth LK, Batzer MA, Brandriff B, Branscomb E, de Jong P, Garcia E, Garnes J A, Gordon LA, Lamerdin JE, Lennon G, et al. An integrated metric physical map of human chromosome 19. Nature Genetics. 1995;11:422–427. doi: 10.1038/ng1295-422. [DOI] [PubMed] [Google Scholar]
- Brandriff BF, Gordon LA, Fertitta A, Olsen AS, Christensen M, Ashworth LK, Nelson DO, Carrano AV, Mohrenweiser HW. Human chromosome 19p: A fluorescence in situ hybridization map with genomic distance estimates for 79 intervals spanning 20 Mb. Genomics. 1994;23:582–591. doi: 10.1006/geno.1994.1546. [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister M. Strategies for mapping large regions of mammalian genomes. In: Burmeister M, et al., editors. Methods in molecular biology, pulsed field gel electrophoresis. Vol. 12. Totowa, N.J: Humana Press; 1992. pp. 259–284. [DOI] [PubMed] [Google Scholar]
- Burmeister M, Bryda EC, Bureau JF, Noben-Trauth K. Mammalian Genome 8: S200–214. 1998. Encyclopedia of the mouse genome VII. Mouse chromosome 10. [DOI] [PubMed] [Google Scholar]
- Burmeister M, Kim S, Price ER, de Lange T, Tantravahi U, Myers RM, Cox D R. A map of the distal region of the long arm of human chromosome 21 constructed by radiation hybrid mapping and pulsed field gel electrophoresis. Genomics. 1991;9:19–30. doi: 10.1016/0888-7543(91)90216-2. [DOI] [PubMed] [Google Scholar]
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Cole SE, Wiltshire T, Rue EE, Morrow D, Hieter P, Brahe C, Fisher EM, Katsanis N, Reeves RH. High-resolution comparative physical mapping of mouse chromosome 10 in the region of homology with human chromosome 21. Mamm Genome. 1999;10:229–234. doi: 10.1007/s003359900978. [DOI] [PubMed] [Google Scholar]
- Craig JM, Bickmore WA. Chromosome bands—flavours to savour. Bioessays. 1993;15:349–354. doi: 10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- de Jong PJ, Yokabata K, Chen C, Lohman F, Pederson L, McNinch J, Van Dilla M. Human chromosome-specific partial digest libraries in lambda and cosmid vectors. Cytogenet Cell Genet. 1989;51:985. [Google Scholar]
- Dietrich WF, Miller JC, Steen RG, Merchant M, Joyce DC, Wessel M, Dredge RD, Marquis A, Stein LD, Goodman N, et al. A genetic map of the mouse with 4006 simple sequence length polymorphisms. Nature Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- Doggett NA, Smith CL, Cantor CR. The effect of DNA concentration on mobility in pulsed field gel electrophoresis. Nucleic Acids Res. 1992;20:859–864. doi: 10.1093/nar/20.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LA, Bergmann A, Christensen M, Danganan L, Lee DA, Ashworth LK, Nelson DO, Olsen AS, Mohrenweiser HW, Carrano AV, et al. A 30-Mb metric fluorescence in situ hybridization map of human chromosome 19q. Genomics. 1995;30:187–194. doi: 10.1006/geno.1995.9886. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi D K, et al. The DNA sequence of human chromosome 21. The chromosome 21 mapping and sequencing consortium. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Barlow DP, Lehrach H. A large inverted duplication allows homologous recombination between chromosomes heterozygous for the proximal t complex inversion. Cell. 1987;48:813–825. doi: 10.1016/0092-8674(87)90078-x. [DOI] [PubMed] [Google Scholar]
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, Muller O, Back W, Zimmer M. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- Johnson EW, Dubovsky J, Rich SS, O'Donovan CA, Orr HT, Anderson VE, Gil-Nagel A, Ahmann P, Dokken CG, Schneider DT, et al. Evidence for a novel gene for familial febrile convulsions, FEB2, linked to chromosome 19p in an extended family from the Midwest. Hum Mol Genet. 1998;7:63–67. doi: 10.1093/hmg/7.1.63. [DOI] [PubMed] [Google Scholar]
- Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, Carskadon SL, Kapfhamer D, Sufalko D, Robinson MS, Noebels JL, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Kapfhamer D, Burmeister M. Genetic map of the region around grizzled (gr) and mocha (mh) on mouse chromosome 10, homologous to human 19p13.3. Genomics. 1994;23:635–642. doi: 10.1006/geno.1994.1552. [DOI] [PubMed] [Google Scholar]
- Kapfhamer D, Sweet HO, Sufalko D, Warren S, Johnson KR, Burmeister M. The neurological mouse mutations jittery and hesitant are allelic and map to the region of mouse chromosome 10 homologous to 19p13.3. Genomics. 1996;35:533–538. doi: 10.1006/geno.1996.0394. [DOI] [PubMed] [Google Scholar]
- Lamerdin JE, Carrano AV. Automated fluorescence-based restriction fragment analysis. Biotechniques. 1993;15:294–303. [PubMed] [Google Scholar]
- Li XY, Mattei MG, Zaleska-Rutczynska Z, van Huijsduijnen RH, Figueroa F, Nadeau J, Benoist C, Mathis D. One subunit of the transcription factor NF-Y maps close to the major histocompatibility complex in murine and human chromosomes. Genomics. 1991;11:630–634. doi: 10.1016/0888-7543(91)90070-u. [DOI] [PubMed] [Google Scholar]
- Lund J, Roe B, Chen F, Budarf M, Galili N, Riblet R, Miller RD, Emanuel BS, Reeves RH. Sequence-ready physical map of the mouse chromosome 16 region with conserved synteny to the human velocardiofacial syndrome region on 22q11.2. Mamm Genome. 1999;10:438–443. doi: 10.1007/s003359901019. [DOI] [PubMed] [Google Scholar]
- Lund J, Chen F, Hua A, Roe B, Budarf M, Emanuel BS, Reeves RH. Comparative sequence analysis of 634 kb of the mouse chromosome 16 region of conserved synteny with the human velocardiofacial syndrome region on chromosome 22q11.2 (in prep) Genomics. 2000;63:374–383. doi: 10.1006/geno.1999.6044. [DOI] [PubMed] [Google Scholar]
- Mohrenweiser H, Olsen A, Archibald A, Beattie C, Burmeister M, Lamerdin J, Lennon G, Stewart E, Stubbs L, Weber JL, et al. Report and abstracts of the third international workshop on human chromosome 19 mapping 1996. Cytogenet Cell Genet. 1996;74:161–186. doi: 10.1159/000134408. [DOI] [PubMed] [Google Scholar]
- Mohrenweiser HW, Tsujimoto S, Gordon L, Olsen AS. Regions of sex-specific hypo- and hyper-recombination identified through integration of 180 genetic markers into the metric physical map of human chromosome 19. Genomics. 1998;47:153–162. doi: 10.1006/geno.1997.5097. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. Counting on comparative maps. Trends Genet. 1998a;14:495–501. doi: 10.1016/s0168-9525(98)01607-2. [DOI] [PubMed] [Google Scholar]
- ————— The lengths of undiscovered conserved segments in comparative maps. Mamm Genome. 1998b;9:491–495. doi: 10.1007/s003359900806. [DOI] [PubMed] [Google Scholar]
- Nusbaum C, Slonim DK, Harris KL, Birren BW, Steen RG, Stein LD, Miller J, Dietrich WF, Nahf R, Wang V, et al. A YAC-based physical map of the mouse genome. Nat Genet. 1999;22:388–393. doi: 10.1038/11967. [DOI] [PubMed] [Google Scholar]
- Nystuen A, Benke PJ, Merren J, Stone EM, Sheffield VC. A cerebellar ataxia locus identified by DNA pooling to search for linkage disequilibrium in an isolated population from the Cayman Islands. Hum Mol Genet. 1996;5:525–531. doi: 10.1093/hmg/5.4.525. [DOI] [PubMed] [Google Scholar]
- Olsen AS, Teglund S, Nelson D, Gordon L, Copeland A, Georgescu A, Carrano A, Hammarstrom S. Gene organization of the pregnancy-specific glycoprotein region on human chromosome 19: Assembly and analysis of a 700-kb cosmid contig spanning the region. Genomics. 1994;23:659–668. doi: 10.1006/geno.1994.1555. [DOI] [PubMed] [Google Scholar]
- Olsen AS, Georgescu A, Johnson S, Carrano AV. Assembly of a 1-Mb restriction-mapped cosmid contig spanning the candidate region for Finnish congenital nephrosis NPHS1 in 19q13.1. Genomics. 1996;34:223–225. doi: 10.1006/geno.1996.0270. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanese JJ, de Jong PJ. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Plon SE, Leppig KA, Do HN, Groudine M. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc Natl Acad Sci. 1993;90:10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, et al. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- Puech A, Saint-Jore B, Funke B, Gilbert DJ, Sirotkin H, Copeland NG, Jenkins NA, Kucherlapati R, Morrow B, Skoultchi AI. Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization. Proc Natl Acad Sci. 1997;94:14608–14613. doi: 10.1073/pnas.94.26.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MS, Thurston SJ, Birkenmeier EH. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Sard L, Tornielli S, Gallinari P, Minoletti F, Jiricny J, Lettieri T, Pierotti MA, Sozzi G, Radice P. Chromosomal localizations and molecular analysis of TDG gene-related sequences. Genomics. 1997;44:222–226. doi: 10.1006/geno.1997.4843. [DOI] [PubMed] [Google Scholar]
- Shiroishi T, Hanzawa N, Sagai T, Ishiura M, Gojobori T, Steinmetz M, Moriwaki K. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics. 1990;31:79–88. doi: 10.1007/BF00661217. [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs L, Carver EA, Shannon ME, Kim J, Geisler J, Generoso EE, Stanford BG, Dunn WC, Mohrenweiser H, Zimmermann W, et al. Detailed comparative map of human chromosome 19q and related regions of the mouse genome. Genomics. 1996;35:499–508. doi: 10.1006/geno.1996.0390. [DOI] [PubMed] [Google Scholar]
- Thomas JW, Lee-Lin SQ, Green ED. Human-mouse comparative mapping of the genomic region containing CDK6: Localization of an evolutionary breakpoint. Mamm Genome. 1999;10:764–767. doi: 10.1007/s003359901088. [DOI] [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, et al. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280:1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- Watkins-Chow DE, Buckwalter MS, Newhouse MM, Lossie AC, Brinkmeier ML, Camper SA. Genetic mapping of 21 genes on mouse chromosome 11 reveals disruptions in linkage conservation with human chromosome 5. Genomics. 1997;40:114–122. doi: 10.1006/geno.1996.4532. [DOI] [PubMed] [Google Scholar]
- Wiltshire T, Pletcher M, Cole SE, Villanueva M, Birren B, Lehoczky J, Dewar K, Reeves R H. Perfect conserved linkage across the entire mouse chromosome 10 region homologous to human chromosome 21. Genome Res. 1999;9:1214–1222. doi: 10.1101/gr.9.12.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ET, Jenne DE, Zimmer M, Porter SD, Gilks CB. Changes in chromatin organization at the neutrophil elastase locus associated with myeloid cell differentiation. Blood. 1999;94:3730–3736. [PubMed] [Google Scholar]