Abstract
Dopamine (DA) undergoes monoamine oxidase catalyzed oxidative deamination to 3,4-dihydroxyphenylacetaldehyde (DOPAL), which is metabolized primarily to 3,4-dihydroxyphenylacetic acid (DOPAC) via aldehyde dehydrogenase (ALDH). Previous studies demonstrated DOPAL to be neurotoxic, more so than DA and other metabolites, and implicated the aldehyde intermediate as a factor in the pathogenesis of Parkinson’s disease (PD). However, the mechanism for generation of DOPAL at aberrant levels and the pathways for toxicity are not conclusively known. Various models for DA catabolism revealed the susceptibility of DOPAL biotransformation (e.g., ALDH) to products of oxidative stress, e.g., 4-hydroxy-2-nonenal, at physiologic/pathologic levels and agents that induce oxidative stress. An elevated concentration of DOPAL correlated with increased protein modification with subsequent work demonstrating significant reactivity of the DA-derived electrophile toward protein nucleophiles compared to DA and other metabolites, e.g., DOPAC. The addition of DOPAL to proteins proceeds via reaction of the aldehyde with Lys residues, yielding a Schiff base; however, post-adduction chemistry occurs for the DOPAL-modification resulting in protein cross-linking. Preliminary work indicates enzymes in DA synthesis and catabolism to be cellular targets for DOPAL. Functional consequences for elevated levels of the DA-derived aldehyde and protein modification may include adverse cellular effects. These data implicate DOPAL as a toxic and reactive intermediate potentially serving as a “chemical trigger” for some stage of PD pathogenesis.
Keywords: dopamine; 3,4-dihydroxyphenylacetaldehyde; Parkinson’s disease; protein modification; biological reactive intermediate
1. Introduction
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by dopaminergic cell death and formation of Lewy bodies. It is a debilitating condition affecting numerous individuals worldwide and a human health concern. The mechanism of pathogenesis is currently unknown; however, results of recent studies indicate exposure to environmental agents and oxidative stress to be significant factors in the disease process [1-3]. To explain the loss of dopaminergic cells observed in PD, it has been hypothesized that a toxicant endogenous to these neurons is generated at aberrant levels (i.e., concentrations higher than the physiologic value of ~2 μM shown to cause toxicity) subsequent to insult [4]. Many candidates for this neurotoxicant have been proposed and include neurotransmitters, metabolites and byproducts of neurotransmitter metabolism [5].
Dopamine (DA) is a neurotransmitter essential for various bodily functions, including the coordination of movement; however, this important neurochemical can undergo auto- and enzymatic oxidation (i.e., biotransformation) processes yielding several damaging biological reactive intermediate (BRI) known to react with cellular nucleophiles [6-8]. One such BRI is 3,4-dihydroxyphenylacetaldehyde (DOPAL), an intermediate of dopamine catabolism resulting from monoamine oxidase-catalyzed oxidative deamination of DA as shown in Figure 1 [9]. As a primary pathway for metabolism, DOPAL is oxidized to 3,4-dihydroxyphenylacetic acid (DOPAC) via numerous aldehyde dehydrogenase (ALDH) enzymes, both cytosolic and mitochondrial as discussed in an excellent review [10]. In addition, cytosolic reductases will catalyze reduction of the aldehyde to an alcohol.
Figure 1.
Dopamine undergoes oxidative deamination via monoamine oxidase to 3,4-dihydroxyphenylacetaldehyde with further oxidation via aldehyde dehydrogenase to 3,4-dihydroxyphenylacetic acid.
Previous work demonstrated DOPAL to be toxic to dopaminergic cells and reactive toward cellular nucleophiles (e.g., proteins) [11, 12]. DA has been implicated as an endogenous neurotoxicant given its ability to auto-oxidize to an orthoquinone and generate superoxide anion [6, 13]; however, for comparison, DOPAL was shown to be more toxic in vitro and in vivo [11]. In addition, evidence was generated suggesting that DOPAL reacts with cellular nucleophiles, e.g., proteins [12, 14]. Such data have implicated the DA-derived BRI as a factor in PD and piqued interest in the endogenous neurotoxicant as part of the effort to discover new therapeutic targets and identify novel biomarkers for earlier disease diagnosis.
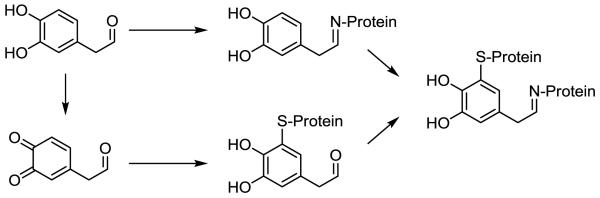
In regards to DOPAL and PD, several key questions remain, including: 1) How is DOPAL, an intermediate in DA catabolism, generated at aberrant levels? Also of interest in regards to this subject is the link between environmental insults correlated with PD, e.g., organochlorine pesticides or oxidative stress [15], and elevation in levels of the DA-derived aldehyde. 2). Is DOPAL a BRI, i.e., able to react with cellular nucleophiles? While it is reasonable to posit the molecule as a BRI given its aldehyde and catechol functional groups (Figure 2), there is paucity of information in regards to the protein reactivity of DOPAL and whether or not post-adduction chemistry occurs. 3). What are the cellular targets of the BRI? In line with this matter, there is a need to elucidate the functional consequence of protein modification by DOPAL and identify its role in the disease process.
Figure 2.
Reactivity of 3,4-dihydroxyphenylacetaldehyde (DOPAL) involves modification of protein amines via the aldehyde. However, catechol oxidation is possible, yielding a thiol-reactive quinone, and such oxidation may explain the protein cross-linking observed for DOPAL. The spontaneous reaction of the DOPAL aldehyde with a protein primary amine (e.g., Lys) may yield a Schiff base as shown or an enamine product. Auto-oxidation of the DOPAL catechol, which is known to occur for DA, or enzyme-mediated oxidation (e.g., tyrosinase, prostaglandin-H-synthase) is predicted to result in formation of an ortho-quinone, which are known to be highly reactive for protein thiols (e.g., Cys).
2. Materials and Methods
2.1. Cell Culture
PC6-3 cells were cultured in RPMI 1640 media supplemented with 10% heat-inactivated horse serum and 5% fetal bovine serum plus penicillin (10 IU/mL) and streptomycin (10 mg/mL) [16]. To induce differentiation, cells were treated with nerve growth factor for four days at a final concentration of 50 ng/mL. For experiments, serum and media were removed and cells were incubated in Krebs-phosphate buffer to minimize loss of DOPAL via media and serum components (e.g., proteins) [15]. To generate DOPAL in situ, cells were treated with 100 μM DA for 15 min followed by a 4 hour incubation with 4-hydroxynonenal (i.e., 50 μM, which is not significantly toxic at 4 hours) to inhibit DOPAL metabolism [15, 27]. PC6-3 cells are a sub-line of PC12 cells, which exhibit dopamine synthesis and metabolism and are a widely used model for dopaminergic cells [17-19].
2.2. DOPAL Synthesis
DOPAL was biosynthesized via monoamine oxidase as previously described [20]. Concentration of DOPAL was determined by HPLC analysis using an Agilent 1100 or 1200 Series capillary HPLC system with photo-diode detector [19].
2.3. SDS-PAGE Analysis
Glyceraldehyde-3-phosphate dehydrogenase (0.3 mg/mL) was incubated with 50 μM DA, DOPAL or DOPAC for 4 hrs at 37 °C in 50 mM sodium phosphate buffer, pH 7.4, before analysis via SDS-PAGE (reducing and denaturing conditions) as previously described [21]. Proteins were stained with Coomassie dye.
2.4. Proteomic Analysis
PC6-3 cells were lysed via sonication in 10 mM potassium phosphate buffer (pH 7.4) containing 1% triton X-100 and centrifuged at 700 × g and 10,000 × g, retaining the supernatant in both cases. The supernatant was treated with 100 μM DOPAL for 4 hrs at 37 °C, and proteins modified by the DA-derived aldehyde were isolated using an aminophenylboronic acid resin, previously shown to selectively bind proteins containing catechol adducts (e.g., DA) [13]. 180 μg of cellular protein was incubated with the resin for 3 hours at room temperature and proteins released via incubation of the resin with 1% and 0.05% trifluoroacetic acid (50 μL). Each release fraction was neutralized by addition of 5 μL 1M sodium phosphate buffer (pH 7.4). Released proteins were separated via a 10% SDS-PAGE gel, stained with Coomassie and identified with the assistance of the University of Iowa Proteomics Facility using in-gel digestion (trypsin) and mass spectrometry via a Thermo LTQ XL ion trap and collision-induced dissociation (University of Iowa Proteomics Facility). The SEQUEST database search engine was utilized to match peptides to known proteins, with a protein match involving 2 or more peptides. Prior to analysis and isolation of cellular protein targets of the DA-derived aldehyde, pilot experiments were performed with bovine serum albumin and glyceraldehyde-3-phosphate dehydrogenase modified by DOPAL (1 mg/mL protein in sodium phosphate buffer, pH 7.4incubated for 4 hours at 37 °C with 100 μM DOPAL) to establish the effectiveness of the aminophenlyboronic acid resin for isolating proteins containing a DOPAL adduct. Proteins were fractionated with a 10% SDS-PAGE gel, stained with Coomassie and matched to molecular weight standards.
3. Generation of DOPAL at Aberrant Levels
PD brains have increased oxidative burden as compared to controls, including elevated levels of iron and decreased concentrations of antioxidants, and such conditions are favorable for oxidative decomposition of lipids [1, 22]. Two major products of lipid peroxidation are 4-hydroxy-2-nonenal (4HNE) and malondialdehyde (MDA), considered to be markers of oxidative stress [23]. Indeed, an earlier report demonstrated elevated levels of 4HNE in PD brains compared to control tissue [24]. Of concern is the reported sensitivity of carbonyl metabolizing enzymes toward lipid peroxidation products, namely MDA and 4HNE, as there is significant evidence demonstrating inhibition of ALDH via such species at physiologic concentrations [25, 26].
As described above, several ALDH enzymes, both cytosolic and mitochondrial, catalyze the oxidation of DOPAL to DOPAC [10], and as a minor pathway, cytosolic reductases participate in the biotransformation of the aldehyde to an alcohol product (i.e., 3,4-dihydroxyphenylethanol, DOPET) [9]. Using various model systems, including rat brain mitochondria, rat striatal synaptosomes and dopaminergic PC6-3 cells, ALDH inhibition was observed in the presence of physiologic concentrations of lipid peroxidation products [16, 20, 27]. The net effect was a time and dose dependent increase in the level of DOPAL. 4HNE-mediated impairment of ALDH activity was compensated via cellular reductases as an increase in DOPET correlated with elevated DOPAL. In contrast, a loss of the reductase activity toward DOPAL resulted from treatment of cells with MDA but not 4HNE.
Other means for generation of DOPAL at an aberrant level are conceivable, quite likely and include impairment of mitochondrial complex I and dysregulation of DA trafficking and uptake. Mitochondrial Complex 1 is responsible for production of NAD necessary for mitochondrial metabolism of DOPAL to DOPAC, and a previous study demonstrated increased concentrations of DOPAL following cellular treatment with the known Complex 1 inhibitor rotenone [28].
DA is synthesized in the cytosol and rapidly sequestered in synaptic vesicles via the action of the vesicular monoamine transporter (VMAT) [9]. Interference with DA trafficking is predicted to yield elevated cytosolic levels of the neurotransmitter and increased MAO-mediated conversion of DA to DOPAL. Recent work demonstrated dieldrin, an organochlorine pesticide with exposure linked to PD, to be toxic to dopaminergic cells and capable of producing oxidative stress and disrupting DA trafficking [15]. Such an organochlorine is predicted to yield elevated levels of reactive intermediates such as DOPAL.
4. Is DOPAL a BRI?
Given the structure of DOPAL, containing a catechol and aldehyde, it is conceivable that the DA metabolite is a BRI and forms adducts with proteins (Figure 2). Spontaneous or enzymemediated (e.g., prostaglandin-H-synthase) catechol oxidation would produce a thiol-reactive ortho-quinone [6, 8], and such a reaction is well documented for DA. In addition, amines are predicted to react with the carbonyl of DOPAL yielding a Schiff base. Furthermore, the possibility exists for DOPAL to serve as a bifunctional electrophile, should the initial protein adduct be electrophilic or readily oxidized. Earlier studies demonstrated the ability of DOPAL to covalently modify tissue and proteins, however, more recent work has shown the DA-derived aldehyde primarily reacts with proteins via Lys [21, 27]. Incubation of DOPAL with N-acetylated amino acids (Cys and Lys) and model peptides (e.g., RKRSRAE) containing Lys produced adducts with mass consistent for a Schiff base (i.e., +134 Da).
The evidence described above demonstrates Lys-reactivity to be primarily responsible for protein modification; however, additional findings suggest post-adduction chemistry occurs for the DOPAL-adduct, potentially involving a quinone intermediate [21]. Interestingly, cross-linking was observed for a model protein (glyceraldehyde-3-phosphate dehydrogenase) modified by DOPAL, with the protein aggregation being time and concentration-dependent. Co-treatment of protein and DOPAL with 5 mM ascorbate afforded protection against cross-linking, suggesting the reaction involves a protein-bound quinone [21]. As shown in Figure 3, treatment of glyceraldehyde-3-phosphate dehydrogenase (~38 kD monomer) with DOPAL yielded loss of the monomeric protein but an increase in protein staining at or higher than 200 kD, indicating cross-linking or aggregation; however, neither DOPAC nor DA, both mono-functional electrophiles, was capable of producing significant protein cross-linking at concentrations equivalent to those used for DOPAL. Both DOPAC and DA contain only the catechol group, which can be oxidized to a thiol-reactive quinone, and no amine-reactive aldehyde. Given this and that ascorbate (a quinone-reducing agent) prevents DOPAL-mediated cross-linking [20], it is likely that proteins react with DOPAL initially via Lys residues yielding a Schiff base product and subsequently via adduct oxidation (i.e., catechol to quinone) and reaction with nucleophiles (e.g., Cys) in proximity. Such a scheme is demonstrated in Figure 2.
Figure 3.
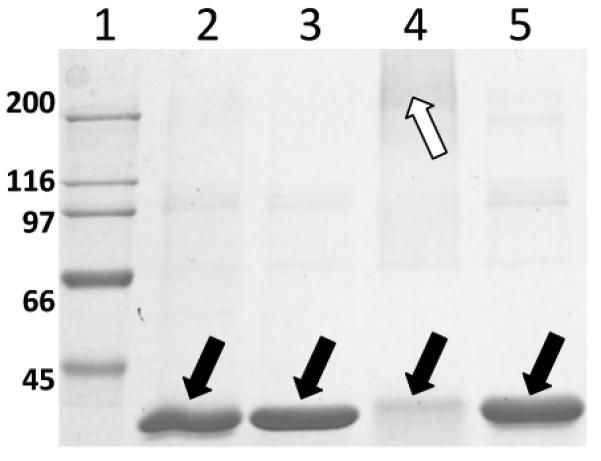
Reaction of 3,4-dihydroxyphenylacetaldehyde (DOPAL) but not dopamine (DA) or 3,4-dihydroxyphenylacetic acid (DOPAC) with glyceraldehyde-3-phosphate dehydrogenase yielded significant protein cross-linking/aggregation. Lane 1, molecular weight marker. Lane 2, control protein. Lane 3, protein with DA. Lane 4, protein with DOPAL. Lane 5, protein with DOPAC. The SDS-PAGE gel was stained with Coomassie. Solid arrows point to the protein monomer (~38 kD) and the open arrow denotes protein aggregates (>200 kD). Note the disappearance of protein monomer in Lane 4 (DOPAL treatment) that correlates with protein aggregation.
The data are clear given DOPAL’s role as a BRI and protein modifier; however, it appears the DOPAL-modification participates in post-adduction chemistry to produce a protein cross-link of unknown structure. In addition, there is a lack of information regarding the quinone form of DOPAL, such as the biochemistry/chemistry of its formation, protein reactivity and chemical stability. Work is in progress to address these issues.
5. Cellular Targets of DOPAL
Given its labile functional groups and propensity to modify proteins, DOPAL most likely reacts with numerous targets in the proteome. However, it is conceivable that selectivity for adduction may exist, and proteins containing catetcholamine binding sites (e.g., tyrosine hydroxylase, VMAT) or highly reactive amines could be preferentially modified. Of interest is the identification of such critical targets and the functional consequence of protein adduction by the BRI DOPAL.
As described above, proteins in PC6-3 cell lysate modified by DOPAL were isolated using an aminophenylboronic acid resin and separated via a 10% SDS-PAGE. Four major bands (approximately 25, 55, 65 and 75 kD) representing proteins containing a catechol adduct were evident following Coomassie staining. Preliminary proteomic analysis of the 25 and 55 kD bands revealed the identity of several key proteins modified by the BRI, including: glutathione-S-transferase, mu 1 (26 kD, 9 peptides identified), ALDH family 2 (56 kD, 6 peptides identified) and tyrosine hydroxylase (56 kD, 3 peptides identified). Both tyrosine hydroxylase and ALDH (family 2) participate in DA biosynthesis and catabolism, respectively, and glutathione-S-transferases of the mu class catalyze glutathione conjugation with oxidized catecholamines (e.g., aminochrome, dopachrome) [29, 30]. Such findings imply specificity of DOPAL adduction for proteins that interact with catechols. In regards to the functional consequence of protein modification, previous studies have demonstrated the sensitivity of ALDH activity to DOPAL and enzyme adduction by the DA-derived aldehyde. Work is in progress to study the interaction of tyrosine hydroxylase with DOPAL.
6. Conclusions and Future Directions
In conclusion, DOPAL is a product of oxidative deamination of the neurotransmitter DA and a neurotoxicant hypothesized to be relevant to PD pathogenesis as described above and in recent reports [31, 32]. Recent data have demonstrated: 1). Mechanisms for generation of the DA-derived metabolite at elevated levels [16, 20, 27, 28]. 2). DOPAL’s role as a BRI, capable of modifying and cross-linking proteins [21, 27]. Preliminary evidence suggests proteins relevant to catechol synthesis or metabolism are targets of the DA-derived aldehyde, as noted in the previous section. Future work will explore further DOPAL’s quinone form, including biological means (e.g., enzyme oxidation) for generation and protein reactivity. In addition, subsequent studies will focus on elucidation of the functional consequence of DOPAL-protein adduction and its role in PD. Initiation or propagation of neurodegenerative disease via DOPAL-protein modification could involve one or more mechanisms, such as: inhibition of enzyme/protein function important for dopaminergic cells, cross-linking proteins in dopamine neurons (e.g., α-synuclein), impairment of the proteasome, increase in oxidative stress. It is predicted that one or more of these mechanisms/endpoints would yield selective dysfunction or toxicity in cells relevant to PD (i.e., dopaminergic neurons). Such work is highly relevant to identifying the role of the BRI DOPAL in PD and may yield novel targets for therapeutic treatment and biomarkers for earlier disease diagnosis.
Abbreviations
- ALDH
aldehyde dehydrogenase
- ALDH2
mitochondrial aldehyde dehydrogenase
- BRI
biological reactive intermediate
- DA
dopamine
- DOPAC
3,4-dihydroxyphenylacetic acid
- DOPAL
3,4-dihydroxyphenylacetaldehyde
- DOPET
3,4-dihydroxphenylethanol
- 4HNE
4-hydroxy-2-nonenal
- MDA
malondialdehyde
- PD
Parkinson’s Disease
- VMAT
vesicular monoamine transporter
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jenner P. Oxidative stress in Parkinson’s disease. Ann Neurol. 2003;53(Suppl 3):S26–36. doi: 10.1002/ana.10483. discussion S36-28. [DOI] [PubMed] [Google Scholar]
- [2].Drechsel DA, Patel M. Role of reactive oxygen species in the neurotoxicity of environmental agents implicated in Parkinson’s disease. Free Radic Biol Med. 2008;44(11):1873–1886. doi: 10.1016/j.freeradbiomed.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Landrigan PJ, Sonawane B, Butler RN, Trasande L, Callan R, Droller D. Early environmental origins of neurodegenerative disease in later life. Environ Health Perspect. 2005;113(9):1230–1233. doi: 10.1289/ehp.7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Burke WJ, Li SW, Williams EA, Nonneman R, Zahm DS. 3,4-Dihydroxyphenylacetaldehyde is the toxic dopamine metabolite in vivo: implications for Parkinson’s disease pathogenesis. Brain Res. 2003;989(2):205–213. doi: 10.1016/s0006-8993(03)03354-7. [DOI] [PubMed] [Google Scholar]
- [5].Burke WJ. 3,4-dihydroxyphenylacetaldehyde: a potential target for neuroprotective therapy in Parkinson’s disease. Curr Drug Targets CNS Neurol Disord. 2003;2(2):143–148. doi: 10.2174/1568007033482913. [DOI] [PubMed] [Google Scholar]
- [6].Graham DG. Oxidative pathways for catecholamines in the genesis of neuromelanin and cytotoxic quinones. Mol Pharmacol. 1978;14(4):633–643. [PubMed] [Google Scholar]
- [7].Hastings TG, Zigmond MJ. Identification of catechol-protein conjugates in neostriatal slices incubated with [3H]dopamine: impact of ascorbic acid and glutathione. J Neurochem. 1994;63(3):1126–1132. doi: 10.1046/j.1471-4159.1994.63031126.x. [DOI] [PubMed] [Google Scholar]
- [8].Mattammal MB, Strong R, Lakshmi VM, Chung HD, Stephenson AH. Prostaglandin H synthetase-mediated metabolism of dopamine: implication for Parkinson’s disease. J Neurochem. 1995;64(4):1645–1654. doi: 10.1046/j.1471-4159.1995.64041645.x. [DOI] [PubMed] [Google Scholar]
- [9].Eisenhofer G, Kopin IJ, Goldstein DS. Catecholamine metabolism: a contemporary view with implications for physiology and medicine. Pharmacol Rev. 2004;56(3):331–349. doi: 10.1124/pr.56.3.1. [DOI] [PubMed] [Google Scholar]
- [10].Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev. 2007;59(2):125–150. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- [11].Burke WJ, Li SW, Chung HD, Ruggiero DA, Kristal BS, Johnson EM, Lampe P, Kumar VB, Franko M, Williams EA, Zahm DS. Neurotoxicity of MAO metabolites of catecholamine neurotransmitters: role in neurodegenerative diseases. Neurotoxicology. 2004;25(1-2):101–115. doi: 10.1016/S0161-813X(03)00090-1. [DOI] [PubMed] [Google Scholar]
- [12].Nilsson GE, Tottmar O. Biogenic aldehydes in brain: on their preparation and reactions with rat brain tissue. J Neurochem. 1987;48(5):1566–1572. doi: 10.1111/j.1471-4159.1987.tb05702.x. [DOI] [PubMed] [Google Scholar]
- [13].LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11(11):1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- [14].Ungar F, Tabakoff B, Alivisatos SG. Inhibition of binding of aldehydes of biogenic amines in tissues. Biochem Pharmacol. 1973;22(15):1905–1913. doi: 10.1016/0006-2952(73)90050-6. [DOI] [PubMed] [Google Scholar]
- [15].Kanthasamy AG, Kitazawa M, Kanthasamy A, Anantharam V. Dieldrin-induced neurotoxicity: relevance to Parkinson’s disease pathogenesis. Neurotoxicology. 2005;26(4):701–719. doi: 10.1016/j.neuro.2004.07.010. [DOI] [PubMed] [Google Scholar]
- [16].Jinsmaa Y, Florang VR, Rees JN, Anderson DG, Strack S, Doorn JA. Products of oxidative stress inhibit aldehyde oxidation and reduction pathways in dopamine catabolism yielding elevated levels of a reactive intermediate. Chem Res Toxicol. 2009;22(5):835–841. doi: 10.1021/tx800405v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shafer TJ, Atchison WD. Transmitter, ion channel and receptor properties of pheochromocytoma (PC12) cells: a model for neurotoxicological studies. Neurotoxicology. 1991;12(3):473–492. [PubMed] [Google Scholar]
- [18].Hirata Y, Adachi K, Kiuchi K. Activation of JNK pathway and induction of apoptosis by manganese in PC12 cells. J Neurochem. 1998;71(4):1607–1615. doi: 10.1046/j.1471-4159.1998.71041607.x. [DOI] [PubMed] [Google Scholar]
- [19].Kitazawa M, Anantharam V, Kanthasamy AG. Dieldrin-induced oxidative stress and neurochemical changes contribute to apoptopic cell death in dopaminergic cells. Free Radic Biol Med. 2001;31(11):1473–1485. doi: 10.1016/s0891-5849(01)00726-2. [DOI] [PubMed] [Google Scholar]
- [20].Florang VR, Rees JN, Brogden NK, Anderson DG, Hurley TD, Doorn JA. Inhibition of the oxidative metabolism of 3,4-dihydroxyphenylacetaldehyde, a reactive intermediate of dopamine metabolism, by 4-hydroxy-2-nonenal. Neurotoxicology. 2007;28(1):76–82. doi: 10.1016/j.neuro.2006.07.018. [DOI] [PubMed] [Google Scholar]
- [21].Rees JN, Florang VR, Eckert LL, Doorn JA. Protein reactivity of 3,4-dihydroxyphenylacetaldehyde, a toxic dopamine metabolite, is dependent on both the aldehyde and the catechol. Chem Res Toxicol. 2009;22(7):1256–1263. doi: 10.1021/tx9000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dexter DT, Carter CJ, Wells FR, Javoy-Agid F, Agid Y, Lees A, Jenner P, Marsden CD. Basal lipid peroxidation in substantia nigra is increased in Parkinson’s disease. J Neurochem. 1989;52(2):381–389. doi: 10.1111/j.1471-4159.1989.tb09133.x. [DOI] [PubMed] [Google Scholar]
- [23].Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11(1):81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- [24].Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman ER, Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci U S A. 1996;93(7):2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mitchell DY, Petersen DR. Inhibition of rat hepatic mitochondrial aldehyde dehydrogenase-mediated acetaldehyde oxidation by trans-4-hydroxy-2-nonenal. Hepatology. 1991;13(4):728–734. doi: 10.1016/0270-9139(91)92572-p. [DOI] [PubMed] [Google Scholar]
- [26].Hjelle JJ, Grubbs JH, Petersen DR. Inhibition of mitochondrial aldehyde dehydrogenase by malondialdehyde. Toxicol Lett. 1982;14(1-2):35–43. doi: 10.1016/0378-4274(82)90007-8. [DOI] [PubMed] [Google Scholar]
- [27].Rees JN, Florang VR, Anderson DG, Doorn JA. Lipid peroxidation products inhibit dopamine catabolism yielding aberrant levels of a reactive intermediate. Chem Res Toxicol. 2007;20(10):1536–1542. doi: 10.1021/tx700248y. [DOI] [PubMed] [Google Scholar]
- [28].Lamensdorf I, Eisenhofer G, Harvey-White J, Hayakawa Y, Kirk K, Kopin IJ. Metabolic stress in PC12 cells induces the formation of the endogenous dopaminergic neurotoxin, 3,4-dihydroxyphenylacetaldehyde. J Neurosci Res. 2000;60(4):552–558. doi: 10.1002/(SICI)1097-4547(20000515)60:4<552::AID-JNR14>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- [29].Segura-Aguilar J, Baez S, Widersten M, Welch CJ, Mannervik B. Human class Mu glutathione transferases, in particular isoenzyme M2-2, catalyze detoxication of the dopamine metabolite aminochrome. J Biol Chem. 1997;272(9):5727–5731. doi: 10.1074/jbc.272.9.5727. [DOI] [PubMed] [Google Scholar]
- [30].Baez S, Segura-Aguilar J, Widersten M, Johansson AS, Mannervik B. Glutathione transferases catalyse the detoxication of oxidized metabolites (o-quinones) of catecholamines and may serve as an antioxidant system preventing degenerative cellular processes. Biochem J. 1997;324(Pt 1):25–28. doi: 10.1042/bj3240025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Panneton WM, Kumar VB, Gan Q, Burke WJ, Galvin JE. The Neurotoxicity of DOPAL: Behavioral and Stereological Evidence for Its Role in Parkinson Disease Pathogenesis. PLoS One. 5(12):e15251. doi: 10.1371/journal.pone.0015251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Goldstein DS, Sullivan P, Holmes C, Kopin IJ, Basile MJ, Mash DC. Catechols in post-mortem brain of patients with Parkinson disease. Eur J Neurol. doi: 10.1111/j.1468-1331.2010.03246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]