Abstract
Using mouse BAC clones spanning an imprinted interval of proximal mouse chromosome 7 and the genomic sequence of the related interval of human chromosome 19q13.4, we have identified a novel mouse gene, Usp29 (ubiquitin-specific processing protease 29), near two known imprinted genes, Peg3 and Zim1. Gene Usp29 is located directly adjacent to Peg3 in a “head-to-head” orientation, and comprises exons distributed over a genomic distance of at least 400 kb. A similar human gene is also found in the homologous location in human chromosome 19q13.4. The mouse Usp29 gene is also imprinted and is transcribed mainly from the paternal allele with highest expression levels in adult brain, especially in the cerebral cortex and hippocampus, and in the forebrain, face, and limb buds of midgestation mouse embryos. Analysis of a full-length 7.6-kb cDNA clone revealed that Usp29 encodes an 869-amino-acid protein that displays significant homology with yeast and nematode ubiquitin carboxyl-terminal hydrolases. These data suggest that, like the candidate Angelman syndrome gene Ube3a (ubiquitin ligase), Usp29 may represent another imprinted gene involved in the ubiquitination pathway. This identification of a third imprinted gene, Usp29, from the Peg3/Zim1-region confirms the presence of a conserved imprinted domain spanning at least 500 kb in the proximal portion of mouse chromosome 7 (Mmu7).
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF229257 and AF229438.]
Although the two alleles of most mammalian autosomal genes are functionally equivalent, the maternal and paternal copies of a subset of genes are distinguished by genomic imprinting, a mechanism by which one allele is epigenetically modified and repressed, depending upon parental origin. The epigenetic modification that determines imprinting is yet to be defined, but several lines of evidence strongly suggest that differences in DNA methylation and chromatin structure may be part of the molecular mechanism (Tilghman 1999). About 30 different imprinted genes have been isolated from human and mouse DNA, and more imprinted genes are predicted to be present in mammalian genomes (Barlow 1997; Morrison and Reeve 1998). The imprinted genes that have been described to date are clustered in discrete chromosomal regions, and the clustering of those genes is conserved in mammals (Barlow 1995; Bartolomei and Tilghman 1997). These data have been interpreted to imply that genome imprinting is a long-range mechanism that controls allele-specific transcription of multiple genes within a defined chromosomal region (Nicholls 1994; Dittrich et al. 1996; Tilghman et al. 1998, 1999).
Imprinted genes exhibit several unique features in addition to parent-of-origin-specific monoallelic expression and differential methylation. The known imprinted genes generally are expressed at early stages of development, and many encode proteins with functions related to embryonic growth (Tilghman 1999). Recent studies also suggest the involvement of imprinted genes in controlling the parental caring behavior of mammals (Lefebvre et al. 1998; Li et al. 1999). Several imprinted genes such as UBE3A (Rougeulle et al. 1998), IGF2 (Moore et al. 1997), ZNF127 (Jong et al. 1999), and IGF2R (Wutz et al. 1997) display bidirectional transcription, producing both sense and antisense transcripts. A number of imprinted genes including H19 and IPW are expressed without any coding capability; the final products of these genes are RNAs rather than proteins (Bartolomei and Tilghman 1997). Imprinted genes also tend to be comosed of relatively small numbers of exons and to contain smaller introns than nonimprinted genes (Hurst et al. 1996), although there are some notable exceptions (e.g., KvLQT1; Lee et al. 1997).
Early mouse genetic studies with translocation mutant mice predicted the presence of nine different imprinted domains distributed onto seven chromosomes (Beechey and Cattanach 1996). The chromosomal locations of most imprinted genes, in fact, are consistent with this initial prediction (Imprinting Map; http://www.mgu.har.mrc.ac.uk/imprinting/imptables.html#impmaps). Mouse chromosome 7 (Mmu7) was predicted in these early studies to contain three different imprinted domains, located in centromeric, central, and distal regions (Searle and Beechey 1990). The central and distal regions of Mmu7 are syntenically homologous to intervals of human chromosome 15q13-11 and 11p15, respectively, that contain genes associated with two imprinting genetic disorders—Prader–Willi/Angelman (Nicholls et al. 1998) and Beckwith–Wiedemann syndromes (Reik and Maher 1997). Each of these domains contains several imprinted genes whose order, sequence, and imprinting status are highly conserved in humans and mice. Until recently, only one imprinted gene, Peg3 (paternally expressed gene 3), had been localized to the centromeric imprinted domain of Mmu7 (Kuroiwa et al. 1996) and the homologous region of human chromosome 19q13.4 (Kim et al. 1997a). We exploited the well-developed physical map surrounding human PEG3 and the known conservation of this interval in humans and mice to identify a second mouse-imprinted gene, Zim1 (imprinted zinc-finger gene 1), located downstream of Peg3 (Kim et al. 1999). More recent studies identified a related and homologously positioned human zinc-finger gene, ZIM2, and showed that the gene shares seven 5′-exons with PEG3, a feature that distinguishes this pair of human genes from their closest murine counterparts (Kim et al. 2000). The significant divergence of Zim1 and ZIM2 gene organization and amino acid sequence suggests that the region surrounding PEG3 has undergone significant changes during the course of mammalian evolution.
Through analysis of clone contigs spanning homologous segments of Mmu7 and sequenced regions of human 19q13.4, we have isolated and characterized another novel mouse gene, Usp29 (ubiquitin-specific processing protease 29) and have identified its human counterpart. Our analyses show that Usp29 is also imprinted and is expressed mainly from the paternal allele during embryogenesis and in adult brain. The sequence of a full-length 7.6-kb mouse cDNA clone and human coding sequences indicate that Usp29 encodes a novel ubiquitin-specific processing protease, and together with Ube3a may represent a second imprinted gene involved in the ubiquitination pathway.
RESULTS
Isolation of Mouse Usp29
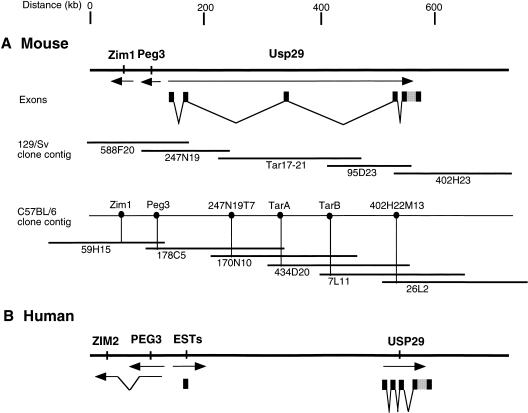
The recent isolation of the maternally expressed gene Zim1 adjacent to Peg3 (Kim et al. 1999), suggested the presence of a larger imprinted domain in the proximal Mmu7 region. To identify additional genes from this conserved region, we analyzed the recently completed human 19q13.4 genomic sequence and information derived from large-insert mouse clones spanning the homologous Mmu7 region. We constructed a physical map of a region spanning approximately 1 Mb around the Peg3 gene from the 129/Sv inbred strain by isolating a series of BAC and PAC clones through chromosome walking; a portion of this physical map is shown in Figure 1. We were unable to clone the genomic region between BAC247N19 and PAC95D23 by chromosome walking because sequences from this region are underrepresented in existing 129/Sv commercial genomic libraries. To obtain genomic clones spanning this region, we utilized the transformation-associated recombination (TAR) technique (Larionov et al. 1996). One 220-kb clone (tar17-21) was successfully obtained. The position of tar17-21 within the Peg3 region was confirmed by verifying that sequences from each end of the TAR clone were present in BACs in the flanking clone contigs. Using low-pass sequencing data from the TAR clone and the surrounding BACs, we developed a series of overgo probes that permitted a complete, parallel set of overlapping BACs to be isolated from the C57BL/6 (RPCI-23) library (Fig. 1). Comparison between the two clone maps provided additional confirmation of the TAR clone's integrity and the length and organization of the 600-kb region.
Figure 1.
Genomic organization of mouse and human USP29. (A) A physical map of a 600-kb region of proximal Mmu7 shows the relative locations and transcription direction of Zim1, Peg3,, and Usp29. The exons of Usp29 are indicated by black boxes, and the coding region of mouse Usp29 is indicated by a gray box. The genomic clones used for 129/Sv map construction include BAC (588F20, 247N19, 402H23), PAC (95D23), and TAR (17-21). A separate clone contig map using C57BL/6 genomic libraries (RPCI-23) is also shown with a subset of BAC clones. (B) The relative physical locations of human PEG3, ZIM2, and USP29 are indicated by arrows.
Genomic subclones derived from the BAC and PAC clones identified several EST matches representing genes derived from this region. In particular, one of the end sequences of BAC402H23, 1.2 kb in length, identified two ESTs (GenBank accession nos. AA549132 and AI503662), indicating the presence of an unknown gene at this site. Database searches with these EST sequences indicated a significant level of homology with a hypothetical yeast protein, a putative ubiquitin carboxyl-terminal hydrolase (GenBank accession no. NP 009614). The novel mouse gene was named Usp29 (ubiquitin-specific processing protease 29). Sequence analysis of the genomic region immediately upstream of mouse Peg3 also identified one EST match (GenBank accession no. W90868). The genomic sequence matching this EST is located about 1 kb upstream of the first exon of Peg3, and the transcriptional direction of the EST sequence is opposite to that of Peg3. These data suggested the presence of an unidentified gene with a transcription start located very near that of the Peg3 transcription unit.
The two distinct groups of ESTs were initially thought to be derived from different genes because of the 400-kb genomic distance that lies between the end of BAC clone 402H23 and the 5′-end of Peg 3 (Fig. 1). Separate screens of a large-insert mouse cDNA library with probes derived from each EST set, however, identified identical sets of cDNA clones. Sequence analysis of these cDNA clones and RT-PCR experiments using primers from the two EST clusters (data not shown) demonstrated that sequences located immediately upstream of Peg3 and the Usp29 region, located 400 kb downstream, represent 5′- and 3′-portions of a single, large transcription unit. One clone isolated from a mouse cerebellum cDNA library, representing a full-length 7.6-kb transcript, was sequenced to provide the complete open reading frame and 5′- and 3′-UTR sequences of the mouse Usp29 gene (GenBank accession no. AF229257).
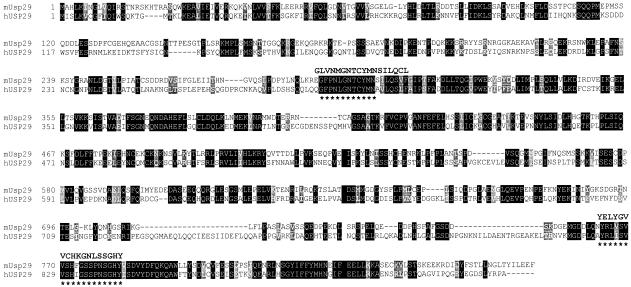
Analysis of this cDNA sequence identified one open reading frame encoding a protein 869 amino acids in length (Fig. 2). Database searches with this predicted protein sequence retrieved a large number of protein entries, most of which display ubiquitin carboxyl-terminal hydrolase activity. Although database searches with the Usp29 cDNA sequence derived a long list of similar proteins, the relatively low level of homology to known proteins suggests that Usp29 is a new member of this mammalian protein family. In fact, the highest homology identified was found with the yeast hypothetical protein (GenBank accession no. NP 009614; 21% sequence identity over 550 amino acid residues), originally identified in BLAST searches with the end sequence of BAC402H23. Comparison of different ubiquitin carboxyl-terminal hydrolases identified two conserved domains that typify the type-2 ubiquitin carboxyl-terminal hydrolases, also known as ubiquitin-specific processing protease (Wilkinson 1997). These two domains, the cys box and the his box, are also present in the predicted sequence of Usp29, indicating that Usp29 belongs to the type-2 ubiquitin carboxyl-terminal hydrolase family (Fig. 2).
Figure 2.
Comparison of mouse and human USP29 amino acid sequences. The upper sequence represents the mouse Usp29 and the lower the human USP29 amino acid sequence. Two conserved domains of the type-2 ubiquitin carboxyl-terminal hydrolase family are indicated by asterisks under the amino acid sequence. The consensus sequences of these two domains constructed from the multiple alignment of yeast sequences are also presented and compared with the sequence alignment of human and mouse USP29. Sequence identity (black background) and conservative substitution with similar properties (gray background) are indicated. The two sequences are aligned for maximal match by the SIM program (http://dot.imgen.bcm.tmc.edu:9331/seq-search/alignment.html).
The exon structure of Usp29 was determined through comparing the cDNA sequence with a draft sequence derived from the 129/Sv genomic clones covering the mouse Peg3-region (Table 1; GenBank accession no. AC020961 and AC020966). Usp29 is composed of 5 exons distributed throughout an ∼400-kb genomic region, with the 5′-end of Usp29 positioned within 150 bp of the 5′-end of Peg3 (Fig. 1). The first and second exons of Usp29 share a low level of homology with the MER repeat family (Smit 1996). The distance between the first and second exons in mouse is estimated to be 7 kb, and the distance between exons 4 and 5 is 900 bp. The relative location of exon 3 is presently unknown because of uncertainty of spacing of contigs in the draft sequence. The entire ORF is located in exon 5, the last exon of mouse Usp29 (Fig. 1).
Table 1.
Position and Size of Exons and Introns of the Mouse Usp29 Gene
| Exon | Position | Length (bp) | Intron | Length (bp) |
|---|---|---|---|---|
| 1 | 1–722 | 722 | 1 | ∼7,000a |
| 2 | 723–943 | 221 | 2 | unknown |
| 3 | 944–1264 | 321 | 3 | unknown |
| 4 | 1265–1452 | 188 | 4 | 949b |
| 5 | 1453–7550 | 6098 |
The length was determined by PCR.
The length was determined by DNA sequencing.
Isolation of Human USP29
To identify the human homolog of mouse Usp29, the 1.2-kb end sequence of 402H23 corresponding to portions of mouse Usp29 coding sequence was hybridized to a Southern blot containing human and other mammalian genomic DNAs. At low stringency, this mouse probe detected positive fragments only from the genomic DNAs of closely related rodent species, such as mouse and rat, but not from human and other mammals (data not shown). These data provided the first suggestion that Usp29 sequences might not be well conserved among mammals. Comparison of the cDNA sequence of mouse Usp29 with the complete, finished genomic sequence of the region surrounding human PEG3 (GenBank accession no. AC025588), however, enabled us to locate a single set of human sequences related to the mouse Usp29 coding region, approximately 400 kb upstream of PEG3 (Fig. 1). The genomic sequence containing part of the potential coding region of human USP29 displays 65%–70% sequence identity at the nucleotide level with the mouse coding sequence. The human USP29 sequence identified a perfect match with one human EST (GenBank accession no. AA909508), confirming the transcription of this sequence. With RT-PCR and RACE, about 3.2 kb of human USP29 transcribed sequence was obtained. Analysis of this sequence identified a single open reading frame, encoding a predicted protein 922 amino acids in length (GenBank accession no. AF229438). Despite the relatively low level of overall sequence conservation between human and mouse predicted proteins (42.5% amino acid sequence identity), the two conserved domains that are unique to the type-2 family, and which define the active sites of these enzymes, are well conserved between human and mouse USP29 (Fig. 2). No other sequence with significant match to mouse Usp29 coding sequence was found in the completed sequence of the human region.
The USP29 cDNA sequence we obtained by RACE is derived from a contiguous 3.2-kb stretch of human genomic sequence, indicating that the coding region of human USP29 is also located in a single exon. Alignment of the 5′-UTR of mouse Usp29, including exons 1–4 (1.3 kb), with the human genomic sequence also revealed the presence of sequences related to the first two exons of mouse Usp29 immediately upstream of human PEG3. As in mouse, this conserved sequence identified two human EST matches (GenBank accession nos. H80201 and H79292), suggesting that this region is also transcribed. But repeated attempts using RT-PCR or RACE did not succeed in linking these upstream sequences to the human USP29 ORF (data not shown). In fact, the human 5′-EST sequences and the USP29 coding exon probe detected different transcripts in human Northern blots; in contrast, the two sets of related mouse sequences detected similar sets of transcripts (see below). These data suggested that the two clusters of cDNA sequences might represent different transcription units in the human genome.
Overlap between cDNA sequences we had isolated and a partial gene recently deposited sequence (GenBank accession no. AF124433) identified the 5′-end of human USP29. RT-PCR using human testis RNA as template confirmed the joining of this 5′-sequence and the USP29 ORF. Assembly of these sequences permitted derivation of a complete 3.5-kb USP29 transcript. Alignment of USP29 cDNA and human genomic sequences identified three additional exons, and also localized the potential 5′-end of the human transcript to a position 9 kb upstream of the USP29 ORF (Fig. 1).
Expression Pattern of Mouse Usp29
Northern Analyses
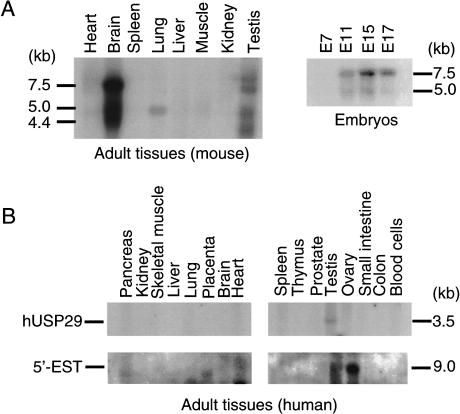
To examine the expression pattern of mouse Usp29, we hybridized two different cDNA probes, derived from the first exon and 3′-UTR of Usp29, respectively, to Northern blots containing poly(A)+ RNAs isolated from different adult tissues and embryos at various developmental stages. The highest levels of adult mouse Usp29 expression were detected in brain. The size of the major transcript detected by both sets of probes was estimated to be about 7.5 kb, consistent with the length of the cDNA clone and sequence we obtained (Fig. 3A). Two minor transcripts, 4.5 and 5 kb in length, respectively, were also detected in brain, and several minor transcripts of different lengths were also detected in testis with the 3′-UTR probe. In contrast, the probe derived from the first exon of mouse Usp29 detected only the 7.5-kb major transcript in brain. Although the significance of these smaller transcripts cannot be established from present data, alternative splicing or alternative promoter usage might provide an explanation for their differential detection. The expression of Usp29 was detected in RNA isolated from mouse embryos as early as 11 days postcoitum (dpc), and the consistent levels of Usp29 expression appear to be maintained throughout the latter half of the gestation period (Fig. 3A).
Figure 3.
Northern blot analyses of mouse and human USP29. (A) The expression pattern of mouse Usp29 was analyzed using poly(A)+ RNAs derived from adult tissues and different-stage embryos. A 500-bp DNA fragment corresponding to the 3′-UTR of Usp29 was used as a probe. A major transcript of mouse Usp29, 7.5 kb in length, and also two minor forms, 5.0 kb and 4.4 kb, were detected in adult brain as well as in embryos. (B) Human USP29 transcripts were detected at a low level only in the adult testis (upper). The estimated size of the transcript is about 3.5 kb. The ESTs matching sequence located in the 5′-side of PEG3 detected a 9-kb mRNA species only in ovary (lower).
In Situ Hybridization
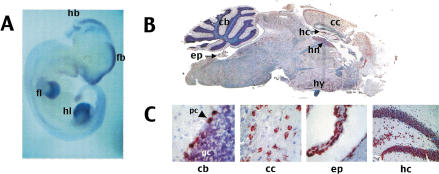
The spatial pattern of Usp29 transcription was analyzed by in situ hybridization of whole-mount midgestation mouse embryos and sectioned adult mouse brain. Both antisense and sense RNA probes of mouse Usp29 were used for this experiment, in which only the antisense probe detected significant expression levels of mouse Usp29. At 10.5 dpc, Usp29 is expressed at highest levels in the forebrain and limb buds, with significant levels of transcript also detected in the hindbrain and in developing structures of the face (Fig. 4A). In adult brain, Usp29 transcripts were detected in many regions, but most notably in the cerebral cortex and hippocampus. These data are consistent with embryonic expression data, since the cerebral cortex and hippocampus are derived from forebrain structures (Fig. 4B). Examination of sections under high-power magnification revealed that Usp29 is transcribed in specific cell types within each region. For example, Usp29 transcripts are present at very high levels in cerebellar Purkinje cells, but not in the underlying, densely packed granule cells (Fig. 4C, left panel). In the cerebral cortex, Usp29 is expressed in the numerous pyramidal cells (Fig. 4C, center left panel). Usp29 expression is also highly concentrated in ependymal cells (Fig. 4C, center right panel) and in the granule layer of the hippocampus (Fig. 4C, right panel).
Figure 4.
Localization of Usp29 transcripts in mouse embryo and adult brain. (A) Whole mount in situ hybridization of Usp29 to 10.5-dpc embryo. Usp29 expression (stained in dark blue) was detected at high levels in the face, forebrain (fb), hindbrain (hb), forelimb (fl), and hindlimb (hl). (B) Sagittal section of whole adult mouse brain hybridized with Usp29 probe.Cells expressing the Usp29 transcript are stained in red; the tissue has been counterstained with hematoxylin, which stains nuclei and therefore regions that are dense in neuronal cell bodies blue. Regions expressing Usp29 at significant levels include cerebellum (cb), hippocampus (hc), habenula (hn), hypothalamus (hy), ependyma (ep), and cerebral cortex (cc). High-power magnification of selected Usp29-expressing regions, showing cell types expressing the gene. Left panel (cb): Section of the cerebellum, showing high levels of Usp29 expression in Purkinje cells (pc) but not in the densely packed granule cells (gc); Center left panel (cc): section of the cerebral cortex, showing high levels of expression in pyramidal cells throughout the cerebral cortex; Center right panel (ep): expression in ependymal cells; and Right panel (hc): showing high levels of Usp29 expression in the granule layer of the hippocampus.
Expression Pattern of Human USP29
The expression of human USP29 was also examined with two cDNA probes derived from the 3′-UTR and coding region of the human gene. As shown in Figure 3B, significant amounts of transcript were detected only in adult testis with these two probes. Although Northern blots carrying poly(A)+ RNA derived from several different human fetal tissues were hybridized, we were unable to detect any significant levels of human USP29 gene expression at these early developmental stages (data not shown). Because the mouse brain Usp29 transcript initiates immediately upstream of Peg3 and the related human upstream region is also transcribed, we hybridized probes derived from matching human EST sequences to a separate Northern blot. Unlike probes from related mouse regions, human “upstream” (adjacent to 5′-PEG3) and USP29 coding sequence probes did not detect the same transcripts. The upstream probes detected a single 9-kb transcript, which is expressed at high levels only in ovary (Fig. 3B). Taken together with the results of RT-PCR and RACE attempts and the different lengths and sites of expression of human and mouse USP29 transcripts, these data suggest that human and mouse genes are transcribed differently.
Imprinting of Mouse Usp29
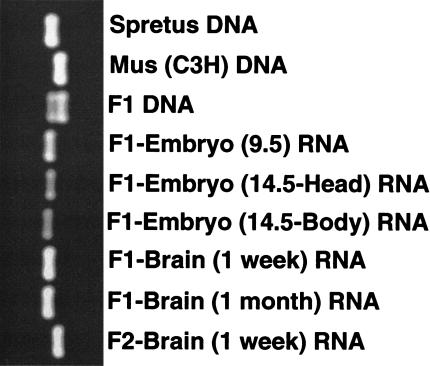
To determine the imprinting status of Usp29, we used hybrid animals derived from matings between Mus musculus and Mus spretus mice (Kim et al. 1999). Several sequence polymorphisms that differentiate alleles of each parental species were identified by comparing the sequences of Usp29 cDNA segments derived from M. musculus and from M. spretus. Among several polymorphisms, two were selected and used for the imprinting test: (1) a base substitution located in the first exon, and (2) a 19-bp insertion/deletion polymorphism located in the 3′-UTR of the gene. Experiments conducted with the two polymorphisms yielded identical results; in both cases, Usp29 transcripts were derived primarily from the paternal allele in the hybrid mice. Data obtained using the insertion/deletion polymorphism located in the 3′-UTR are presented in Figure 5. To confirm these results, a reciprocal analysis was performed with interspecific backcross progeny (the offspring of an interspecific hybrid female and a C3Hf male; Fig. 5, lane 9). The result of this test confirmed results obtained with F1 hybrid mice, demonstrating that Usp29 transcripts are derived primarily from the paternal allele.
Figure 5.
Paternal expression of mouse Usp29. PCR amplification was performed across the 19-bp insertion/deletion polymorphism located in the 3′ untranslated region of mouse Usp29. RNAs derived from the F1 hybrid (male M. spretus × female M. musculus) and the F2 hybrid (male M. musculus × female F1) were used for this imprinting test. Three genomic DNAs—M. spretus (lane 1), M. musculus (lane 2), and F1 hybrid (lane 3)—are also included in this test. Exclusive paternal expression of mouse Usp29 is observed when using RNAs derived from embryos (lanes 4–6), neonatal (lane 7), and adult (lane 8) brains of F1 offspring, and also the neonatal brain (lane 9) of F2 animals.
Analysis of Usp29 transcripts derived from mouse embryos at different stages of gestation confirmed monoallelic expression. Usp29 transcripts detected in 9.5-dpc and 14.5-dpc embryos are derived mainly from the paternal allele (Fig. 5, lanes 4–6). Since the major site of mouse Usp29 expression is brain, we focused primarily on analysis of cDNA prepared from this tissue. Usp29 transcripts in neonatal (7 day old) and adult (1 month old) mouse brains were also derived mainly from the paternal allele (Fig. 5, lanes 7 and 8), indicating that Usp29 expression is stably imprinted throughout development.
DISCUSSION
Data presented here identify a new imprinted mouse gene, Usp29, located adjacent to mouse Peg3 and describe the location, sequence, and expression patterns of the homologous human sequence. Mouse Usp29 is expressed primarily from the paternal allele in the mouse brain from at least midgestation into adulthood; significant levels of embryonic expression were also detected in the developing limbs. The predicted protein is likely to function as a ubiquitin carboxyl-terminal hydrolase based on sequence similarity to other known genes of this class. Because it does not share significant amino acid sequence homology with identified mammalian proteins, Usp29 appears to represent a novel member of this diverse family. Along with Peg3 and Zim1, Usp29 represents the third imprinted gene identified in a conserved 500-kb interval of proximal Mmu7. Our results indicate that this relatively unexplored imprinted domain may occupy a large genomic segment containing a number of imprinted genes, as has been observed for other known imprinted regions.
Although the actual biochemical role of Usp29 is currently unknown, the sequence similarity of Usp29 with members of the type-2 ubiquitin carboxyl-terminal hydrolase family suggest that Usp29 might be a deubiquitinating enzyme (Wilkinson 1997). This raises the possibility that the gene might be involved in regulating the turnover rate of many proteins. As shown for Ube3a, an imprinted gene that is also thought to be involved in the ubiquitin pathway (Kishino et al. 1997), the absence of Usp29 might be expected to be associated with pleiotrophic effects on growth, development, and fitness with significant detriment to the animal. Although the biochemical functions of both proteins must be demonstrated formally, it is tempting to speculate that these two oppositely imprinted genes—one functioning as a ligase and the other as a hydrolase of ubiquitin–protein complexes—might serve opposing functions that must be properly balanced for successful development of the mammalian nervous system. The high level of Usp29 expression in mouse brain, a feature that is shared with Ube3a, suggests that the Usp29 gene might play an especially significant role in mouse neurological development. However, although both mouse and human genes are expressed in testis, human USP29 is not transcribed at significant levels in brain. Although Northern blots are notoriously insensitive, Northern data were supported by the fact that we could not amplify USP29 coding region sequences from human brain cDNA templates by RT-PCR (data not shown). Low levels of human USP29 transcription or expression in very limited brain regions cannot be ruled out at this point. But these data may also indicate that the mouse and human USP29 genes have diverged significantly in biological function.
The different expression patterns of human and mouse USP29 genes is not entirely unprecedented. For example, although human and mouse PEG3 genes are both expressed in adult brain, the human gene is expressed at highest levels in ovary. By contrast, trace levels of mouse Peg3 were detected in ovary only by sensitive methods such as RNase protection (Kim et al. 1997a). Significant differences in tissue-specific expression of orthologs have also been noted by other groups, including differences linked to the species-specific usage of different promoters (Wang et al. 1996; Kissinger and Raff 1998; Fougerousse et al. 2000). Our data also suggest that exon structures of human and mouse USP29 differ significantly. Although human and mouse USP29 transcripts are both expressed in testis, human USP29 mRNA is not present in significant levels in brain—the primary site of mouse expression. Our data indicate that the human and mouse major transcripts, at least, originate at significantly different genomic sites and most likely use different promoters. Whether these differences reflect real change in gene function is difficult to predict at this time, and must await further study.
Comparison of cDNA and genomic sequence shows that the 5′-ends of Peg3 and Usp29 genes are located less than 150 bp apart. It therefore seems likely that Peg3 and Usp29 share regulatory elements that control sites and levels of each gene's transcription, and possibly allele-specific expression. Consistent with this notion, the expression patterns of mouse Usp29 and Peg3 are very similar: both genes are expressed in embryos and at significant levels only in adult brain. Recent studies also indicate that the region surrounding the transcription start sites of the two genes is associated with a CpG-rich region that is differentially methylated in parent-of-origin-specific manner (Li et al. 2000; J. Kim et al., unpub.). These observations support the idea that the transcription and imprinting of Peg3 and Usp29 might be jointly controlled through a small, shared genomic region. Further insights to the regulation and function of this newly defined imprinted domain should soon be forthcoming, as the boundaries of the domain, additional resident genes, and candidate regulatory elements are discovered through comparative sequence analysis in the upcoming year.
METHODS
Mouse Genomic Clone Isolation and Analysis
For the construction of a physical map of the Peg3/Zim1-region, several genomic clones were identified and characterized. The two BAC clones 588F20 and 247N19 were obtained through screening high-density mouse 129/Sv BAC library filters (Research Genetics, Huntsville, Alabama) with a pool of three genomic DNA fragments corresponding to the transcribed region of Peg3 (Kuroiwa et al. 1996). The BAC clone 402H23 was isolated by screening the same library with two evolutionarily conserved probes (25670krab and ZNF134 finger probes; Kim et al. 1997a, 1999) derived from the coding region of human ZNFs located in H19q13.4. Because of the lack of clones covering the genomic region between the two groups of BAC clones described in the above, we utilized the TAR (Transformation-Associated Recombination) cloning procedure (Larionov et al. 1996) with one of the end sequences of BAC247N19 and a 200-bp B2 sequence as bites using 129/Sv genomic DNA. This technique derived one genomic clone, tar17-21. The PAC clone 95D23, connecting the tar 17-21 and BAC 402H23 clones, was obtained by screening mouse 129/Sv PAC library filters with two end clones of tar17-21 and 402H23. A separate screening of genomic clones covering the Peg3/Zim1-region was performed using C57BL/6 BAC library filters (RPCI-23 library, obtained from Roswell Park Cancer Institute, Buffalo, New York). About 30 BAC clones were obtained using six pooled overgo probes spanning the region (Fig. 1). The probes were designed and labeled as described previously (Ross et al. 1999). All BAC-end clones were isolated using a single-primer PCR approach (Kim et al. 1997b). The DNA samples of BAC, PAC, and TAR clones were prepared with the alkaline lysis protocol (Sambrook et al. 1989), digested with rare-cutting restriction enzymes including NotI, EagI, and ClaI, separated on pulse-field gels (Chef Mapper instrument, BioRad). DNAs were transferred to nylon membranes (Hybond, Amersham), and hybridized according to standard protocols (Stubbs et al. 1990).
cDNA Isolation and 5′- and 3′-RACE
Mouse Usp29 cDNA clones were isolated from a cerebellum cDNA library (Doyle et al. 1997) with two probes corresponding to the first exon and the 3′-UTR of Usp29. Six of the 14 cDNA plaques positive with the two probes were randomly selected, plaque-purified, and analyzed further. To extend further the 5′-end of Usp29, we performed the RACE (Rapid Amplification of cDNA End) technique (Frohman et al. 1988). A cDNA template derived from adult brain RNA was used with the following two oligonucleotides: Mim2 (5′-ATGGGAAGGTGCGACATGAA-3′) and Mim5 (5′-CTACAAACTTCGGCAACGGGTT-3′). Human USP29 cDNA clones were also obtained through 5′- and 3′-RACE. A cDNA template derived from adult testis RNA was used with the following two sets of oligonucleotides: HUSP29-1 (5′-ACACCTAGATCCCAGAACTCAG-3′) and HUSP29-3 (5′-CCCTGCAAGATTAGAATGGTGC-3′) for 3′-RACE, and HUSP29-2 (5′-GATACTCCACACATTACCAGTG-3′) and HUSP29-4 (5′-CATGAGCACTAGAATCTCCATC-3′) for 5′-RACE. Amplified RACE products were separated on 0.8% agarose gel, and the major fragments in terms of mass were isolated from the gels using a gel extraction column (QIAquick gel extraction kit, QIAGEN). The fragments were subcloned into the TA cloning vector (TA cloning kit, Invitrogen).
Imprinting Tests
The imprinting (or monoallelic expression) of mouse Usp29 was tested with hybrid offspring (F1) produced by crossing M. musculus (C3Hf) females with M. spretus male mice. The reciprocal imprinting test was performed with the offspring (F2) produced by backcrossing (M. musculus × M. spretus) hybrid females with C3Hf males. Embryos of 2 different stages (E9.5, E14.5) and tissues from 7-day-old and 1-month-old animals were collected from each cross. RNA was isolated using a commercially available kit (Rapid total RNA isolation kit; 5′-3′, Inc, Boulder, CO). RNA samples were treated with RNase-free DNase I (Strategene) for 30 min at 37°C, and 50 μg were used for the synthesis of cDNA (using the cDNA synthesis module, Amersham). The final volume of each reverse transcription reaction was 40 μl, and 1 μl of this material was taken for each PCR reaction. For imprinting tests of mouse Usp29, the 3′-UTR of mouse Usp29 was amplified with the following two oligonucleotides: mUsp29-7 (5′-TGCAAGGTCCTTAGCACATC-3′) and mUsp29-8 (5′-GTGCATAGCCTGCATCAGCA-3′). PCR amplification of the 3′-UTR of Usp29 was performed using the following program in an MJ Research PTC-250 instrument: 95°C, 30 sec; 60°C, 1 min; 72°C, 1 min for 30 cycles, and 72°C, 5 min for 1 cycle. PCR reactions were carried out in a 50 μl reaction mixture containing 300 ng of each primer, 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 1.5 mm MgCl2, 1 mm dNTPs, 1% Triton X-100, and 1.25 U Taq DNA polymerase. Ten μl of each PCR reaction mixture were separated on 1.8% agarose gels to visualize the amplified PCR products.
Sequencing and Sequence Analysis
Subcloned genomic and cDNA fragments were sequenced from both directions using a fluorescence-based cycle-sequencing DNA sequencing kit (Dye terminator sequencing core kit, PE Applied Biosystems), and reactions were analyzed on an ABI 377 automated sequencer. Sequence alignments and database search were performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Northern Blot Analysis
A set of commercially available poly(A)+ RNA blots were used for expression analyses of human and mouse USP29: mouse multiple northern blot, mouse embryo blot, human multiple tissue northern blot, and human fetal northern blot (Clontech). For mouse Usp29, two probes representing different Usp29 transcribed regions were used: the first exon and 3′-UTR of Usp29. For human USP29, two probes corresponding to the coding region and 3′-UTR of the gene, respectively, were used. For the human ESTs derived from the 5′-side of PEG3, a 1-kb DNA fragment isolated from the EST clone was used. Procedures and conditions for generating probes and for performing hybridizations were as described previously (Stubbs et al. 1990).
Whole Mount and Section in Situ Hybridization
To generate an antisense and a sense riboprobe for the Usp29 in situ hybridization, the transcribed region of mouse Usp29 (Genbank accession no. AF229257, nt 2979-3638) was first amplified with the following two oligonucleotides: mUsp29-1 (5′-CCAGGGTCCTCATTGTTCAT-3′) and mUsp29-2 (5′-TTGCAAGGGATGCCGGTAGA-3′). The initial PCR product was further amplified with the following two sets of oligonucleotides: mUsp29-1 and mUsp29-2-T7 (5′-TAATACGACTCACTATAGGGTTGCAAGGGATGCCGGTAGA-3′) for an antisense probe and mUsp29-1-T7 (5′-TAATACGACTCACTATAGGGCCAGGGTCCTCATTGTTCAT-3′) and mUsp29-2 for a sense probe. Reamplified PCR products were treated once with phenol/chloroform, washed with TE on a microcon-100 (Amicon), and concentrated to 1 μg/μl concentration. One μg of each template DNA was used for each in vitro transcription reaction with T7 polymerase. For whole mount in situ hybridizations, antisense and sense probes were labeled by digoxigenin-UTP with an RNA labeling kit (DIG RNA labeling kit, Boehringer Mannheim), anti-DIG-AP (alkaline phosphatase) from sheep, and BM Purple AP substrate (Boehringer Mannheim), which were used according to the manufacturers instructions. The hybridization experiments were performed according to a standard protocol (Hogan et al. 1994) with minimum modification. For section in situ hybridizations, brain tissues derived from 2-month-old mice were fixed by cardiac perfusion with 4% paraformaldehyde. Serial sections, in sagittal planes, were cut at 5 μm. The probes used for the section in situ hybridization were labeled with biotin-16-UTP (Boehringer Mannheim) and were detected by the DAKO GenPoint kit (DAKO). After color development of the probes, the sectioned tissues were also counterstained with hematoxylin according to the manufacturer's instructions (DAKO).
Acknowledgments
We thank Dr. Elbert Branscomb and Mark Shannon for helpful discussion and critical reading of the manuscript. This work was performed under the auspices of the U.S. Department of Energy by LLNL under contract no. W-7405-ENG-48.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: stubbs5@llnl.gov; FAX (925) 422-2282.
REFERENCES
- Barlow DP. Gametic imprinting in mammals. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- Barlow DP. Competition—A common motif for the imprinting mechanism? EMBO J. 1997;16:6899–6905. doi: 10.1093/emboj/16.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- Beechey CV, Cattanach BM. Genetic imprinting map. Mouse Genet. 1996;94:96–99. [Google Scholar]
- Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, et al. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nature Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- Doyle J, Ren X, Lennon G, Stubbs L. Mutations in the Cacnl1a4 calcium channel gene are associated with seizures, cerebellar degeneration, and ataxia in tottering and leaner mutant mice. Mamm Genome. 1997;8:113–120. doi: 10.1007/s003359900369. [DOI] [PubMed] [Google Scholar]
- Fougerousse F, Bullen P, Herasse M, Lindsay S, Richard I, Wilson D, Suel L, Durand M, Robson S, Abitbol M, et al. Human–mouse differences in the embryonic expression patterns of developmental control genes and disease genes. Hum Mol Genet. 2000;9:165–173. doi: 10.1093/hmg/9.2.165. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Hurst LD, McVean G, Moore T. Imprinted genes have few and small introns. Nature Genet. 1996;12:234–237. doi: 10.1038/ng0396-234. [DOI] [PubMed] [Google Scholar]
- Jong MTC, Gray TA, Ji Y, Glenn CC, Saitoh S, Driscoll DJ, Nicholls RD. A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet. 1999;8:783–793. doi: 10.1093/hmg/8.5.783. [DOI] [PubMed] [Google Scholar]
- Kim J, Ashworth L, Branscomb E, Stubbs L. The human homolog of a mouse-imprinted gene, Peg3, maps to a zinc finger gene-rich region of human chromosome 19q13.4. Genome Res. 1997a;7:532–540. doi: 10.1101/gr.7.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bergmann A, Stubbs L. Exon sharing of a novel human zinc-finger gene, ZIM2, and paternally expressed gene 3 (PEG3) Genomics. 2000;64:114–118. doi: 10.1006/geno.1999.6112. [DOI] [PubMed] [Google Scholar]
- Kim J, Carver EA, Stubbs L. Amplification and sequencing of end fragments from bacterial artificial chromosome clones by single-primer polymerase chain reaction. Anal Biochem. 1997b;253:272–275. doi: 10.1006/abio.1997.2404. [DOI] [PubMed] [Google Scholar]
- Kim J, Lu X, Stubbs L. Zim1, a maternally expressed mouse Kruppel-type zinc-finger gene located in proximal chromosome 7. Hum Mol Genet. 1999;8:847–854. doi: 10.1093/hmg/8.5.847. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nature Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Kissinger JC, Raff RA. Evolutionary changes in sites and timing of actin gene expression in embryos of the direct- and indirect-developing sea urchins, Heliocidaris erythrogramma and H. tuberculata. Dev Genes Evol. 1998;208:82–93. doi: 10.1007/s004270050157. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li L-L, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, et al. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nature Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen XN, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci USA. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, Hu RJ, Johnson LA, Feinberg AP. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nature Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nature Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- Li L, Szeto IY, Cattanach BM, Ishino F, Surani AM. Organization and Parent-of-origin-specific methylation of imprinted peg3 gene on mouse proximal chromosome 7. Genomics. 2000;63:333–340. doi: 10.1006/geno.1999.6103. [DOI] [PubMed] [Google Scholar]
- Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison IM, Reeve AE. A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum Mol Genet. 1998;7:1599–1609. doi: 10.1093/hmg/7.10.1599. [DOI] [PubMed] [Google Scholar]
- Nicholls RD. New insights reveal complex mechanisms involved in genomic imprinting. Am J Hum Genet. 1994;54:733–740. [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Saitoh S, Horsthemke B. Imprinting in Prader-Willi and Angelman syndromes. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- Reik W, Maher ER. Imprinting in clusters: Lessons from Beckwith-Wiedemann syndrome. Trends Genet. 1997;13:330–334. doi: 10.1016/s0168-9525(97)01200-6. [DOI] [PubMed] [Google Scholar]
- Ross MT, LaBrie S, McPherson J, Stanton VP., Jr . Screening large-insert libraries by hybridization. In: Boyl Ann., editor. Current Protocols in Human Genetics. New York: Wiley Press; 1999. [Google Scholar]
- Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nature Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Searle AG, Beechey CV. Genome imprinting phenomena on mouse chromosome 7. Genet Res. 1990;56:237–244. doi: 10.1017/s0016672300035333. [DOI] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Stubbs L, Huxley C, Hogan B, Evans T, Fried M, Duboule D, Lehrach H. The hox-5 and surfeit gene clusters are linked in the proximal portion of mouse chromosome 2. Genomics. 1990;6:645–650. doi: 10.1016/0888-7543(90)90499-k. [DOI] [PubMed] [Google Scholar]
- Tilghman SM, Caspary T, Ingram RS. Competitive edge at the imprinted Prader-Willi/Angelman region? Nature Genet. 1998;18:206–208. doi: 10.1038/ng0398-206. [DOI] [PubMed] [Google Scholar]
- Tilghman SM. The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- Wang D, Marsh JL, Ayala FJ. Evolutionary changes in the expression pattern of a developmentally essential gene in three Drosophila species. Proc Natl Acad Sci USA. 1996;93:7103–7107. doi: 10.1073/pnas.93.14.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- Wutz A, Smrzka O, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]