Abstract
Bacteria in the genus Providencia are pathogens of many organisms, including humans and insects. We and colleagues have isolated five different strains belonging to four distinct Providencia species as natural infections of Drosophila melanogaster captured in the wild. We found that these isolates vary considerably in pathology to infected D. melanogaster, differing in the level of mortality they cause, their ability to replicate within the host and the level that the fly’s immune response is elicited. One interesting bacterium was Providencia sneebia, which causes nearly complete mortality and reaches large numbers in the fly but does not elicit a comparably strong immune response. Through coinfection experiments, we determined that P. sneebia avoids recognition by the immune system. We tested for biofilm formation and replication within D. melanogaster cells as possible mechanisms for P. sneebia escape from host immunity, but did not find evidence for either. D. melanogaster and Providencia provide a powerful system for studying general host-pathogen interactions, and for understanding how the well-studied immune model host D. melanogaster interacts with its natural bacterial pathogens.
Keywords: Providencia, Drosophila melanogaster, Pathogens, Host-Pathogen Interactions, Virulence, Innate Immunity
1. Introduction
Closely related bacterial pathogens may utilize a wide range of mechanisms to infect hosts, in part because virulence mechanisms are genetically labile and are often horizontally transferred between reasonably distantly related microbes [1]. Understanding differences in pathology between closely related bacteria highlights recent shifts in virulence, and can ultimately lead to the identification of the underlying genetic basis. Several strains and species of the γ-Proteobacterial genus Providencia have recently been isolated from field infections of wild caught Drosophila melanogaster [2, 3, P. Juneja and S. M. Short unpublished], and in the present work we contrast the pathological interactions of these bacterial species within their Drosophila host. D. melanogaster is a well established model host for studying innate immunity [4] and the pathology of virulent bacteria [e.g. 5–8], but few natural pathogens of D. melanogaster have been identified or extensively studied [but see 9–11]. We believe that Drosophila and Providencia comprise a powerful natural system for the study of variation in virulence and host-pathogen interactions. Because many microbial virulence strategies focus on conserved aspects of eukaryotic physiology and cell biology, inferences from this system can apply across broad host ranges, including from insects to humans.
Bacteria of the genus Providencia are Gram-negative opportunistic pathogens that have been isolated from a wide variety of environments and organisms ranging from humans to insects to sea turtles and shark mouths [12–15]. Providencia rettgeri, Providencia alcalifaciens, and Providencia stuartii have all been isolated from human stool samples both as part of the natural human gut flora and as the cause of gastric upset such as traveler’s diarrhea [16–18]. Some strains of P. alcalifaciens, but no strains of P. rettgeri or P. stuartii, have been found to be intracellularly invasive in human cell lines [16, 17, 19, 20]. Providencia also cause urinary tract and other nosocomial infections in humans [12, 13]. Numerous studies surveying bacteria associated with insects such as blowflies, stable flies and Mexican fruit flies have isolated Providencia species either from the whole insect or specifically from the gut [e.g. 21–23], although it is unclear whether these and other associations have meant the bacteria were acting as pathogens or were simply present in the insects’ environment. Providencia have been recurrently found in association with D. melanogaster, including in a survey for bacterial associates in a natural population [24], in the hemolymph of laboratory cultures of domino mutant larvae that are void of hemocytes and generally sick with bacterial infections [25], and as natural infections in wild-caught D. melanogaster [2, 3, P. Juneja and S. M. Short unpublished].
The D. melanogaster innate immune system has been well described, primarily from experiments measuring the response to injection of avirulent bacteria or generic immune elicitors [reviewed in 4]. D. melanogaster is also an excellent model for studying the pathology of virulent bacteria, since many virulence mechanisms are effective across a broad range of hosts. As a result, Drosophila has successfully been used as an experimental host to model clinical pathogenesis in humans and animals, insect vectoring of human disease and microbiological control of insect agricultural and medical pests. For example, Drosophila have been used to study opportunistic human infectors such as Serratia marcescens and the pathologies of Pseudomonas aeruginosa communities found in cystic fibrosis patients [7, 26]. D. melanogaster has also stood in as a model host for other arthropods such as ticks and mosquitoes that bear Francisella tularensis infections, ticks that host Ehrlichia chaffeensis, and caterpillars infected with Photorhabdus luminescens vectored by entomopathogenic nematodes [27–29].
Despite previous studies of bacterial pathogens of other animals using D. melanogaster as a model host, very little is known about the bacteria that infect D. melanogaster itself in its natural habitat. In some of the few efforts to identify bacterial pathogens of wild Drosophila, four different species belonging to the genus Providencia were recovered along with isolates of other bacteria from the hemolymph of wild caught D. melanogaster [2, 3, P. Juneja and S. M. Short unpublished]. Since the hemolymph of a healthy fly should be sterile, the presence of bacteria can be considered to constitute an infection. Two of the recovered Providencia species are the previously described Providencia rettgeri and Providencia alcalifaciens [12]. The other two species were identified as novel species named Providencia sneebia and Providencia burhodogranariea, the latter of which has two distinct strains designated B and D [3].
In this paper, we determine the pathology of Providencia species and strains in D. melanogaster, where pathology is defined as the proportion of mortality caused by the bacteria, the bacterial ability to proliferate within the fly, and the levels of host immunity induced by infection as measured by the expression of antimicrobial peptide (AMP) genes. We find Providencia to be highly variable in all three phenotypes. The ability of the bacteria to proliferate within the fly, the amount of AMP expression, and the level of mortality the bacterial cause are often all positively correlated, with the most deadly bacteria reaching the highest amount within the fly and inducing the highest levels of AMP expression. A notable and interesting variation to this pattern is P. sneebia, which kills about 90% of infected flies and reaches very large numbers in these flies but induces less AMP expression than other Providencia species, even those that cause significantly lower mortality and do not proliferate as effectively within flies. Through coinfections with P. sneebia and P. rettgeri, we concluded that P. sneebia is able to actively avoid recognition by the fly’s immune system and is resistant to ectopic immune induction. Two possible hypotheses to explain these observations are that P. sneebia invades and replicates within insect cells or forms a biofilm during infection, but we do not find evidence supporting either hypotheses in vitro, suggesting that P. sneebia virulence mechanisms are more complicated. The diversity of virulence profiles we observe among these Providencia isolates indicates they will be a rich substrate for future study of Providencia infection dynamics in a natural and experimentally tractable host.
2. Methods and materials
2.1. Fly stocks and bacteria strains
D. melanogaster fly stocks that were used were either wild type Oregon R (OreR), OR;imd10191;OR [30], Toll 1-RxA,ry,h,st,e/Tm3 Ser [31], or expressing green fluorescent protein (GFP) under the promoter of the AMP Diptericin A (DptA), DptA-GFP [32]. They were maintained on standard glucose medium (12 g agar, 100 g glucose and 100 g Brewer's yeast per 1.2 L of water, plus a final concentration of 0.04% phosphoric acid and 0.4% propionic acid added to inhibit microbial growth in the food) and kept at room temperature (22–24 °C). Table 1 provides a complete list of Providencia bacterial strains. All Providencia strains were grown in LB media at 37 °C overnight with shaking, except for P. burhodogranariea strains, which were grown at 25 °C. Listeria monocytogenes 10403S was grown at 37 °C in BHI medium with shaking. E. coli Mach1-T1, a cloning strain (Invitrogen Corp), was grown at 37 °C in LB medium with shaking.
Table 1.
Bacterial Strains Used. (*) indicates the strains that are the main focus of this work.
| Species | Strain | DSM # | Isolated from | Citation |
|---|---|---|---|---|
| Providencia burhodogranariea | Type/B* | 19968 | wild D. melanogaster hemolymph | [2, 3] |
| B97 | wild D. melanogaster hemolymph | [2, 3] | ||
| B18 | wild D. melanogaster hemolymph | [2, 3] | ||
| D* | wild D. melanogaster hemolymph | [2, 3] | ||
| Providencia rettgeri | Dmel* | wild D. melanogaster hemolymph | [2, 3] | |
| Type | 4542 | fowl cholera | [12] | |
| Providencia alcalifaciens | Dmel* | wild D. melanogaster hemolymph | P. Juneja and S. M. Short, unpublished | |
| Type | 30120 | human infant dysentery | [12] | |
| Providencia sneebia | Type* | 19967 | wild D. melanogaster hemolymph | [2, 3] |
| A16 | wild D. melanogaster hemolymph | [2] | ||
| A36 | wild D. melanogaster hemolymph | [2] | ||
| A75 | wild D. melanogaster hemolymph | [2, 3] | ||
| A83 | wild D. melanogaster hemolymph | [2] | ||
| A91 | wild D. melanogaster hemolymph | [2, 3] | ||
| A101 | wild D. melanogaster hemolymph | [2, 3] | ||
| A102 | wild D. melanogaster hemolymph | [2, 3] | ||
| A104 | wild D. melanogaster hemolymph | [2] | ||
| Providencia heimbachae | Type | 3591 | penguin feces | [12] |
| Providencia stuartii | Type | 4539 | human | [12] |
| Providencia vermicola | Type | 17385 | entomopathogenic nematode | [37] |
| Providencia rustigiannii | Type | 4541 | human feces | [13] |
2.2. Mortality
Overnight cultures used for infecting flies were grown to saturation and then diluted to an A600nm of 1.0. To deliver infections, a 0.15 mm minuten pin (Fine Science Tools) mounted on a 200 μL pipet tip was dipped into the diluted overnight culture and poked into the thorax of a CO2 anesthetized fly. This delivers about 103 to 104 bacteria to each fly. Sterilely wounded flies were pricked with a needle that was sterilized in 95% ethanol. Anesthetized control flies were handled in the same way as the others but were not wounded. Flies were maintained in vials with food at room temperature and surviving flies were counted once a day for 6 days after infection. Infection with each bacterium was performed on at least 2 days with controls done on each day. Product limit survival estimates and homogeneity by log-rank tests were conducted using proc lifetest in SAS version 9.1 (SAS Institute). P-values were corrected for multiple tests in some cases by a Bonferroni correction with a cut off value of p=0.0025 for comparing all strains that are the focus of the paper, p=0.00625 for comparing among P. sneebia isolates only, and p= 0.025 for comparing only among P. burhodogranariea strains. In contrasts of different strains of P. burhodogranariea, only those infections that were preformed on the same day were compared.
2.3. Bacterial load
To measure systemic bacterial load in infected flies, single OreR flies were infected by pinprick as described in section 2.2, then homogenized in 500 μL LB and plated by robotic spiral platers (manufactured by Don Whitley Scientific and Spiral Biotech) on LB agar plates at 2, 4, 6, 10, 18, 24, and 32 hours post infection. Flies were kept in vials with food at room temperature between infection and homogenization. The LB agar plates were incubated overnight at 25 °C for P. burhodogranariea or 37 °C for P. rettgeri, P. alcalifaciens, P. sneebia and sterile wound. Gut commensal bacteria grow more slowly than Providencia under these conditions, so by limiting incubation to overnight we exclude any commensal bacteria from our assay. The number of colony forming units (CFU) on each plate was recorded using a counter associated with the spiral platers, allowing the concentration of viable bacteria in each homogenate to be calculated based on the number and position of colonies on the plates. Bacterial loads for flies infected with P. alcalifaciens and P. sneebia were compared at each individual time point using proc glm in SAS version 9.1 with the model: ln(CFU+1)=bacterial treatment + sex. The boxplot was generated using the function boxplot in R. A small number of surviving flies from each treatment were also homogenized at 7–10 days post infection as described above.
2.4. Antimicrobial peptide expression
We first examined DptA-GFP flies to determine how much AMP expression occurred during infection. We infected flies on replicate days as described in section 2.2 and kept them in vials with food until the time examined. Other AMP promoters examined which had undetectable levels of fluorescence were Defensin, Drosocin, Attacin and Cecropin [32]. At 6, 24, and 32 hours post infection flies were anesthetized and examined under a dissecting scope and scored for the intensity of GFP fluorescence blind of the treatment. This assay was restricted to females because males were found to have too much background fluorescence.
For quantification of AMP expression by QPCR, OreR flies were either infected with a bacterium or sterilely wounded as described in section 2.2 then were frozen at −80 °C in pools of 8 flies at 0, 2, 4, 6, 10, 18, 24, and 32 hours post treatment. Flies were maintained in vials with food at room temperature between infection and freezing. Each treatment was performed on at least two different days. Total RNA was extracted with Trizol (Invitrogen Corp) using the manufacturer’s suggested protocol, then reverse transcribed to cDNA from poly-T primers using standard procedures. The abundances of the AMPs Diptericin A (DptA), Drosomycin (Drs) and Defensin (Def) and the housekeeping gene rp49 were quantified by QPCR on an ABI 7000 Sequence Detection System (Applied Biosystems) using specific TaqMan primers and the manufacturer’s suggested protocol (primer and probe sequences available upon request). For statistical analysis, gene expression at each hour was examined separately in proc glm in SAS version 9.1 using the model: AMP Ct = Rp49 Ct + treatment + date infected. Correction for multiple tests was achieved using the Tukey-Kramer method. Least squares means were recovered at the mean Rp49 Ct. Fold induction was calculated as 2 to the power of the difference between the Ct of the sterile wound control and the Ct of the infection treatment for each time post-infection.
2.5. Coinfection
For coinfections, overnight cultures of P. rettgeri and P. sneebia were grown to saturation and then diluted to an A600nm of 2.0. The bacteria were then mixed at proportions 1:1, 1:3 or 3:1 with either the alternate bacteria or LB. Flies were then infected in the thorax with a small needle dipped in the culture as described in section 2.2, replicated on two different days. Although three different proportions of each bacterium were examined, we found that the results were the same for each infection class (singly infected P. rettgeri, singly infected P. sneebia, or coinfected) regardless of the mixing proportion, allowing us to pool all proportions in final analyses. We only examined male flies for AMP expression and bacterial load in the coinfection because we had found no difference between the sexes in our primary examination of mono-infections. At 6, 24, and 32 hours post infection flies were frozen at −80 °C. RNA extraction, QPCRs, and statistical analysis for AMP expression were performed as described in section 2.4. Fisher’s combined probability was used to summarize the independent expression experiments.
For the examination of AMP expression in DptA-GFP flies, infected or control flies were placed in vials with standard fly food and examined blind of treatment at 6, 24, and 32 hours post infection with a dissecting scope. Here, only female flies were examined due to male background fluorescence. The survival of these same flies was monitored up to six days post infection and statistically analyzed as in section 2.2.
Determination of the bacterial load of coinfected flies and statistical analysis was carried out as described in section 2.3. To distinguish between P. sneebia and P. rettgeri, we took advantage of P. rettgeri’s natural resistance to tetracycline. All samples were plated on LB plates without antibiotic and on plates with a tetracycline concentration of 10 μg/mL. The number of CFU on the tetracycline plates was inferred to be the count of P. rettgeri and the difference in CFU between the paired plates was assumed to be the P. sneebia count. PCR and restriction enzyme digestion of the 16S gene looking for species-specific digestion pattern was done to check that the proper species were growing on the correct plates. This experiment was carried out twice on different days.
2.6. Biofilm formation
Overnight bacteria cultures were diluted to an A600 of 1.0, then gently centrifuged into a pellet and washed three times with 1X PBS, and ultimately concentrated to 20X. 5 μl of bacteria or PBS, as a control, were added to 200 μL of Schneider’s media with 10% fetal calf serum in a 96-well plate. Bacteria that received the antibiotic treatment sat in media alone for approximately 1 hour before the antibiotics were added to the well. The antibiotics ceftazidime and kanamyacin were added to a final concentration of 1mg/mL and 200 μg/mL, respectively. At 6 and 24 hours after the bacteria or antibiotics were added to the media, the wells were washed three times with sterile water before the addition of 0.1% crystal violet, which was incubated for 15 minutes. The wells were then washed twice with water before drying for 5 hours. 30% acetic acid was added to the wells to solubilize the crystal violet. The A540nm was read using Multiskan Spectrum plate reader (Thermo Scientific). The final absorbance was calculated as the difference from the PBS control well at that time.
2.7. Antibiotic protection assay
D. melanogaster S2 cells were maintained in Schneider’s media with 10% fetal calf serum at 25 °C. For the antibiotic protection assay, cells were seeded in 6 well plates the day before the assay was carried out so that there would be approximately 105 cells/mL the next day. Overnight cultures of bacteria were washed three times with PBS before addition to the wells containing S2 cells at a multiplicity of infection of 10. After two hours the media was removed and the cells, which lightly adhere to the bottom of the wells, were washed while still in the wells three times with PBS. Schneider’s media with 10% fetal calf serum containing 1 mg/mL ceftazidime and 20 μg/mL kanamycin was then added to the wells. Neither ceftazidime nor kanamycin should penetrate eukaryotic cell membranes, so only extracellular bacteria should be killed. The cells were incubated with the antibiotics for 2 hours to kill extracellular bacteria. At 0, 6 and 24 hours following this two-hour incubation, the media only was removed from the wells and centrifuged. The pellet was then washed with water before being plated on BHI or LB plates, depending on the bacteria, to provide an estimate of the number of viable bacteria in suspension (this number should be near zero because of the presence of antibiotics). The S2 cells were then washed off with water and spun down and washed again with water. The pellet was then resuspended in BHI media before being plated on either BHI or LB. CFUs were manually counted to yield the number of viable bacteria residing inside the S2 cells.
3. Results
3.1. Mortality
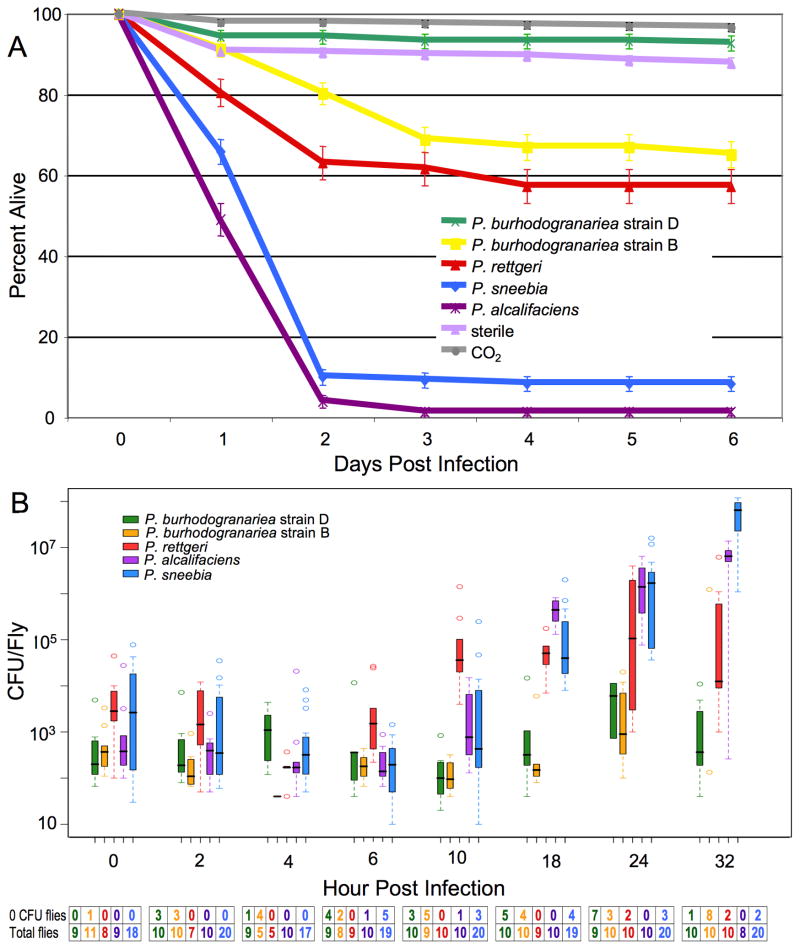
Given that closely related bacteria often vary in their virulence to a given host, we hypothesized that the different strains of Providencia isolated from wild caught Drosophila melanogaster might also vary in pathology (Table 1). There was minimal mortality (5–10%) among control flies either only anesthetized on CO2 or wounded with a sterile needle. When flies are infected with P. burhodogranariea strain D, less than 10% of infected flies died by six days post infection (Fig. 1A). This is not significantly different from the amount of mortality from either control (Fig. 1A; P. burhodogranariea strain D contrasted to CO2 control: p=0.0612, P. burhodogranariea strain D-sterile wound: p= 0.0436, not significant after correcting for multiple tests). About 40% of flies infected with P. burhodogranariea strain B die from the infection, which is highly significantly different from P. burhodogranariea strain D (p <0.0001), although for unknown reasons, P. burhodogranariea strain B infections displayed more day-to-day variation in mortality than infections with any other bacteria with mortality rates ranging from 20% to 60%. P. rettgeri strain Dmel likewise caused moderate mortality, with fewer than 50% of the flies dying. The amounts of mortality caused by P. rettgeri and P. burhodogranariea strain B are not significantly different from each other (p=0.0303), although both infections caused significantly higher mortality than is observed in controls (all p <0.0001). P. sneebia strain Type and P. alcalifaciens strain Dmel each caused much greater mortality than any of the other species. Both within the first two days of infection, P. sneebia kills about 90% of infected flies and P. alcalifaciens causes mortality in 99% of infected flies. Mortality from infections with each P. sneebia and P. alcalifaciens is significantly different from all other treatments, including each other (all p <0.0001). Thus, there are three major classes of virulence among our isolated Providencia as defined by mortality: P. burhodogranariea strain D causes minimal mortality, P. rettgeri and P. burhodogranariea strain B cause moderate amounts of mortality, and P. sneebia and P. alcalifaciens are highly virulent.
Figure 1.
Mortality of and bacterial proliferation in D. melanogaster. (A) Mortality of D. melanogaster from Providencia Infection. Wild type D. melanogaster were infected through pinprick infections with different strains of Providencia. All treatments result in highly significant differences in mortality (all pairwise contrasts p < 0.0001), except the difference between sterile needle and CO2 controls, between infection with P. burhodogranariea strain D and either control, and between P. rettgeri and P. burhodogranariea strain B (in all cases p > 0.0025, the Bonferroni corrected cut off value). (B) Providencia Bacterial Load in D. melanogaster. Boxplot of the number of CFU present in D. melanogaster during the first 32 hours post infection. Note that the y-axis is a log scale. Whiskers approximate two times the standard deviation. The table under the graph has the number of flies that had no CFU at each time point for each treatment, as well as the total infected flies per treatment at each time point. Flies with no CFU present were not included in the boxplot. Sterilely wounded control flies never had any CFU at any time point.
Multiple isolates of P. sneebia and P. burhodogranariea strain B have been recovered from the hemolymph of wild caught D. melanogaster (Table 1) [2, 3]. We infected flies with each of these to test whether there is heterogeneity among isolates in the mortality caused by these strains. Of the two other isolates of P. burhodogranariea strain B, only isolate B97 is significantly different than the Type strain B, with B97 causing less mortality (Suppl. Fig. 1; Suppl. Table 1; p = 0.0003). Eight P. sneebia isolates were tested and all caused greater than 80% mortality, although some of them cause slightly but significantly different mortality than the Type strain (Suppl. Table 1). This suggests that while there is some variation among isolates, P. sneebia can be considered to always be highly virulent while P. burhodogranariea is never highly virulent.
The Drosophila humoral immune response is activated by two major signaling pathways, the Toll pathway and the Imd pathway [4]. The Imd pathway tends to be more responsive to Gram-negative bacteria, whereas the Toll pathway preferentially activated by Gram-positive bacteria. We therefore hypothesized that the Imd pathway would be most important in fighting Providencia. We measured the mortality of flies that were mutationally deficient in either the Toll or Imd pathway after infection with Providencia. We found Toll pathway mutants showed no significant difference in mortality compared to wild type flies after infection with either strain of P. burhodogranariea or with P. rettgeri (p>0.05, in all cases). P. sneebia and P. alcalifaciens did cause significantly greater mortality in the Toll mutant flies compared to the wild type flies (p<0.05, in both cases), but flies of both genotypes suffered severe mortality within 2 days of infection with these bacteria (Suppl. Fig. 2A). In contrast, Imd mutant flies infected with any strain of Providencia suffered very high mortality within 2 days post infection (Suppl. Fig. 2B). Notably, we observed high mortality in flies infected with the P. rettgeri and P. burhodogranariea strains, which cause only moderate to low mortality in wild type flies (Suppl. Fig. 2B). All Providencia infections in Imd mutant flies were significantly different than those seen in infected wild type flies (p<0.05, in all cases). These data indicate that the Imd pathway is essential to fighting Providencia infection, and that P. rettgeri and P. burhodogranariea infections are controlled by the host immune system and not simply limited by inherent failure of the bacteria to be able to colonize the fly.
We were intrigued by the recurrent isolation of diverse Providencia species from Drosophila, so we examined the amount of mortality caused in D. melanogaster by the Type strains of 6 Providencia species isolated in other contexts, including P. rettgeri and P. alcalifaciens isolates not derived from Drosophila (Table 1; Suppl. Fig. 3). Except for P. alcalifaciens strain Type, all species caused less than 20% fly mortality in wild type flies. The Type strain of P. alcalifaciens caused less mortality than our Dmel strain (Suppl. Fig. 2; p<0.0001), which suggests there are genetic differences between the strains. The Type strain of P. rettgeri also caused less mortality than our Dmel strain (Suppl. Fig. 2; p<0.0001). These data indicate that the high amount of Providencia induced D. melanogaster mortality is specific to those strains that were isolated from wild flies.
3.2. Bacterial load
For a given host and pathogen pair, bacterial proliferation and host mortality may or may not be correlated. To test our hypothesis that the Providencia species that cause the highest mortality are those that are best able to replicate in flies, we measured the number of bacteria present in D. melanogaster at multiple time points for the first 32 hours after infection. Plates from control flies that were sham-infected with a sterile needle did not have any bacteria growth after the overnight incubation period (data not shown), indicating that the control flies did not have any Providencia within or on them. Commensal bacteria from the gut begin to appear on all plates after they have been incubated for at least 24 hours. Infections with the five bacteria start to diverge in CFU counts around 10 hours post infection (Fig. 1B). There are a few individual flies that are able to clear the infection during the first few hours. It is unclear why some flies are able to clear their infections and others are not, although we suspect it reflects minor heterogeneities in the infection process.
Flies infected with either strain of P. burhodogranariea cleared their infections or maintained stable bacterial loads around the level of the initial introduction over the first 32 hours of infection (Fig. 1B). These bacteria are eventually cleared from all surviving flies, as survivors have no bacteria present 7–10 days post infection (data not shown). P. rettgeri, P. sneebia and P. alcalifaciens all show an increase in the number of CFU per fly after 6 hours of infection. Among the flies infected with P. rettgeri, there is a large amount of variation in the number of bacteria present in individual flies at 24 and 32 hours post infection, ranging from 103 to 107 CFU per fly (Fig. 1B). It seems likely that this variation reflects divergence in the infection trajectory among individual flies, where those with the highest bacterial loads probably succumb to the infection and the others survive. Flies that survived their infections for 7–10 days post infection carried either no CFU or between 102 and 3×104 CFU per fly (data not shown). Both P. sneebia and P. alcalifaciens are able to rapidly proliferate to very high numbers in the fly by 32 hours post infection, which is shortly before flies die from these infections. The number of bacteria present in the infected flies is significantly different between P. sneebia and P. alcalifaciens at 18 hours post infection (p=0.0115), but not at 24 and 32 hours (both p>0.05). Approximately 10% of the total P. sneebia infected flies had no bacteria present at their time of sampling. These flies were most likely able to clear the bacteria within the first few hours of infection and probably represent the small percent of flies that survive in the mortality assays (Fig. 1A). This hypothesis is supported by the observation that flies infected with P. sneebia that survive 7–10 days post infection are free of Providencia (data not shown). In total, across all species, these data demonstrate that the Providencia species that are best able to proliferate within the fly are those that cause the highest mortality.
3.3. D. melanogaster immune response to infection
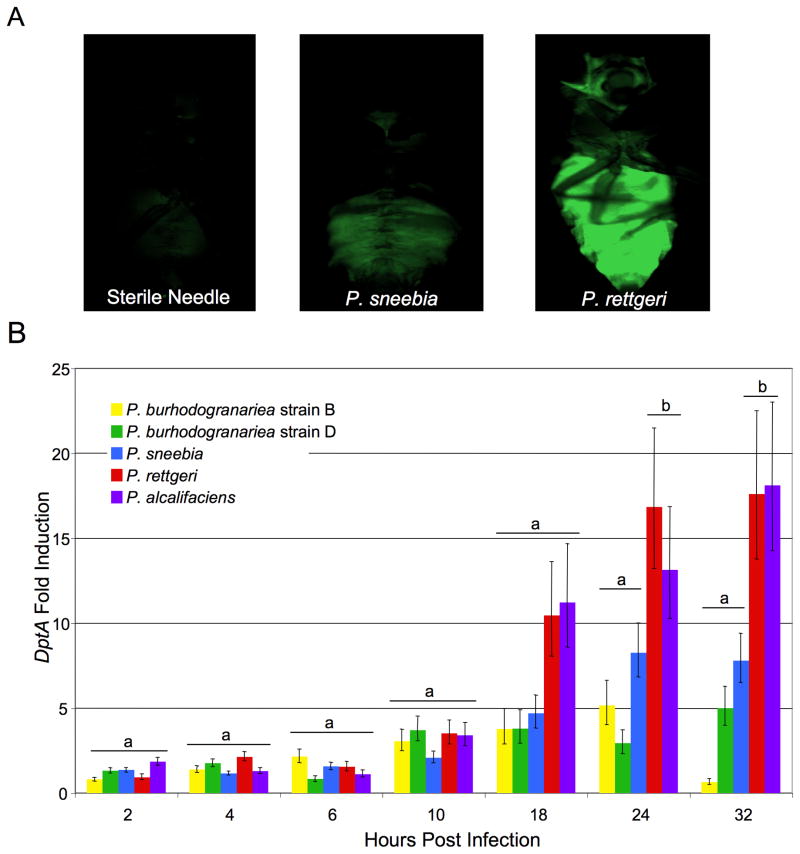
Insects respond to the presence of bacteria by activating their humoral immune system, which results in the production of antimicrobial peptides (AMPs). Induction of AMP gene expression varies among different microbes and immune elicitors [33], and we hypothesized that the Providencia bacteria that were most proliferative during infection would cause the highest induction of the immune response. To initially test this hypothesis, we infected transgenic flies that express GFP driven by AMP promoters [32] then examined individual flies by eye at 6, 24, and 32 hours post infection. Although the expression patterns of several different AMPs were examined (see section 2.4), only DptA produced a strong fluorescence after infection with most Providencia. Both P. burhodogranariea strains failed to drive detectable fluorescence signal even with DptA. As expected, DptA-GFP expression was localized to the immune responsive fat body. We repeatedly saw that flies that were infected with P. sneebia showed lower fluorescence than flies infected with P. alcalifaciens or P. rettgeri, both in the intensity of the GFP expression as well as the proportion of a single fly expressing GFP (Fig. 2A). This contrasted with our expectation based on the high levels of P. sneebia proliferation within flies and host mortality caused from infection.
Figure 2.
DptA Expression in Flies Infected with Providencia. (A) DptA-GFP flies infected with (left to right) a sterile needle, P. sneebia, or P. rettgeri at 32 hours post infection. (B) Graph of DptA expression as measured by QPCR. The fold induction was calculated as the level of expression above that caused by a sterile wound alone. Error bars represent the standard error. At each time point, treatments labeled with “a” are not significantly different from the sterile wound alone while those with “b” are significantly different from the sterile wound (corrected for multiple tests by Tukey-Kramer method, cut off p=0.05).
We used QPCR of AMP mRNAs to better quantify the immune response of infected flies for the first 32 hours of infection relative to control flies that were wounded with a sterile needle. By calculating the fold induction over the sterile wound, we could determine the amount of AMP expression that was specifically attributable to the bacteria and not to the wound in delivering the infection (Fig. 2B). Consistent with our observations of the DptA-GFP flies, P. sneebia infections consistently resulted in lower expression of DptA than P. rettgeri and P. alcalifaciens did at later times in the infection progression. At 24 and 32 hours after infection with P. sneebia, DptA expression was not significantly different from expression in response to the sterile wound alone (Fig. 2B; both p>0.05). In contrast, P. rettgeri and P. alcalifaciens induced significantly higher levels of expression than the sterile wound at 24 and 32 hours after infection (Fig. 2B; in all cases p<0.05). None of the bacterial infections drove DptA expression above the level seen from sterile wound alone prior to 24 hours post infection, and flies infected with either strain of P. burhodogranariea never showed DptA expression above what is seen for the sterile wound treatment at any time point (Fig. 2B).
Providencia induction of Drs over sterile wound was generally much smaller, and none of the infection treatments differ significantly from the sterile wound until 32 hours post infection (Suppl. Fig. 4A). The pattern of Def expression was more complex, with strong induction in response to P. rettgeri and P. alcalifaciens infections at 18 and 24 hours post infection (Suppl. Fig. 4B). The induction of Def in response to P. sneebia is much delayed relative to P. alcalifaciens and P. rettgeri infection, with strong induction not appearing until 32 hours post infection.
In summary, we observed that some of Providencia species that proliferate the most within the fly and cause the greatest host mortality also drive higher AMP expression. An interesting departure from this trend is P. sneebia, which is highly virulent and reaches the highest abundance within the fly, but expression of DptA caused by P. sneebia infection is never significantly higher than that caused by a sterile wound (Fig. 2B).
3.4. Coinfections with P. rettgeri and P. sneebia
P. sneebia could avoid inducing a strong immune response by actively evading detection by the host or by actively suppressing the immune response. To distinguish between these two possibilities, we took advantage of the differences in mortality and immune induction resulting from P. rettgeri and P. sneebia infections. We coinfected flies with both bacteria simultaneously and then measured AMP expression, host mortality, and bacterial load. We hypothesized that if P. sneebia actively suppresses the immune response, we would see low levels of AMP expression even in the presence of P. rettgeri. Alternatively, if P. sneebia is not detected by the immune system, we would expect to see high levels of AMP expression induced by the presence of P. rettgeri in the coinfection.
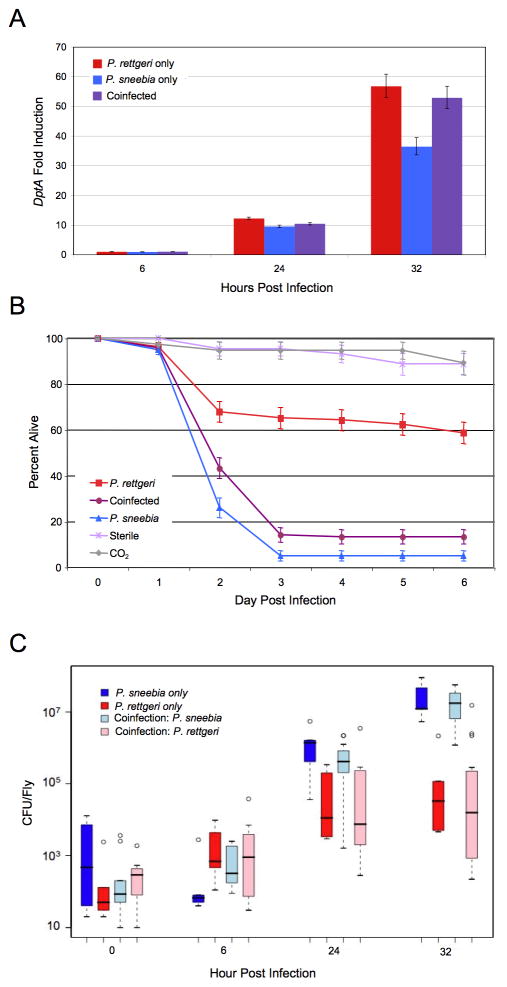
We measured DptA, Drs and Def levels in groups of flies either coinfected or infected with an individual bacteria at 6, 24, and 32 hours post infection by QPCR (Fig. 3A; Suppl. Fig. 5). Across all 3 AMPs, infection with P. sneebia alone caused a lower immune response than infection with P. rettgeri alone at 32 hours post infection (Fisher’s combined probability, p = 0.00061), consistent with the results presented in the previous section. Expression of the 3 AMPs at 32 hours post infection in coinfected flies was not significantly different from expression in flies singly infected with P. rettgeri (Fisher’s combined probability, p = 0.528), but coinfected flies had significantly higher expression than flies infected with P. sneebia (Fisher’s combined probability, p = 0.014). This result was further supported by visually examining the level of GFP expression in individual DptA-GFP flies, in which coinfected and P. rettgeri infected flies fluoresced more intensely than flies infected with P. sneebia. These data show that the lower expression of AMPs in flies infected with P. sneebia alone is not due to suppression of the immune response.
Figure 3.
D. melanogaster Coinfected with Both P. sneebia and P. rettgeri. (A) Graph of DptA expression from infection with P. sneebia, P. rettgeri or both measured by QPCR. The fold induction was calculated as the level of expression over that caused by a sterile wound alone. Error bars represent the standard error. The AMP genes Def and Drs showed similar patterns (Suppl. Fig. 4.) (B) Survival of D. melanogaster from infections with P. sneebia, P. rettgeri or both. (C) Bacterial load of D. melanogaster infected with P. sneebia, P. rettgeri or both. The two bacteria are plotted separately in pale colors for the coinfected flies. Whiskers approximate two times the standard deviation. Coinfected flies show full induction of the immune system, but succumb to their infections and permit bacterial growth that is not different than what is observed in single infections.
Consistent with previous mortality measurements (Fig. 1A), approximately 40% of the flies infected with P. rettgeri alone died from their infections, whereas about 95% of the flies infected with P. sneebia died within 72 hours (Fig. 3B). Coinfected flies exhibited 85% mortality. While all three treatments are significantly different than each other (in all cases p < 0.01), the overall mortality of coinfected flies is clearly more similar to that of flies infected with P. sneebia alone (Fig. 3B). When bacterial load was measured at 32 hours post infection, we observed that the abundance of each individual bacterium in the coinfected flies was not significantly different than their levels in flies that are singly infected (Fig. 3C; in all cases p > 0.05). Thus, it appears that the growth trajectories of the bacteria are completely independent of each other, and coinfected flies carry a bacterial load equivalent to the sum of each single infection. Considering all three coinfection phenotypes together, it is evident that P. sneebia is not able to suppress the host immune response, but is able to proliferate and cause host mortality even in the presence of an immune response triggered by P. rettgeri.
3.5. Biofilm formation
One way that P. sneebia could protect itself from recognition and the microbicidal AMPs is by forming a biofilm within the fly [34]. The fly would only be able to detect bacteria at the perimeter of the biofilm, and thus the magnitude of the immune response would not be proportional to the total number of bacteria present. Additionally the bacteria within the biofilm would be able to freely multiply without being affected by expressed AMPs. We tested all of our Providencia isolates for their ability to form biofilms in vitro in 96-well plates. E. coli was used as a control that can form a biofilm [35]. None of our Providencia isolates formed a biofilm (Table 2). However, we cannot definitively rule out the possibility that P. sneebia might form a biofilm within the fly, since there could be host-specific molecules that act as signal to trigger P. sneebia biofilm formation in vivo.
Table 2.
Providencia isolated from D. melanogaster are not able to form biofilms. Measurements of absorbance of crystal violet at 540nm of replicate wells after biofilm formation in vitro. E. coli, which is capable of forming a biofilm, has much higher absorbance than any of the Providencia species.
| Relative biomass (A540nm) | ||||||||
|---|---|---|---|---|---|---|---|---|
| without antibiotics | with antibiotics | |||||||
| 6 hours | 24 hours | 6 hours | 24 hours | |||||
| Bacteria | average | std dev | average | std dev | average | std dev | average | std dev |
| E. coli | 0.0717 | +/− 0.0277 | 0.2471 | +/− 0.0929 | 0.0208 | +/− 0.0054 | 0.0219 | +/− 0.0021 |
| P. sneebia | −0.0027 | +/− 0.0047 | 0.0343 | +/− 0.0036 | −0.0023 | +/− 0.0084 | 0.0125 | +/− 0.0010 |
| P. alcalifaciens | 0.0039 | +/− 0.0108 | 0.0261 | +/− 0.0161 | 0.0007 | +/− 0.0031 | 0.0058 | +/− 0.0006 |
| P. rettgeri | 0.0056 | +/− 0.0098 | 0.0277 | +/− 0.0020 | 0.0117 | +/− 0.0112 | 0.0104 | +/− 0.0041 |
| P. burhodogranariea strain B | 0.0043 | +/− 0.0002 | 0.0258 | +/− 0.0063 | 0.0030 | +/− 0.0074 | 0.0065 | +/− 0.0001 |
| P. burhodogranariea strain D | 0.0048 | +/− 0.0083 | 0.0127 | +/− 0.0028 | 0.0060 | +/− 0.0006 | 0.0121 | +/− 0.0040 |
3.6. Antibiotic protection assay
Another way that P. sneebia could evade detection and proliferate would be if it were able to invade cells and replicate within them. Strains of P. alcalifaciens that were isolated from human patients with diarrhea have been shown to invade human cells, demonstrating that some Providencia are able to do the first step in this process [16, 17, 19, 20]. We used an antibiotic protection assay to test whether P. sneebia is able to divide within D. melanogaster cells. We also tested whether P. alcalifaciens and P. rettgeri are able to divide within D. melanogaster cells since they proliferate within the fly during infection. E. coli was used as a negative control bacteria that would be passively phagocytosed by the cells but is unable to replicate within. Listeria monocytogenes was used as a positive control that is able to replicate within insect cells [36]. Bacteria were exposed to a phagocytic D. melanogaster cell line for two hours before antibiotics were added to the media to kill all extracellular bacteria. At 0, 6 and 24 hours post antibiotic killing of extracellular bacteria, both the media and the cells were plated separately and CFU were counted. The CFU found in the media were minimal by comparison to those within the cells. Since E. coli will only be passively phagocytosed by the cells, it was used as a standard for determining if any of our strains are actively invading the cells. Among all replicates, there were consistently fewer P. sneebia and P. rettgeri than E. coli inside host cells at the zero hour time point (Table 3), suggesting that these bacteria have some resistance to phagocytosis by these D. melanogaster cells. By contrast, P. alcalifaciens had higher numbers of CFU than E. coli at the initial time point suggesting that our strain of P. alcalifaciens is invasive. The positive control, L. monocytogenes, was able to replicate to high numbers within the cells. P. rettgeri, P. sneebia, P. alcalifaciens, and the negative control, E. coli, all had fewer intracellular CFU 24 hours after addition of antibiotic than at 0 hours, indicating that none of them are able to replicate within the insect cells. These data suggest that P. sneebia does not avoid recognition by the immune response by invading and proliferating in D. melanogaster cells.
Table 3.
Providencia isolated from D. melanogaster are not intracellular pathogens. Number of CFU within D. melanogaster S2 cells at 0, 6, and 24 hours post antibiotic killing of extracellular bacteria. Two replicate wells were measured each day and the experiment was carried out on multiple days with similar results. Listeria monocytogenes, which is capable of intracellular invasion and proliferation, grows to high density within host cells, whereas Providencia and E. coli, which is not capable of intracellular invasion, are progressively eliminated.
| well | 0 hour CFU | 6 hour CFU | 24 hour CFU | |
|---|---|---|---|---|
| P. sneebia | 1 | 543 | 468 | 28 |
| 2 | 466 | 460 | 22 | |
| P. rettgeri | 1 | 285 | 110 | 49 |
| 2 | 138 | 120 | 88 | |
| P. alcalifaciens | 1 | 2541 | 2988 | 1620 |
| 2 | 2265 | 2061 | 1170 | |
| E. coli | 1 | 1734 | 942 | 154 |
| 2 | 1500 | 465 | 105 | |
| L. monocytogenes | 1 | 7570 | 8617 | 109680 |
| 2 | 7145 | 14749 | 141200 | |
| PBS | 0 | 0 | 0 |
4. Discussion
We have established that closely related bacteria in the genus Providencia vary in their pathology in a natural host, Drosophila melanogaster, as measured by the amount of mortality they cause, their ability to replicate within the host and the magnitude of the host immune response to their presence. Those bacteria which are able to grow most effectively in the fly often also trigger the most robust immune response and result in the most host death. However, one of these bacteria, P. sneebia, causes nearly complete mortality and quickly replicates to high numbers within the fly but does not induce a strong immune response. Through coinfections with the less virulent P. rettgeri, we concluded that P. sneebia is able to actively avoid detection by the immune system as well as protect itself from the immune response. We did not find evidence that P. sneebia forms a biofilm or replicates intracellularly in vitro. Although we were unable to determine the exact virulence mechanisms used by P. sneebia during infection of D. melanogaster, our data imply that P. sneebia implements more complicated or multiple strategies to subvert the immune system.
We note that the proportion of flies that die from each bacterial infection in the mortality assays is approximately equivalent to the proportion of flies that sustain high numbers of bacteria in the load experiments, suggesting that the individual flies in which Providencia is able to replicate are those that succumb to the infection. The data we have for flies infected with P. burhodogranariea strain B does not conform to this hypothesis, as that bacterium causes a moderate amount of mortality despite not replicating within the fly as much as the similarly virulent P. rettgeri. This suggests that P. burhodogranariea strain B might do proportionally more damage to the fly, possibly by producing a harmful compound, at a lower density than the other bacteria. This also points at a distinction between the two P. burhodogranariea strains, as they both have similar levels of bacteria present during the first 32 hours of infection but strain D causes significantly less mortality. The two strains are defined as distinct based on differences in metabolic profiles and in sequence of some housekeeping genes [3]. Our data suggest there are likely to be further genetic differences between the strains, including in genes involved in the phenotypes examined here.
Although we are primarily interested in Providencia species that are natural pathogens of D. melanogaster, we also examined mortality due to infection by other species in the genus, which have been isolated as clinical infections of humans and other animals, or in one case, P. vermicola, as an associate of entomopathogenic nematodes [12, 13, 37]. The only bacteria we found to cause high mortality in infected Drosophila are those which were originally isolated from wild caught D. melanogaster. Two of these species were also previously described as clinical pathogens of humans, but in both of these species, P. alcalifaciens and P. rettgeri, the Dmel strain isolated from D. melanogaster caused greater mortality than the Type strain of the species. These results suggest that Providencia strains may become highly adapted to the host species they infect, and that the isolates recovered from D. melanogaster may be genetically suited to infect Drosophila and its close relatives. More detailed genomic and pathological examination of Providencia should reveal genes specifically involved in virulence to Drosophila.
Because we are specifically interested in the D. melanogaster-Providencia interaction after infection has occurred, we have relied on artificial infections to deliver the bacteria. Nevertheless, it is worth considering how the bacteria may establish infections in the wild. Our Providencia isolates do not cause mortality after being fed to flies in reasonable doses in the laboratory (data not shown), so it is not likely that they orally infect flies in the wild unless they are aided by coinfectors [e.g. 38, 39]. There is good reason to believe, however, that our method of infecting through a pinprick wound may mimic infections that wild D. melanogaster can receive [40]. Wild caught flies often have melanization independent of natural pigmentation patterns, indicating healed wounds, and frequently carry ectoparasitic mites that could be the cause of some of these wounds (unpublished observation). Mite wounds in honey bees have been shown to be secondarily colonized by environmental bacteria [41]. P. burhodogranariea strain B has been isolated from a mite pulled from a wild caught D. melanogaster (P. Juneja, personal communication), suggesting that mites may also directly vector bacterial infections, although fly-to-fly transmission of Providencia via mites has not been experimentally demonstrated.
We anticipate that D. melanogaster-Providencia system will be an excellent one for continued examination of many aspects of host and pathogen interactions. There is ample phenotypic diversity in the host-pathogen interaction, with clear variation among Providencia species in pathological phenotypes. Both the bacteria and the insect host can be easily and inexpensively manipulated in the lab, providing a valuable setting to conduct research that will not only give insight into interactions specific to this host-pathogen pairing, but also into generic virulence mechanisms and their genetic basis. D. melanogaster has been extensively studied as a generic host for pathogenic bacteria and a model for innate immune system function, and these Providencia isolates now provide an opportunity to study how flies fight those bacteria that infect them in their natural environment.
Supplementary Material
Supplementary Figure 1. Mortality of D. melanogaster from Infection with Other Isolates of P. sneebia and P. burhodogranariea strain B. Wild type D. melanogaster were infected through pin prick infections with different strains of Providencia. All isolates of P. burhodogranariea strain B cause low to moderate mortality, while infection with any isolate of P. sneebia results in high mortality.
Supplementary Figure 2. Mortality of Immune Mutant D. melanogaster. (A) Toll (B) Imd pathway deficient flies. When the Imd pathway is non-functional, all bacteria are highly virulent, while when the Toll pathway is mutated, the bacteria cause the same degree of mortality as seen in the wild type flies.
Supplementary Figure 3. Mortality of D. melanogaster After Infection with Type strains of other Providencia species. Only those Providencia strains that were isolated from wild Drosophila are able to cause a high mortality. The Type strains of P. rettgeri and P. alcalifaciens are both significantly different from the Dmel strains (P. rettgeri strain Dmel contrasted to P. rettgeri strain Type: p<0.0001. P. alcalifaciens strain Dmel contrasted with P. alcalifaciens strain Type: p<0.0001).
Supplementary Figure 4. AMP Expression in Flies Infected with Providencia as measured by QPCR. (A) Drs (B) Def. The fold induction was calculated as the level of expression above that caused by a sterile wound alone. Error bars represent the standard error. (*) indicates those treatments that were significantly different from wounding with a sterile needle (corrected for multiple tests by Tukey-Kramer method, cut off p=0.05).
Supplementary Figure 5. AMP Expression in Flies Coinfected with P. sneebia and P. rettgeri as measured by QPCR. (A) Def (B) Drs. The fold induction was calculated as the level of expression above that caused by a sterile wound alone. Error bars represent standard error.
Acknowledgments
We would like to thank Punita Juneja, Sarah Short, Jacob Crawford, Susan Rottschaefer, and Jennifer Comstock for helpful comments on the manuscript, discussion and help collecting the data. This work was supported by NSF grant DEB-0415851 and NIH grant AI083932.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 2.Lazzaro BP. PhD thesis. USA: Pennsylvania State University; 2002. A population and quantitative genetic analysis of the Drosophila melanogaster antibacterial immune response. [Google Scholar]
- 3.Juneja P, Lazzaro BP. Providencia sneebia sp. nov. and Providencia burhodogranariea sp. nov., isolated from wild Drosophila melanogaster. Int J Syst Evol Microbiol. 2009;59:1108–1111. doi: 10.1099/ijs.0.000117-0. [DOI] [PubMed] [Google Scholar]
- 4.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 5.Brandt SM, Dionne MS, Khush RS, Pham LN, Vigdal TJ, Schneider DS. Secreted bacterial effectors and host-produced Eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol. 2004;2:e418. doi: 10.1371/journal.pbio.0020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vodovar N, Acosta C, Lemaitre B, Boccard F. Drosophila: a polyvalent model to decipher host- pathogen interactions. Trends Microbiol. 2004;12:235–242. doi: 10.1016/j.tim.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Nehme NT, Liégeois S, Kele B, Giammarinaro P, Pradel E, Hoffmann JA, Ewbank JJ, Ferrandon D. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 2007;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkey CD, Blow N, Watnick PI. Genetic analysis of Drosophila melanogaster susceptibility to intestinal Vibrio cholerae infection. Cell Microbiol. 2009;11:461–474. doi: 10.1111/j.1462-5822.2008.01267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci U S A. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vodovar N, Vallenet D, Cruveiller S, Rouy Z, Barbe V, Acosta C, Cattolico L, Jubin C, Lajus A, Segurens B, Vacherie B, Wincker P, Weissenbach J, Lemaitre B, Medigue C, Boccard F. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- 11.Liehl P, Blight M, Vodovar N, Boccard F. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2006;17:193–199. doi: 10.1371/journal.ppat.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Hara CM, Brenner FW, Miller JM. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev. 2000;13:534–546. doi: 10.1128/cmr.13.4.534-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manos J, Belas R. The Genera Proteus, Providencia, and Morganella. In: Dworken M, Falkow S, Rosenberg E, Schleifer KH, Stackenbrandt E, editors. Prokaryotes. Vol. 6. Springer; Singapore: 2006. pp. 245–269. [Google Scholar]
- 14.Foti M, Giacopello C, Bottari T, Fisichella V, Rinaldo D, Mammina C. Antibiotic Resistance of Gram Negatives isolates from loggerhead sea turtles (Caretta caretta) in the central Mediterranean Sea. Mar Pollut Bull. 2009;58:1363–1366. doi: 10.1016/j.marpolbul.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 15.Interaminense JA, Nascimento DC, Ventura RF, Batista JE, Souza MM, Hazin FH, Pontes-Filho NT, Lima-Filho JV. Recovery and screening for antibiotic susceptibility of potential bacterial pathogens from the oral cavity of shark species involved in attacks on humans in Recife, Brazil. J Med Microbiol. 2010;59:941–947. doi: 10.1099/jmm.0.020453-0. [DOI] [PubMed] [Google Scholar]
- 16.Albert MJ, Alam K, Ansaruzzaman M, Islam MM, Rahman AS, Haider K, Bhuiyan NA, Nahar S, Ryan N, Montanaro J, Mathan MM. Pathogenesis of Providencia alcalifaciens-induced diarrhea. Infect Immun. 1992;60:5017–5024. doi: 10.1128/iai.60.12.5017-5024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobreira M, Leal NC, Magalhães M, Guth BE, Almeida AM. Molecular analysis of clinical isolates of Providencia alcalifaciens. J Med Microbiol. 2001;50:29–34. doi: 10.1099/0022-1317-50-1-29. [DOI] [PubMed] [Google Scholar]
- 18.Yoh M, Matsuyama J, Ohnishi M, Takagi K, Miyagi H, Mori K, Park K, Ono T, Honda T. Importance of Providencia species as a major cause of travellers’ diarrhoea. J Med Microbiol. 2005;54:1077–1082. doi: 10.1099/jmm.0.45846-0. [DOI] [PubMed] [Google Scholar]
- 19.Janda JM, Abbott SL, Woodward D, Khashe S. Invasion of HEp-2 and other eukaryotic cell lines by Providenciae: further evidence supporting the role of Providencia alcalifaciens in bacterial gastroenteritis. Curr Microbiol. 1998;37:159–165. doi: 10.1007/s002849900357. [DOI] [PubMed] [Google Scholar]
- 20.Maszewska A, Torzewska A, Stączek P, Różalski A. Enterocyte-like Caco-2 cells as a model for in vitro studies of diarrhoeagenic Providencia alcalifaciens invasion. Microb Pathog. 2010:1–9. doi: 10.1016/j.micpath.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Kuzina LV, Peloquin JJ, Vacek DC, Miller TA. Isolation and Identification of Bacteria Associated with Adult Laboratory Mexican Fruit Flies, Anastrepha ludens ( Diptera: Tephritidae) Curr Microbiol. 2001;42:290–294. doi: 10.1007/s002840110219. [DOI] [PubMed] [Google Scholar]
- 22.Ahmad A, Broce A, Zurek L. Evaluation of significance of bacteria in larval development of Cochliomyia macellaria (Diptera: Calliphoridae) J Med Entomol. 2006;43:1129–1133. doi: 10.1603/0022-2585(2006)43[1129:EOSOBI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Mramba F, Broce A, Zurek L. Isolation of Enterobacter sakazakii from stable flies, Stomoxys calcitrans L. (Diptera: Muscidae) J Food Prot. 2006;69:671–673. doi: 10.4315/0362-028x-69.3.671. [DOI] [PubMed] [Google Scholar]
- 24.Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, Promislow DE. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl Environ Microbiol. 2007;73:3470–3479. doi: 10.1128/AEM.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun a, Hoffmann JA, Meister M. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc Natl Acad Sci U S A. 1998;95:14337–14342. doi: 10.1073/pnas.95.24.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vonkavaara M, Telepnev MV, Rydén P, Sjöstedt A, Stöven S. Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis. Cell Microbiol. 2008;10:1327–1338. doi: 10.1111/j.1462-5822.2008.01129.x. [DOI] [PubMed] [Google Scholar]
- 28.Luce-Fedrow A, Von Ohlen T, Chapes SK. Ehrlichia chaffeensis infections in Drosophila melanogaster. Infect Immun. 2009;77:4815–4826. doi: 10.1128/IAI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallem EA, Rengarajan M, Ciche TA, Sternberg PW. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr Biol. 2007;17:898–904. doi: 10.1016/j.cub.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 30.Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathogens. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson KV, Jürgens G, Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell. 1985;42:779–89. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- 32.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci U S A. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart J, Lemaitre B, Hoffmann JA, Imler J. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 34.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Chavant P, Gaillard-Martinie B, Talon R, Hébraud M, Bernardi T. A new device for rapid evaluation of biofilm formation potential by bacteria. J Microbiol Methods. 2007;68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- 37.Somvanshi VS, Lang E, Sträubler B, Spröer C, Schumann P, Ganguly S, Saxena AK, Stackebrandt E. Providencia vermicola sp. nov., isolated from infective juveniles of the entomopathogenic nematode Steinernema thermophilum. Int J Syst Evol Microbiol. 2006;56:629–633. doi: 10.1099/ijs.0.63973-0. [DOI] [PubMed] [Google Scholar]
- 38.de Oliveira D, de Souza TD, Murate LS, Jankevicius JV, Gaziri LC, Jankevicius SI. Protease and phospholipase inhibition protect Veneza zonata (Hemiptera Coreidae) against septicemia caused by parasite trypanosomatid 563DT. J Invertebr Pathol. 2004;85:9–17. doi: 10.1016/j.jip.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Broderick NA, Raffa KF, Handelsman J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc Natl Acad Sci U S A. 2006;103:15196–15199. doi: 10.1073/pnas.0604865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defences. Nat Rev Microbiol. 2008;6:302–313. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- 41.Kanbar G, Engels W. Ultrastructure and bacterial infection of wounds in honey bee (Apis mellifera) pupae punctured by Varroa mites. Parasitol Res. 2003;90:349–354. doi: 10.1007/s00436-003-0827-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Mortality of D. melanogaster from Infection with Other Isolates of P. sneebia and P. burhodogranariea strain B. Wild type D. melanogaster were infected through pin prick infections with different strains of Providencia. All isolates of P. burhodogranariea strain B cause low to moderate mortality, while infection with any isolate of P. sneebia results in high mortality.
Supplementary Figure 2. Mortality of Immune Mutant D. melanogaster. (A) Toll (B) Imd pathway deficient flies. When the Imd pathway is non-functional, all bacteria are highly virulent, while when the Toll pathway is mutated, the bacteria cause the same degree of mortality as seen in the wild type flies.
Supplementary Figure 3. Mortality of D. melanogaster After Infection with Type strains of other Providencia species. Only those Providencia strains that were isolated from wild Drosophila are able to cause a high mortality. The Type strains of P. rettgeri and P. alcalifaciens are both significantly different from the Dmel strains (P. rettgeri strain Dmel contrasted to P. rettgeri strain Type: p<0.0001. P. alcalifaciens strain Dmel contrasted with P. alcalifaciens strain Type: p<0.0001).
Supplementary Figure 4. AMP Expression in Flies Infected with Providencia as measured by QPCR. (A) Drs (B) Def. The fold induction was calculated as the level of expression above that caused by a sterile wound alone. Error bars represent the standard error. (*) indicates those treatments that were significantly different from wounding with a sterile needle (corrected for multiple tests by Tukey-Kramer method, cut off p=0.05).
Supplementary Figure 5. AMP Expression in Flies Coinfected with P. sneebia and P. rettgeri as measured by QPCR. (A) Def (B) Drs. The fold induction was calculated as the level of expression above that caused by a sterile wound alone. Error bars represent standard error.