Abstract
We describe a novel analytical approach to gene recognition based on cross-species comparison. We first undertook a comparison of orthologous genomic loci from human and mouse, studying the extent of similarity in the number, size and sequence of exons and introns. We then developed an approach for recognizing genes within such orthologous regions by first aligning the regions using an iterative global alignment system and then identifying genes based on conservation of exonic features at aligned positions in both species. The alignment and gene recognition are performed by new programs called GLASS and ROSETTA, respectively. ROSETTA performed well at exact identification of coding exons in 117 orthologous pairs tested.
A fundamental task in analyzing genomes is to identify the genes. This is relatively straightforward for organisms with compact genomes (such as bacteria, yeast, flies and worms) because exons tend to be large and the introns are either non-existent or tend to be short. The challenge is much greater for large genomes (such as those of mammals and higher plants), because the exonic 'signal' is scattered in a vast sea of non-genic 'noise'. While coding sequences comprise 75% of the yeast genome, they represent only about 3% of the human genome. Computational approaches have been developed for gene recognition in large genomes, with most employing various statistical tools to identify likely splice sites and to detect tell-tale differences in sequence composition between coding and non-coding DNA (Burset & Guigo 1996). Some programs perform e novo recognition, in that they directly use only information about the input sequence itself. One of the best programs of this sort is GENSCAN (Burge 1997), which uses a Hidden Markov Model to scan large genomic sequences. Other programs employ “homology” approaches, in which exons are identified by comparing a conceptual translation of DNA sequences to databases of known protein sequences (Pachter et al. 1999; Gelfand et al. 1996).
In this paper, we explore a powerful new approach to gene recognition by using cross-species sequence comparison, i.e., by simultaneously analyzing homologous loci from two related species. Specifically, we focus on the ability to accurately identify coding exons by comparison of syntenic human and mouse genomic sequences.
It is well known that cross-species sequence comparison can help highlight important functional elements such as exons, because such elements tend to be more strongly conserved by evolution than random genomic sequences. If a protein encoded by a gene is already known in one organism, it is relatively simple to search genomic DNA from another organism to identify genes encoding a similar protein (using such computer packages such as Wise2 (http://www.sanger.ac.uk/Software/Wise2). A more challenging problem is to idenitfy exons directly from cross-species comparisons of genomic DNA. Computer programs are available that identify regions of sequence conservation, using simple “dot plots” or more sophisticated “pip plots” (Jang et al. 1999), which can then be individually analyzed in an ad hoc fashion to see whether they may contain such features as exons or regulatory elements. However, these programs simply identify conserved regions and do not systematically use the cross-species information to perform exon recogntion.
We sought to develop an automatic approach to exon recognition by using cross-species sequence comparison to identify and align relevant regions and then searching for the presence of exonic features at corresponding positions in both species. We began by undertaking a systematic comparison of the genomic structure of 117 orthologous gene pairs from human and mouse to understand the extent of conservation of the number, length, and sequence of exons and introns. We then used these results to develop algorithms for cross-species gene recognition, consisting of GLASS, a new alignment program designed to provide good global alignments of large genomic regions by using a hierarchical alignment approach, and ROSETTA, a program that identifies coding exons in both species based on coincidence of genomic structure (splice sites, exon number, exon length, coding frame, and sequence similarity).
ROSETTA performed extremely well in identifying coding exons, showing 95% sensitivity and 97% specificity at the nucleotide level. The performance was superior to programs that use much more sophisticated signals and statistical analysis but analyze only a single genome (Burset and Guigo 1996, Burge 1997). To our knowledge, ROSETTA is the first program for gene recognition based on cross-species comparison of genomic DNA from two organisms. The approach can be readily generalized to other pairs of organisms, as well as to the study of three or more organisms simultaneously.
With the current explosion of knowledge regarding the human and mouse genomic sequences, cross-species comparison is likely to provide one of the most powerful approaches for extracting the information in mammalian genomes.
RESULTS
Comparison of Human and Mouse Genomic Loci
Comparisons of mRNA sequences of 1196 orthologous human and mouse gene pairs were recently reported (Makalowski et al. 1996), showing that coding regions tend to show approximately 85% identity at the nucleotide and protein levels. We sought to extend this analysis by comparing genomic structures, where known. The mRNA sequences from the orthologous gene pairs were searched against GenBank Release 109 (October 1998), to identify those for which the genomic sequence was available in both species. Entries were required to contain the complete genomic sequence encompassing all coding exons, although not necessarily including the introns between non-coding exons.
A total of 117 orthologous gene pairs were identified and studied (Table 1). For the purpose of comparing the genomic structure of the gene pairs, we used dynamic programming algorithms (employing both nucleotide similarity and codon similarity using the PAM20 matrix (Dayhoff et al. 1978)) to align the sequences. We carefully inspected the alignments to ensure that they correctly aligned the exons.
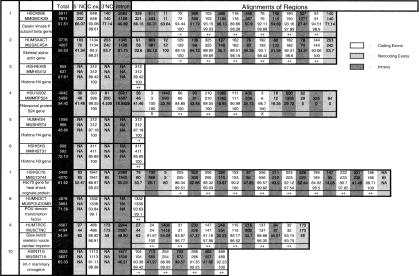
Table 1.
Comparative Analysis of Human and Mouse Loci
(The complete table is available online as supplemental material at the Genome Research Website: www.genome.org.) In this table we report a structural comparison of 117 orthologous human and mouse genomic loci. We also report the exon prediction performance of ROSETTA on each of these loci. Each entry in the table, numbered 1–117, is a pair of orthologous loci. In the first column, the GenBank LOCUS of the human entry, followed by the GenBank LOCUS of the mouse entry, followed by short descriptions of the genes, are given. The following columns have the following meanings, depending on the rows: (1) first row corresponds to the human entry; (2) second row corresponds to the mouse entry; (3) third row corresponds to nucleotide sequence similarity; (4) fourth row, when applicable, corresponds to amino acid similarity; and (5) fifth row, when applicable, corresponds to ROSETTA predictions. Thus the columns have the following meaning: (1) third column, colored dark, corresponds to the total size for human and mouse and the total sequence similarity using the GLASS alignment; (2) fourth column corresponds to the sizes and nucleotide similarity of the 5′-UTRs; (3) fifth column corresponds to the sizes, nucleotide, and protein similarity of the translated regions; (4) sixth column corresponds to the sizes and nucleotide similarity of the 3′-UTRs; (5) seventh column corresponds to the sizes and nucleotide similarity of the introns; and (6) the rest of the columns correspond to the sizes, nucleotide similarity, and protein similarity plus ROSETTA predictions whenever applicable. The color shading the regions indicates the type of the regions: coding exons (white); noncoding exons (light gray); and introns (medium dark gray). The ROSETTA predictions are indicated as follows: (++) coding exon predicted correct on both ends; (+−) coding exon predicted correct only on the 3′-end; (−+) coding exon predicted correct only on the 5′-end. (−−) coding exon was not missed totally, but both 3′- and 5′-boundaries were wrongly predicted; and (X) coding exon was missed altogether. Structurally unusual cases, such as when two coding exons in human correspond to one in mouse, can readily be seen in the table. For instance, entry 30 has such a situation. Coding exons 5 and 6 in human can be seen to correspond to coding exon 5 in mouse.
The comparison defined the striking extent of evolutionary conservation:
Exon Number
The number of exons was identical for 95% of the genes studied. There were six instances in which the number of exons differed.
In two cases, a single internal coding exon in mouse is reported to correspond to two internal coding exons in human. In the spermidine synthase gene (Table 1, gene 30), mouse exon 5 corresponds to human exons 5 and 6, with the total exonic lengths agreeing perfectly. In the lymphotoxin beta gene (Table 1, gene 85), mouse exon 2 corresponds to human exons 2 and 3. Interestingly, the mouse exon 2 is 316 bp while the sum of the lengths of human exon 2, intron 2 and exon 3 is only 301 bp.
In the next three cases, the correspondence broke down for terminal exons. In the keratin 13 gene (Table 1, gene 40) and the adenosine deaminase gene (Table 1, gene 66), the coding sequences show substantial sequence divergence at the 3′-end and one of the organisms has an extra exon. In the proteosome LMP2 gene (Table 1, gene 46), the extra human exon shows striking sequence similarity to a portion of the 3′-untranslated region (UTR) in the mouse. The final case was also in the LMP2 gene. The first two coding exons in the human correspond to one exon in the mouse. There is no apparent relationship between their lengths (even including the intron). It is possible that some of the apparent differences are due to error in annotation in the databases.
Exon Length
The length of corresponding exons was strongly conserved. The lengths were identical in 73% of cases. Those differences that did occur were quite small: the mean ratio of the larger to smaller length was 1.05.
Moreover, the differences were nearly always a multiple of three. The length difference was a multiple of three for 95% of all exons and 99% of all internal coding exons. This is readily understood in terms of the effects of evolutionary selection: length differences divisible by three alter an integral number of codons, while other length differences would require a second compensatory change in a succeeding exon to restore the translational reading frame and would thus be less likely.
Only three instances were found in which corresponding internal exons had lengths differing by other than a multiple of three.
In the skeletal muscle specific myogenic gene (Table 1, gene 49), the respective lengths of exons 2 and 3 are 81 bp and 123 bp in the human and 82 and 122 in the mouse. Remarkably, two instances occur in the gene encoding the Flt3 ligand (Table 1, gene 100). The respective lengths of exons 2 and 3 are 111 bp and 54 bp in the human and 122 bp and 46 bp in the mouse, while the respective lengths of exons 5 and 6 are 139 bp and 179 bp in the human and 144 bp and 189 bp in the mouse.
Intron Lengths
While exon lengths tended to be well-preserved, intron lengths varied considerably. The mean ratio of the larger to the small length was 1.5. As would be expected, there was no tendency for intron lengths to differ by a multiple of three. Human introns tended to be larger than mouse introns (68% of cases), but this could represent a selection bias reflecting the fact that the less extensive sequencing of the mouse genome may lead to an underrepresentation of instances in which the mouse genomic locus is larger. This question will need to be revisited in the presence of larger amounts of genomic sequence.
Sequence Similarity
Coding regions showed strong sequence similarity, with approximately 85% identity as previously reported (Makalowski et al. 1996; Makalowski and Boguski 1998, 1998a; Lamerdin et al. 1995; Koop and Hood 1994). In contrast, introns showed only weak sequence similarity with approximately 35% sequence identity, which is not much higher than the background rate of sequence identity in gapped alignments of random sequences.
The degree of conservation varied considerably among genes. For example, the gene encoding the ribosomal protein S24 (Table 1, gene 4) showed 88% identity at the DNA level and 100% identity at the amino acid level in coding exons, but only 27% identity at the DNA level in introns. The perfect identity at the amino acid level is consistent with the protein being highly constrained, as might be expected for a component of the ribosome. In contrast, some introns exhibited a striking degree of similarity. In the tumor necrosis factor-beta gene (Table 1, gene 93), the first intron has 75% nucleotide identity and nearly perfect agreement in length (86 bp in human, 83 bp in mouse). Interestingly, the flanking exons are less well-conserved, showing only 70% nucleotide identity and 60% amino acid identity.
Global Sequence Alignment, GLASS
To recognize genes based on the coincidence of biological signals in two organisms, it is important to start with an accurate global alignment of the genomic sequences. Existing global alignment techniques were not well-suited for our purposes.
Standard dynamic programming (SDP) methods (based on the Needleman-Wunsch (1970) or Smith-Waterman (1981) algorithms) were unsuitable for two reasons. First, their running time scales in proportion to O(NM) (where N and M are the lengths of the genomic sequences compared, which can be very large for genomic sequence comparisons). Second, they are not sensitive to finding short regions of good alignment (such as a 50-base exon) flanked by much longer regions of poor alignment (such as long introns).
Faster local alignment methods (such as BLAST) are better suited, but still insufficient. First, they provide only lists of local alignments ranked by quality rather than a global correspondence map of two long genomic sequences. Second, they detect alignments by looking for perfect matches of a predetermined length (e.g., 11 bases) and thereby may miss important conserved regions.
Accordingly, we designed a new alignment system called GLASS (GLobal Alignment SyStem), suitable for aligning hundreds of kilobases of genomic sequence. GLASS works by iteratively aligning matching segments (Fig. 1). First, a rough alignment map is constructed by finding long segments that match exactly, and whose flanking regions have high similarity. The procedure is repeated on the intervening regions using successive smaller matching segments. Finally, the remaining short unaligned regions are aligned using standard alignment techniques.
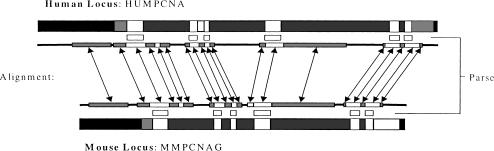
Figure 1.
Regions of the human and mouse homologous genes: Coding exons (white), noncoding exons (gray}, introns (dark gray), and intergenic regions (black). Corresponding strong (white) and weak (gray) alignment regions of GLASS are shown connected with arrows. Dark lines connecting the alignment regions denote very weak or no alignment. The predicted coding regions of ROSETTA in human, and the corresponding regins in mouse, are shown (white) between the genes and the alignment regions.
More precisely, GLASS works as follows. The program takes as input two genomic segments and returns a global alignment for the segments. The global alignment is computed recursively. The basic steps are as follows:
For an initial value of k, find all matching k-mers i.e., k-mers that appear in both sequences.
Treating each matching k-mer as a unique “k-mer character”, convert both the human and mouse sequences into strings of such characters, corresponding to occurrences of the matching k-mers. (Only the matching k-mers are represented in these strings).
Align these two strings using the following non-standard dynamic programming procedure. Matching k-mers receive a score equal to the sum of the alignment scores obtained by applying SDP to the short region flanking the occurrence of the k-mer in the human and mouse sequence (specifically, SDP is applied to the 12 nucleotides to the left and 12 nucleotides to the right.) Mismatches and gaps in the alignment of the k-mer string receive a score of 0.
In the above alignment, identify those pairs of matching k-mers that lie within regions of good local alignment between the human and mouse sequences; that is, those that have a score exceeding a threshold T (T is typically 4).
From this list of pairs of matching k-mers, remove those that are inconsistent with the underlying human and mouse genomic sequences. Specifically, two k-mers are inconsistent if they correspond to positions that overlap by i>0 bases in one species but not in the other species.
Using the remaining list of matching k-mers, fix the alignment between the nucleotides in the underlying human and mouse sequences contained in these k-mers.
Recursively align the regions between aligned nucleotides, by repeating steps 1–6 using a smaller value of k. As currently implemented, GLASS recursively employs k-mers with k = 20, 15, 12, 9, 8, 7, 6 and 5.
Once the last recursive alignment is performed, extend all pairs of aligned segments by short local alignments to the left and right by SDP.
Finally, align the remaining (usually short) unaligned regions using SDP.
Various parameters used in the GLASS program were adjusted on the basis of a test set consisting of 12 orthologous gene pairs (Table 2). For the SDP of nucleotide sequences in the above steps, the respective scores for match, mismatch, gap open, and gap extension were 1, -1, -6, and -2.
Table 2.
Training Set of Human/Mouse Homologs
An example of an alignment between two orthologous genomic loci is shown in Fig. 1.
Once the genomic sequences are aligned, the sequences are processed to mask repeats (using RepeatMasker, http://ftp.genome.washington.edu/RM/RepeatMasker.html) and poorly aligned regions (defined as those containing too many gaps or too many mismatches). The remaining well-aligned sequence was then used for generecognition.
Gene Recognition, ROSETTA
To perform gene recognition, we began by specifying the 'gene model' to be recognized in a genomic region.
A “coding exon” was defined to be the translated portion of an exon, together with a designated strand and reading frame. Coding exons can be initial (consisting of the region from the start of translation to either a splice site or the in-frame stop codon), internal (consisting of the region between two splice sites), or terminal (consisting of the region from either a splice site or a start of translation to the in-frame stop codon). Coding exons thus differ from actual exons in that they exclude the nucleotides in the 5′- and 3′-UTRs.
A “parse” of a genomic region is a sequence ([a1, b1, t1, s1, f1], [a2, b2, t2, s2, f2],. . . [an, bn, tn, sn, fn]) where ei = (ai, bi, ti, si, fi) denotes consecutive exons with starting and stopping points (ai, bi), type ti ɛ {initial, internal, terminal}, designated strand si ɛ { +, -} and reading frame fi ɛ {0,1,2}. The parse is valid provided that the following properties hold for each pair of consecutive coding exons ei and ei +1: (i) If ei is terminal, then ei +1 is initial, and vice versa, and (ii) if ei is not terminal, then ei and ei +1 have consistent strands and reading frames and are both open in the designated reading frame.
Currently, strands are handled separately and parses in the two strands are merged in a post-processing step. Details are described in Methods.
Our automatic procedure involved using a dynamic programming approach to find the optimal valid parse with respect to a given scoring procedure. Each parse ([a1, b1, t1, s1, f1], [a2, b2, t2, s2, f2], … [an, bn, tn, sn, fn]) of the human genomic sequence corresponds to a parse ([a′1, b′1, t′1, s′1, f′1], [a′2, b′2, t′2, s′2, f′2], … [a′n, b′n, t′n, s′n, f′n]) of the mouse genomic sequence, by means of the cross-species sequence alignment. Each parse is assigned a score consisting of the sum of scores for the individual coding exons. The score for each coding exon consisted of several components, reflecting the presence of appropriate splice sites, codon usage, amino acid alignment and length.
Splice Sites
Splice site scores were calculated by using a hybrid method that combines the GENSCAN splice site detector (Burge 1997) and a directionality effect (Pachter et al. 1999). The splice site scores for the splice acceptor and splice donor sites in both the human and mouse sequences were summed to obtain an overall score for each putative coding exon. For initial or terminal exons, splice site scores were only computed at the appropriate end.
Codon Usage
A codon usage score was computed for both the human and mouse exons, and the two scores were added together. Each score was calculated by summing the log odds ratio for each codon, based on published codon frequencies for the organism (Delphin et al. 1999).
Amino Acid Similarity
A amino acid similarity score was calculated by comparing corresponding codons in the two exons and using the PAM20 matrix to score matches, mismatches and gaps. This score reflected the tendency of particular amino acid substitutions to occur between human and mouse (Dayhoff et al. 1978).
Exon Length
An exon length score was calculated, consisting of two components. The first component reflected agreement with the known length distribution of initial, internal and terminal exons. The second component penalized exons pairs that differed in length, particularly when the difference was not a multiple of 3.
Various parameters were optimized, based on an analysis of the test set of 12 orthologous genes (Table 2). The precise definitions of the scores are available on our web site (http://theory.lcs.mit.edu/crossspecies/).
Gene Recognition: Results
We applied ROSETTA to our collection of 117 orthologous gene pairs and evaluated its performance on the 105 genes that were not part of the training set. The program performed extremely well at identifying internal coding exons. Of internal coding exons, 94% were predicted perfectly at both ends and another 4% at one of the two ends. When one end is incorrectly predicted, the error typically involves only a few bases and typically is due to an alternative choice of splice site that more closely matches the expected pattern.
Only six of the internal coding exons (3%) were completely missed, and the reasons for the failures are instructive. (i) Three of these were in the galactose-1-phosphate uridyl transferase gene (Table 1, gene 37). They resulted from the failure to recognize mouse intron 4, because the 5′-splice site has a GC rather than the canonical GT (Leslie et al. 1992). As a result, the gene is predicted to end at a downstream stop codon and a new gene is predicted to begin at an ATG codon upstream of exon 8. Exons 5, 6, and 7 are thus missed. (ii) Another exon is missed in the 21-hydroxylase gene (Table 1, gene 95), because the 5′-splice site is regarded as unlikely by our splice site detector: G-GTGCCTC in human, and T-GTTACCC in mouse. (iii) The two other internal coding exons that were missed are the instances in which two exons in one species correspond to a single exon in the other (in the Flt3 ligand (Table 1, gene 100) and lymphotoxin beta (Table 1, gene 85) genes, as noted above). The program's rules do not currently handle this special case.
The program was somewhat less accurate for initial and terminal coding exons. A total of 71% of such exons were correctly predicted at both ends. An additional 19% were correctly predicted at one end, with the incorrect end almost always being the initiation codon of an initial exon or the stop codon of a terminal exon. The errors typically involve predicting a splice site rather than the initiation or stop codon. In 17 cases, these splice sites are in fact annotated splice sites of the 5′ and 3′ UTRs.
A total of 2 initial and 9 terminal coding exons were completely missed. The initial coding exons were missed because they had length 3, consisting only of the ATG, which gave too weak a signal to detect. The terminal coding exons were missed because the coding exon was extremely short in one case (3 bp in human, 6 bp in mouse) or because the sequences were highly divergent between human and mouse.
Overall, the exon predictions were very accurate at the nucleotide level: 95% of nucleotides lying within coding exons were correctly predicted as such, and 97% of nucleotides predicted to lie within coding exons in fact did so. ROSETTA thus had 95% sensitivity and 97% specificity at the nucleotide level. ROSETTA predicted 26 coding exons that failed to overlap with any known exon.
We also compared our results with the performance of GENSCAN (Burge 1997). On our dataset, GENSCAN had similar nucleotide sensitivity (98%) but considerably lower nucleotide specificity (89%). Moreover, GENSCAN predicted 68 regions not overlapping any known coding exon, whereas ROSETTA predicted only 26 such instances.
DISCUSSION
The analysis of large genomes is challenging because the important functional elements comprise only a small portion of the sequence: the problem is to extract signal from noise. Feature detectors that perform well enough in small genomes may become overwhelmed by large genomes and yield too many false positives.
A powerful solution is to first increase the signal-to-noise ratio by using evolutionary conservation among species. One can thereby focus attention on the portion of the sequence that is conserved (thereby decreasing noise) and search for features that are present in both species (thereby increasing the specificity of the signal).
Such strategies, of course, require that the elements to be found are indeed conserved by evolution. This certainly is the case for coding exons in human and mouse. Our study of the genomic structure of 117 orthologous gene pairs provides a quantitative description of the high degree of conservation in the number, length and sequence of coding exons.
The basic notion of using cross-species sequence comparison to identify important functional elements is well known, and has been used to study particular human and mouse regions (Hardison et. al 1997, Oeltjen et al. 1997, Jang et al.1999). Gene recognition, however, does not emerge by simple inspection from the pattern of conservation: many non-genic elements are also well-conserved, sometimes more so than genic elements. On average, the coding exons represented only a subset of the total well-aligned sequence.
We sought to develop an automatic method for recognizing genes on the basis of orthologous sequences from two different species. The approach involves aligning the genomic sequences and then parsing the sequences to find a gene model in which the proposed exons are supported by features (splice sites, codon usage, etc.) present in both species. Alignment is performed with the GLASS program and gene recognition with the ROSETTA program. Both programs are available for use on a public web server (http://theory.lcs.mit.edu/crossspecies/) and the programs themselves are available from the authors.
The resulting program identifies the location of coding exons with high specificity and sensitivity. The vast majority of coding exons are identified perfectly. The overall results were robust across genes, including instances such as the tumor necrosis factor beta gene in which the first intron shows higher conservation than the flanking exons. The remaining errors largely result from highly unusual features such as rare splice signals or fused exons.
ROSETTA represents only a first attempt at systematically using cross-species information for gene recognition. It should be possible to refine the program by incorporating feature detectors used in single-species gene recognition programs (such as those for promoters, poly-adenylation sites, etc., as well as more sophisticated statistical tests), by refining the way in which the existing detectors are combined and by incorporating rules to detect special cases (such as fused exons or non-canonical splice sites). The program is designed to recognize a single optimal gene model; a further challenge would be to recognize conserved patterns of alternative splicing by exploiting backtracking features of dynamic programming.
ROSETTA assumes that one has already identified apparently syntenic regions between two species. This is not a difficult task, in that syntenic regions tend to be large and can be preliminarily identified using relatively coarse similarity searches.
Our list of 117 orthologous pairs studied is necessarily biased toward genes with smaller genomic loci, owing to the fact that genomic sequences from such loci are over-represented in current databases. Such a bias towards shorter genes could potentially enhance ROSETTA's performance because of a higher signal-to-noise ratio in such genes. Moreover, all our loci contain single genes and therefore we have not tested ROSETTA's performance in larger genomic regions that may contain multiple genes, genes on both strands, and/or large non-coding intergenic regions.
An interesting question is whether the mouse is a suitable organism to select for exon prediction in human genes. Organisms whose sequence has not drifted sufficiently far from that of humans will not increase the signal-to-noise ratio sufficiently, while organisms that are too distant may make it difficult to recognize important signals. Interestingly, ROSETTA produced approximately equal amounts of over-prediction and under-prediction, which may suggest that the human and mouse are at a felicitous distance for the purpose of coding exon prediction.
With the explosion in the sequencing of the human and mouse genomes, cross-species sequence comparison should become an increasingly important technique for extracting information from the mammalian genome. We demonstrate here a systematic technique for extracting the vast majority of the information about coding exons. The next challenge will be to create similar systematic techniques to extract information about non-coding exons, promoters, regulatory elements and other important functional features of the genome.
METHODS
Database Construction
A database of 1196 corresponding human/mouse mRNA pairs that had been previously compiled (Makalowski et al. 1996) was used to compile a database of 117 orthologous and annotated (with respect to gene structure) human and mouse genes. This was done by matching the human and mouse mRNA entries of the database to all human and mouse DNA entries in GenBank Release 109 (October 1998). Genes in the human or mouse that did not have a corresponding entry in the other organism were rejected. Entries were accepted only if they contained all of the coding part of the gene, as well as the introns that lie between coding exons. Therefore, entries were accepted even if they did not constitute a complete gene, provided that they contained the coding part of the gene. In some cases, this caused us to accept genes without annotations or sequence for non-coding exons. Even though structural comparative information in non-coding regions could not be compiled for these entries, they were very useful for evaluating the quality of the ROSETTA coding region prediction method.
Our training set consisted of twelve pairs of human and mouse homologous genes, shown in Table 2. Training involved several steps. (1) Tuning parameters of GLASS in order to perfect the construction of global alignments of homologous genomic loci. (2) Choosing a PAM matrix for the protein alignments of pairs of potential exons. (3) Defining appropriate likelihood penalties for exons that were not preserved as is typical (for example, exons whose length difference was not a multiple of 3). We do not report any results on the training set.
Sequence Alignments and Comparative Analysis
When two corresponding regions in the human and mouse are not very similar, GLASS does not necessarily produce the exact map between the regions. For that reason, the corresponding introns, coding, and non-coding exon fragments in human and mouse sequences were further realigned using the standard dynamic programming (SDP) alignment algorithm in order to compute more accurate local alignments for the purpose of compiling nucleotide similarity statistics. Furthermore, the corresponding coding fragments were translated into protein and aligned using a PAM20 matrix obtained from the NCBI website (http://www.ncbi.nlm.nih.gov/) for the purpose of compiling protein similarity statistics.
The nucleotide similarity statistics in Table 1 for corresponding regions were computed using our similarity count SC(*,*) function. SC is a non-symmetric function that, given two sequences s1, s2 (in our case in human and mouse, respectively) and an alignment between s1 and s2, returns the number of valid matching positions of s1 into s2. The number of valid matching positions is the number of positions j in s1 that are mapped with a match to s2 and such that either (1) j is the first or last position in s1, or (2) j – 1 and j + 1 are not mapped to gaps in s2. Thus, spurious matches in predominantly gapped regions do not add to the similarity count. This way the similarity count is not higher in the cases where the region s2, in our case the mouse region, is much longer than s1 and therefore s1 can be aligned with many gaps and a large number of spurious matches. The similarity counts were divided by the lengths of the human regions. Amino acid similarity statistics for corresponding coding regions were computed by counting the number of matching positions in the amino acid alignment of the regions, and dividing it by the length of the human exon
Total sequence nucleotide similarity statistics were computed using the global alignment of the sequences derived by GLASS. A window of good alignment was defined to be a window of size 51 containing at least 20 matches. Any matches not contained inside a window of good alignment were discarded, and the number of remaining matches was divided by the length of the human locus.
Computational Prediction of Coding Regions
Masked Regions
Before finding the optimal parse, we preprocessed the human and mouse sequences to mask repeats using the RepeatMasker program (http://ftp.genome.washington.edu/RM/RepeatMasker.html), and regions of weak alignment. RepeatMasker was applied with the -s option that makes it around 5% more accurate. We used the -rod option when masking the mouse sequences. We used the default option when masking the human sequences. A position was defined to be in a region of weak alignment if it was either (1) situated in a gap of length at least 30, or (2) in the middle of a window of size 37 that contained fewer than 10 matches. Nucleotides in masked regions were disqualified from being predicted as coding.
Splice Site Scores
Splice site scores were computed using the directional rule modification to the GENSCAN splice site detector, as explained in (Pachter et al. 1999). Donor splice site scores were multiplied by 0.5 and acceptor splice site scores were multiplied by 3.5. These values were obtained by requiring that the mean scores for donor and acceptor splice sites be equal in our training set. Potential exons with a combined score of less than -10 for flanking splice sites were disqualified from being predicted as coding exons.
Coding Exon Length
Corresponding potential coding exons with different lengths in the human and mouse sequences were penalized as follows: initial and terminal exons with different lengths were given a penalty of –3 if lengths were equal mod 3, and –9 otherwise. For internal coding exons, the corresponding penalties were –9 and –27. These values were chosen heuristically, and were found to combine well with the other components of the scoring, most importantly the PAM20 matrix and the splice site scores. For instance, a PAM20 gap penalty is –19, while a PAM20 base substitution “penalty” ranges from +1 to –17, with typical values in the –8 range.
Merging Forward and Reverse Complement Strand Parses
Currently ROSETTA handles forward and reverse complement strands separately. Parses in the two strands are subsequently merged in a post-processing step. For each predicted exon e, a window extending 2000 positions in each direction from the endpoints of e, is used to count the forward and reverse complement coverage of the genomic region. That is, the number of predicted coding positions in each direction, included in the window, is calculated. If the direction of e is the direction with the highest count, e is accepted. Otherwise e is rejected. Future versions of ROSETTA may include a sophisticated genomic region model, where parses in both strands are simultaneously optimized.
For further details on the parameter selection for ROSETTA and GLASS we refer to our web site (http://theory.lcs.mit.edu/crossspecies/).
WWW Resources
http://www.ncbi.nlm.nih.gov/. 1998. National Center for Biology Information webpage.
http://ftp.genome.washington.edu/RM/RepeatMasker.html. RepeatMasker webpage.
http://www.sanger.ac.uk/Software/Wise2/. Wise 2 webpage.
http://www.theory.lcs.mit.edu/crossspecies. Rosetta and Glass webpage.
Acknowledgments
S.B., B.B., and this work were supported in part by Merck. L.P. was supported in part by a graduate fellowship from the Program in Mathematics and Molecular Biology and by a National Institutes of Health training grant. E.S.L. and J.M. were supported in part by a grant from the National Human Genome Research Institute. We thank Bruce Birren, Ken Dewar, and Daniel Kleitman for helpful discussions. We thank Eric Banks for support with software development.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: lander@wi.mit.edu; FAX (617) 252-1902.
REFERENCES
- Allard W J, Sigal IS, Dixon RAF. Sequence of the gene encoding the human M1 muscarinic acetylcholine receptor. Nucl Acids Res. 1987;15:10604. doi: 10.1093/nar/15.24.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski M S, Cox DR, Myers RM. Genomes and evolution – Overview. Curr Op Genet Dev. 1996;6:683–685. doi: 10.1016/s0959-437x(96)80020-7. [DOI] [PubMed] [Google Scholar]
- Burge C. Identification of genes in human genomic DNA. Ph.D. dissertation. Stanford University, Department of Mathematics; 1997. [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Bio. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Burset M, Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- Dayhoff M, Schwartz R M, Orcutt B C. A model of evolutionary change in proteins. Atlas Prot Seq Struct. 1978;5:345–352. [Google Scholar]
- Delphin M E, Stockwell P A, Tate W P, Brown C M. Transterm, the translational signal database, extended to include full coding sequence and untranslated regions. Nuc Acids Res. 1999;27:293–294. doi: 10.1093/nar/27.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand M S, Mironov A A, Pevzner P A. Gene recognition via spliced sequence alignment. Proc Natl Acad Sci USA. 1996;93:9061–9066. doi: 10.1073/pnas.93.17.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R C, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Jang W, Hua A, Spilson S V, Miller W, Roe B A, Meisler M H. Comparative sequence of human and mouse BAC clones from the mnd2 region of chromosome 2p13. Genome Res. 1999;9:53–61. [PMC free article] [PubMed] [Google Scholar]
- Koop B F, Hood L. Striking sequence similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nat Genet. 1994;7:48–53. doi: 10.1038/ng0594-48. [DOI] [PubMed] [Google Scholar]
- Lamerdin J E, Montgomery MA, Stilwagen SA, Scheidecker LK, Tebbs RS, Brookman KW, Thompson LH, Carrano AV. Genomic Sequence Comparison of the Human and Mouse XRCC1 DNA Repair Gene Regions. Genomics. 1995;25:547–554. doi: 10.1016/0888-7543(95)80056-r. [DOI] [PubMed] [Google Scholar]
- Leslie N D, Immerman EB, Flach JE, Florez M, Fridovich-Keil JL, Elsas L. The human galactose-1 phosphate uridyltransferase gene. Genomics. 1992;14:474–480. doi: 10.1016/s0888-7543(05)80244-7. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Synonymous and Nonsynonymous Substitution Distances are Correlated in Mouse and Rat Genes. J Mol Evol. 1998;47:119–121. doi: 10.1007/pl00006367. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Zhang J, Boguski M S. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Research. 1996;6:846–857. doi: 10.1101/gr.6.9.846. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci. 1998a;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Tsuchiya M, Asano S, Yamamoto O, Hirata Y, Kubota N, Oheda M, Nomura H, Yamazaki T. The chromosomal gene structure and the mRNAs for human granulocyte colony-stimulating factor. EMBO J. 1986;5:575–581. doi: 10.1002/j.1460-2075.1986.tb04249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunch CD. A general method applicable to the search for similarities in the amino acid sequences of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Oeltjen J C, Malley T, Muzny D M, Miller W, Gibbs R A, Belmont J W. Large scale comparative sequence analysis of the human and murine bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Pachter L, Batzoglou S, Spitkovsky V I, Banks E, Lander E S, Kleitman D J, Berger B. A dictionary based approach for gene annotation. J Comp Biol. 1999;1999 Fall-Winter (6) 3–4:419–430. doi: 10.1089/106652799318364. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]