Abstract
A cattle–human whole-genome comparative map was constructed using parallel radiation hybrid (RH) mapping in conjunction with EST sequencing, database mining for unmapped cattle genes, and a predictive bioinformatics approach (COMPASS) for targeting specific homologous regions. A total of 768 genes were placed on the RH map in addition to 319 microsatellites used as anchor markers. Of these, 638 had human orthologs with mapping data, thus permitting construction of an ordered comparative map. The large number of ordered loci revealed ⋝ 105 conserved segments between the two genomes. The comparative map suggests that 41 translocation events, a minimum of 54 internal rearrangements, and repositioning of all but one centromere can account for the observed organizations of the cattle and human genomes. In addition, the COMPASS in silico mapping tool was shown to be 95% accurate in its ability to predict cattle chromosome location from random sequence data, demonstrating this tool to be valuable for efficient targeting of specific regions for detailed mapping. The comparative map generated will be a cornerstone for elucidating mammalian chromosome phylogeny and the identification of genes of agricultural importance.“Ought we, for instance, to begin by discussing each separate species—in virtue of some common element of their nature, and proceed from this as a basis for the consideration of them separately?” from Aristotle, On the Parts of Animals, 350 B.C.E.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AW244888-AW244897, AW261132-AW261195, AW266849-AW267161, AW289175-AW289430, AW428566-AW428607, AW621146, AW621147.]
Comparative genomics has its roots in Aristotle, who understood that the commonalities among species would facilitate comprehension of the underlying “differentiae” that distinguish animals with common features. More than 1500 years later, after Mendel expounded the principles of inheritance and Darwin provided the intellectual framework for revealing a common molecular ancestry among species, the first example of linkage conservation in vertebrates was found among mice and rats for the albino coat color allele and pink eye dilution (Feldman 1924). Hence, mammalian form and physiology were understood to have common evolutionary origins arising from chromosome phylogeny. The historical threads to the present detailed gene maps of mammals run through a series of technical breakthroughs, from the use of isozymes, to somatic cell hybrid genetics, to the current explosion in gene mapping brought about by radiation hybrid (RH) technology (Cox et al. 1990). This progress is best represented by > 30,000 mapped human genes (Deloukas et al. 1998) and 2983 mouse–human gene homologies (Mouse Genome Database, Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine; http://www.informatics.jax.org, February, 2000). The development of RH cell panels for a number of species has led to a renaissance in comparative mapping that will soon erase the lead of “model organisms” in genes mapped, thereby revolutionizing our understanding of mammalian chromosome evolution (O'Brien et al. 1999). The phylogenomic approach to studying comparative genome organization and evolution (Eisen 1998; Bouzat et al., 2000) will eventually extend down the Woesian Tree of Life (Woese et al. 1990) until the commonalities among species are reduced to life's essentials and the “differentiae” of earth's biota are understood in molecular terms.
Among mammals, cattle have well-developed synteny and linkage maps (Eggen and Fries 1995; Womack and Kata 1995). There are now nearly 500 structural genes with cattle chromosome assignments (U.S. Bovine ArkDB http://bos.cvm.tamu.edu/bovgbase.html). Most of these genes have been mapped by physical methods, such as somatic cell hybrid analysis and in situ hybridization, leading to the identification of conserved synteny among a diverse spectrum of vertebrate genomes (Wakefield and Graves 1996; O'Brien et al. 1999). Interspecies chromosome painting has been applied to comparative mapping of human and cattle chromosomes (Solinas-Toldo et al. 1995), thus marking the major boundaries of conserved synteny on a genome-wide basis. Although chromosome painting provides a general view of comparative chromosome organization, the ability to draw meaningful inference about chromosomal evolution is limited by a paucity of ordered structural genes on the cattle gene map, i.e., there are < 200 genes on all the published cattle linkage maps (Ma et al. 1996; Barendse et al. 1997; Kappes et al. 1997). Adding genes to an ordered cattle gene map is critically important for the eventual isolation and characterization of genes affecting economically important traits of livestock and for understanding the evolution of vertebrate genomes (Womack and Kata 1995).
Mass production of expressed sequence tags (ESTs) is a powerful method for gene identification (Adams et al. 1991), and the combination of ESTs with RH mapping has proven invaluable for the development of a human gene map (Deloukas et al. 1998). Similarly, the development of a 5000 rad cattle–hamster RH panel opened the door to large-scale gene mapping in cattle (Womack et al. 1997). We recently demonstrated the power of RH mapping of cattle ESTs for comparative genomics (Band et al. 1998; Ma et al. 1998; Ozawa et al. 2000) and have shown that existing knowledge of comparative chromosome organization can be used to predict the map location of ESTs accurately in the cattle genome in silico. This in silico method for comparative genome analysis was termed comparative mapping by annotation and sequence similarity (COMPASS). The COMPASS approach differs from other approaches for comparative mapping, such as comparative anchor tagged sequences (CATS: Lyons et al. 1997), in that COMPASS relies on generating homologous DNA sequence information (e.g., ESTs), followed by similarity search to identify putative orthologs, and then predicting the chromosome location of the sequences on the basis of existing comparative maps. By contrast, CATs utilizes available sequence data. Using the combined approach of COMPASS and RH mapping of ESTs on bovine chromosome 5, we have shown COMPASS to be a useful predictive approach for gene mapping (Ozawa et al. 2000).
Herein, we used ESTs derived from cattle ovary and spleen cDNA libraries and sequences of cattle genes in public domain databases to create a whole-genome RH map. A COMPASS software tool facilitated the map-building process. The whole-genome cattle RH map was anchored with microsatellite markers from the existing cattle linkage maps. Construction of RH maps for all bovine autosomes and the X chromosome allowed us to create detailed cattle–human comparative maps. Our goals were to reveal the spectrum of chromosome rearrangements as compared with the human genome, and to create practical resources for the livestock genomics community. The whole-genome cattle–human comparative map will serve as a cornerstone for efforts to identify genes of agricultural importance and as an essential resource for understanding genome evolution in vertebrates.
RESULTS
A Cattle RH Map
A total of 1314 markers were scored on the 5000 rad cattle–hamster RH panel. Of these, 1087 markers were placed in 61 linkage groups assigned to all 29 autosomes and BTAX, with 468 markers ordered in 1:1000 framework maps (see enclosed poster insert). The remaining markers were either unlinked (n = 113) or linked with ambiguous placement (n = 114; see Methods for description). Failure of these markers to be included in the map may be the result of genotyping errors, amplification of paralogous sequences (resulting in much higher than expected retention frequencies), and mapping outside terminal framework markers or within gaps. Markers that were unlinked and those linked with ambiguous placement are not shown on the map; details concerning these markers can be found at http://cagst.animal.uiuc.edu. Among the 1087 mapped markers, 768 are genes (supplement Table 1, available online at http://www.genome.org) and 319 are microsatellites (supplement Table 2, available online at http://www.genome.org) that were used as anchor markers to orient the linkage groups properly. Among the 768 mapped genes, 358 are cattle ESTs, 156 from ovary and 202 derived from spleen cDNA libraries. The remaining gene sequences were extracted from GenBank and included 387 cattle mRNA sequences, 11 goat ESTs, and 12 human mRNA sequences.
Thirteen chromosomes were formed by one contiguous linkage group each. The most fragmented chromosomes, BTA9 and BTA14, each had five linkage groups containing 24 and 50 markers, respectively (Table 1). The average chromosome length is 311 cR5000, ranging from 637 cR for BTA19 to 125 cR for BTA29 (Table 1). Total length of the RH map is 9330 cR5000, with an approximate genome-wide ratio of 3 cR:1 cm. This ratio is probably an underestimate because of 31 gaps in the map that could not be closed, even with specifically targeted microsatellite markers and ESTs. Genome coverage is ∼ 92% (No. unlinked/No. linked = 113/1201), as defined by the probability that a random marker typed on the RH panel will be linked to another marker in a known linkage group (Hukriede et al 1999).
Table 1.
Summary Statistics of RH Map by Chromosome
| Chr | Markers | Retention | Length | Linkage Groups | |||
|---|---|---|---|---|---|---|---|
| Type I | Type II | Total | Framework | ||||
| 1 | 31 | 16 | 47 | 22 | 0.290 | 340 | 2 |
| 2 | 29 | 10 | 39 | 23 | 0.189 | 373 | 3 |
| 3 | 37 | 14 | 51 | 9 | 0.185 | 290 | 1 |
| 4 | 22 | 12 | 34 | 15 | 0.205 | 362 | 1 |
| 5 | 51 | 34 | 85 | 27 | 0.183 | 439 | 1 |
| 6 | 21 | 13 | 34 | 13 | 0.160 | 356 | 1 |
| 7 | 39 | 14 | 53 | 17 | 0.157 | 457 | 3 |
| 8 | 23 | 14 | 37 | 19 | 0.198 | 409 | 2 |
| 9 | 13 | 11 | 24 | 7 | 0.133 | 189 | 5 |
| 10 | 37 | 8 | 45 | 17 | 0.227 | 332 | 1 |
| 11 | 37 | 9 | 46 | 19 | 0.263 | 324 | 1 |
| 12 | 14 | 9 | 23 | 9 | 0.182 | 258 | 4 |
| 13 | 29 | 8 | 37 | 21 | 0.190 | 352 | 1 |
| 14 | 29 | 21 | 50 | 22 | 0.252 | 283 | 5 |
| 15 | 26 | 8 | 34 | 12 | 0.176 | 352 | 3 |
| 16 | 29 | 9 | 38 | 24 | 0.283 | 387 | 2 |
| 17 | 23 | 10 | 33 | 17 | 0.217 | 394 | 3 |
| 18 | 33 | 6 | 39 | 17 | 0.221 | 310 | 2 |
| 19 | 49 | 6 | 55 | 30 | 0.453 | 637 | 1 |
| 20 | 13 | 8 | 21 | 5 | 0.209 | 206 | 1 |
| 21 | 20 | 9 | 29 | 14 | 0.181 | 256 | 2 |
| 22 | 19 | 5 | 24 | 13 | 0.256 | 220 | 2 |
| 23 | 29 | 13 | 42 | 22 | 0.270 | 272 | 1 |
| 24 | 17 | 5 | 22 | 10 | 0.223 | 188 | 2 |
| 25 | 22 | 7 | 29 | 13 | 0.244 | 210 | 3 |
| 26 | 21 | 5 | 26 | 11 | 0.233 | 294 | 1 |
| 27 | 10 | 6 | 16 | 7 | 0.144 | 138 | 1 |
| 28 | 13 | 4 | 17 | 11 | 0.343 | 168 | 1 |
| 29 | 12 | 5 | 17 | 8 | 0.308 | 125 | 2 |
| X | 20 | 20 | 40 | 14 | 0.162 | 409 | 3 |
| Total | 768 | 319 | 1087 | 468 | 0.224 | 9330 | 61 |
The average retention frequency (RF) of the mapped markers is 22.4% ranging from 45.3% for BTA19, which contains the selectable marker thymidine kinase, to 13.3% for BTA9 (Table 1). The relatively low RF for markers on BTAX (16.2%) was expected, because X chromosome markers are present in the RH cell lines in the hemizygous state (the cattle parental line was created from a male). Large variation of RF among individual chromosomes resulted in widely different resolution for the different chromosomes.
A Whole-Genome Cattle–Human Comparative Map
A whole genome comparative map was created using a parallel RH mapping approach (Yang and Womack 1998). The construction of comparative chromosome maps was dependent largely on the existing RH map information for humans in the public domain databases. Among the 768 genes on the cattle RH map, 687 (89.5%) had putative human orthologs identified by similarity searches against the UniGene database; the remaining 81 (10.5%) were ESTs or database sequences that had no significant human hits in UniGene. Among the 687 mapped genes with UniGene hits, 548 had human GB4 RH mapping information, 22 were mapped exclusively on the G3 panel, 68 had human cytogenetic assignments only, and 49 had no human mapping information.
Comparative maps of each chromosome were constructed by aligning the cattle RH maps with human chromosome segments containing the same putative orthologs. The human RH map coordinates permitted the identification of conserved chromosome segments in the two genomes (see map enclosed with this issue). Local differences in gene order within conserved segments were tolerated because such differences could be explained by mapping errors in either species or small rearrangements below the level of resolution of either the cattle or human mapping panels. Some of these local differences in order could represent new segments or rearrangements, but we chose to represent the number of rearrangements in the most conservative fashion. Despite the limitations inherent in RH map resolution, the alignments allowed us to determine the boundaries and orientation of conserved chromosome segments. A total of 105 conserved chromosome segments containing two or more genes were defined. Two new conserved segments each containing two genes with GB4 data were identified on BTA20 (HSA5 position 632 cR) and BTA11 (HSA11 position 271 cR). Two additional conserved segments were defined by at least two loci having GB4, cytogenetic, or G3 data (BTA25, HSA7 segment at position 50 cR; BTA21, HSA15 segment at position 145 cR). In addition, 28 conserved segments were defined putatively by single genes or internal rearrangements that could not be identified unambiguously due to low map resolution within specific regions. There are also 15 single genes on the map that are located within conserved segments on chromosomes that contradict COMPASS predictions (see below). These genes might represent unidentified paralogs, i.e., where the paralog maps in the “correct” location predicted from the comparative maps. Although not yet confirmed with ≥ 2 genes there are potentially an additional 43 conserved segments in the comparative map. On the basis of currently available data for flanking genes, human centromeres were assigned to their location within conserved segments (see enclosed map). All cattle chromosomes with the possible exception of BTA9 and BTA23 have undergone centromere repositioning relative to human chromosomes.
Four cattle chromosomes show complete conservation of synteny with their human homologs: BTA12 and HSA13, BTA19 and HSA17, BTA24 and HSA18, and BTAX and HSAX. However, for all of these chromosomes multiple internal rearrangements are observed. BTA3 is the only cattle chromosome for which there was no statistical support for the occurrence of internal rearrangements when compared with the homologous segment on HSA1. By examination of conserved segments, 41 putative translocations leading to the present organization of the cattle and human chromosomes can be identified (see enclosed map). Translocations were counted by summing the number of human syntenies that were found to be homologous with cattle chromosomes (e.g., three human chromosome syntenies have homologous regions on BTA17: HSA4, HSA12, and HSA22), excluding those for which homologs appear to be completely conserved (e.g., BTA19 and HSA17). Fifteen cattle chromosomes appear to be comprised of genes found on only one human chromosome.
Novel Sequences
The 81 ESTs and database sequences that had no significant hits against human UniGene were examined more fully by similarity searches against other DNA databases. Among these 81 sequences, 33 have hits in nonredundant GenBank or dbEST. The remaining 48 sequences may represent novel genes (not yet discovered in another species), rapidly diverging orthologs, or genomic DNA contaminants in the library (all 3′ ESTs had poly(A) tails). These genes are listed as ESTs with no UniGene hit for sequence similarity (see supplement Table 1, available online at www.genome.org).
Chromosome Distribution of Cattle Genes
The chromosome distribution of 465 cattle genes was examined. These genes represent a random set derived from cattle ovary ESTs and GenBank sequences. Spleen ESTs were not used because they were chosen using COMPASS specifically to fill gaps in the comparative map (see below). The observed numbers of genes per chromosome differed from that expected based on chromosome physical length. The test for heterogeneity among the deviations of the observed from the expected values was χ2 = 93.2, P = 1.16 × 10−8, df = 29. The Bonferroni-corrected probabilities for each chromosome revealed that BTA18 and BTA19 have more genes than expected (P < 0.05).
Accuracy of COMPASS Predictions
The large number of human and cattle genes mapped in parallel permitted an estimate of the accuracy of the COMPASS predictive tool on a set of 465 randomly selected genes. Only random genes chosen from cattle ovary and GenBank sequences were utilized, and predictions were made on the basis of preexisting comparative mapping information drawn largely from synteny mapping data (Bovine Genome Database, http://bos.cvm.tamu.edu/bovgbase.html). The spleen ESTs were not used for estimating the accuracy of COMPASS because they were selected from a larger set to fill in gaps in the comparative map on the basis of COMPASS predictions. Among the 465 randomly chosen genes, 333 (71.6%) had GB4 data that could be used for COMPASS prediction of chromosome assignments. Of these, COMPASS predicted a single correct chromosome assignment for 254 genes; 60 genes had two possible chromosome assignments, of which one of the two predictions was correct. The COMPASS prediction of two cattle chromosome assignments is due to “gaps” in the comparative chromosome maps. For all but two of these dual assignments, RH mapping subsequently confirmed one of the two predicted locations, thereby refining the location of evolutionary breakpoints by shrinking the gaps in the comparative map. Among the 19 inconsistent predictions, six had human cytogenetic assignments that produced COMPASS predictions consistent with actual cattle RH map location. These inconsistencies are thus most likely attributable to GB4 mapping errors. Of the remaining 13 inconsistent predictions, 11 were unconfirmed singletons and two were part of new conserved segments (see enclosed map). The 11 unconfirmed singletons most likely represent undiscovered human paralogs and mapping errors. Thus, the overall accuracy of COMPASS, including the dual assignments, is 94.7% (314/333). In addition to the predictive power of COMPASS for assigning ESTs (or any DNA sequence) to the cattle gene map, COMPASS was also useful for predicting map locations of human genes when the cattle gene was mapped but the human gene was not. For example, the human ortholog of UBE2D3 should map to HSA4 on the basis of its map position on BTA16. These genes, 48 in total, are indicated on the map with underlining (see enclosed map).
COMPASS was also used to target genes for mapping from the spleen cDNA library. A total of 138 spleen ESTs with UniGene hits were selected for mapping from among 867 unique genes identified from this library (data not shown). Among these, 27 were targeted to fill gaps (had multiple chromosome predictions); all 27 mapped to one of the predicted chromosomes. The remaining 110 spleen ESTs that were selected to fill in sparse regions on the map had chromosome location predicted with 96.5% accuracy.
DISCUSSION
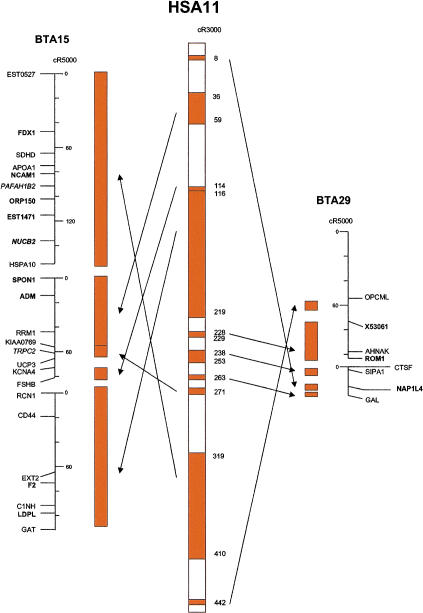
RH mapping was used in conjunction with EST sequencing, public domain DNA databases, and bioinformatics tools to create a first generation-ordered cattle–human whole-genome comparative map containing 638 common reference loci. The RH map, including microsatellite markers, provides coverage of ∼ 90% of the cattle genome. The cattle–human comparative map, although quite extensive by comparison with existing information, has many uncharacterized gaps that remain to be filled. For example, we did not present information on the Y chromosome because the number of genes was insufficient for building a good RH map. As another example, BTA15 and BTA29 are comprised of genes found on HSA11, yet only 41% of the map length of HSA11 can be accounted for on these two bovine autosomes (Fig. 1). On the basis of GB4 cR of each human chromosome accounted for on the cattle genome, we estimate a minimum of 50% comparative genome-wide coverage on our map (data not shown). If we assume 5% additional coverage because of centromere region expansions in the human RH map (all the cattle chromosomes are acrocentric, except BTAX), and 5% additional coverage from cytogenetically assigned markers (with no GB4 mapping data), we estimate ∼ 60% of the human genome to be accounted for on the comparative map. Using COMPASS for targeted mapping should lead rapidly to a human–cattle comparative map with complete genome coverage.
Figure 1.
Cattle-on-human comparative map of HSA11. Clear space between segments represents regions of the human genome for which no cattle orthologs have been mapped. Maps were simplified to show only comparatively mapped genes. Coverage of HSA11 on BTA15 and BTA29 is 41% (see text).
Many factors can affect the resolution of RH maps, including experimental factors and choice of mapping software used to perform the analysis. Maps produced with different software result in similar gene orders and numbers of framework markers but show large variation in cR distance (Hukriede et al. 1999). This directly affects the estimate of map resolution, as is apparent when comparing chromosome maps created with RHMAP (Yang and Womack 1998; Gu et al. 1999; Rexroad et al. 1999) or RHMAPPER (Band et al. 1998; Ozawa et al. 2000). The whole genome map created with RHMAPPER generated an average value of 3 cR/cm, yielding a ratio of 330 Kb/cR5000 assuming ∼ 1Mb/cm. Although it is difficult to compare RH panels between different species, the average retention rate and resolution of the cattle 5000 rad panel are similar to those of the zebrafish 5000 rad LN54 RH panel (Hukriede et al. 1999). The RH panels for most other species have higher resolutions: 70 Kb/cR7000 for pig (Hawken et al. 1999), 100 Kb/cR3000 for mouse (Van Etten et al. 1999), 166 Kb/cR5000 for dog (Priat et al. 1998), and 106 Kb/cR3000 for rat (Watanabe et al. 1999). With the creation of the first whole genome cattle RH map it is now possible to target new markers and/or candidate genes for fine resolution mapping with a recently developed 12,000 rad panel (Rexroad et al. 2000).
The cattle RH map consists of 61 linkage groups with 31 gaps. Although we estimate ∼ 90% coverage, as discussed above, large regions of many human chromosomes are not yet represented on the cattle RH map (coverage ranges from 18% for HSA18 to 80% for HSA1). In these uncharted chromosome segments, expressed genes in the homologous cattle regions appear to be underrepresented, at least in the cDNA libraries from which we are sequencing. An alternate explanation for the large gaps could be that certain regions of the cattle genome are not retained in the hybrid lines, or that there is a high frequency of radiation-induced breakage in certain areas of the cattle genome. Wherever possible, microsatellite markers were added to create a more complete map. In general, we found that an insufficient number of markers are available for complete coverage of these regions. For example, an initial gap was identified between markers TGLA53 and C4BPB on BTA16. Three additional microsatellites were typed within this gap: BM1311, BM121, and BMS1348. All were added to the distal linkage group of BTA16; however despite being 1.3 cm apart on the genetic linkage map (Kappes et al. 1997), on the RH map, linkage could not be detected between BM1348 and C4BPB, apparently because of the high frequency of breakage between these loci. BTA14 is another example where a paucity of known markers affects mapping efficiency. The RH map of BTA14 contains five linkage groups even though recombination data shows tight linkage between markers from adjacent groups. It may be necessary to use other physical mapping methods in addition to COMPASS to close these gaps in the RH maps.
In general, the cattle–human comparative RH map correlates well to chromosome paints (Hayes 1995; Solinas-Toldo et al. 1995; Chowdhary et al. 1996). The enhanced detail of the RH comparative map enables clarification of some discrepancies among the maps created by synteny mapping, in situ hybridization, linkage analysis, and chromosome painting. For example, the homology of the telomeric end of BTA1 with a segment of HSA21 on our map confirms the chromosome paint analysis by Hayes (1995). In addition, the presence of conserved segments from three different chromosomes on BTA17 was confirmed, as was the conserved segment of HSA4 on BTA27. In contrast with chromosome painting, no evidence of homology between BTA10 and HSA5 was found. Similarly, segments of HSA20 (proximal to the centromere) and HSA4 were not confirmed on BTA13 and BTA24, respectively. It is noteworthy that many singletons on the comparative map detected by synteny mapping were not confirmed by RH mapping. Interestingly, the gene BS69 on BTA13 shows similarity with two UniGene clusters, one on HSA10, the other on HSA20, both of which have conserved segments on BTA13. This may be evidence for an ancestral duplication followed by a translocation event. An example of identification of a new conserved segment detected on the RH map but not found on a chromosome paint is the HSA1 segment homologous to the centromeric portion of BTA28. A different example of change in map resolution is shown on the RH maps of BTA10 and BTA21 that show previously undescribed rearrangements between homologous segments of regions on HSA14 and HSA15.
In all, we observed 105 conserved segments with two or more genes and a potential for 149 total segments between the human and cattle genomes. Schibler and coworkers (1998) observed 107 (62 with > 2 mapped genes) conserved segments between goat and human by fluorescence in situ hybridization (FISH) mapping of goat BACs containing human orthologs. Both gene order and the number of breakpoints confirm the similarities between the two ruminant genomes. The fact that only four new conserved segments between the cattle and human genomes have been revealed in our work suggests that the cattle–human comparative map includes a high percentage of the total number of conserved segments. However as the number of known syntenies increases, segment size tends to decrease for the segments not yet revealed (Nadeau and Sankoff 1998). Thus we may expect to find many new segments by targeting the remaining 30%–40% of the comparative map.
Examination of human-on-cattle centromere positions (see enclosed map) shows that human centromere sites are associated with translocations and internal rearrangements. In several cases, comparative map distances are distorted around the position of human centromeres, where the cattle RH map distances are much smaller. For example on BTA11, the 117 cR conserved segment on HSA2 that contains the human centromere shows a very large distance on the human RH map relative to the tight linkage on the cattle RH map. This indicates either sensitivity to radiation around the centromere or loss/gain of genetic material when the centromere is repositioned. The only human chromosome that appears to show conservation of relative centromere position is HSA6 (see enclosed map). BTA23 and BTA9 could have arisen by centric fission of an ancestral chromosome homologous to HSA6. Alternatively, HSA6 may have arisen from a centric fusion of ancestral chromosomes homologous to BTA23 and BTA9.
Conservation of synteny for the X chromosome has been shown for several mammalian species (Ohno 1973; Murphy et al. 1999; Watanabe et al. 1999), with the exception of certain mouse orthologs of genes within the human pseudoautosomal region (PAR) (Carver and Stubbs 1997). The RH map of BTAX includes 20 genes, 16 with mapped human orthologs, thus providing valuable additional data for comparative mapping. The comparative map of BTAX confirms the conservation of synteny with HSAX; however, we note an inversion of the cattle p-arm relative to the human chromosome. A combination of linkage and FISH data (Solinas-Toldo et al. 1995) placed the cattle centromere between markers XBM111 and XBM361. The RH map shows that the cattle q-arm has conserved order with HSAXpter-Xq21. These data imply a shift in position of the centromere relative to HSAX, without any evidence of a causative rearrangement. Centromere repositioning independent of surrounding markers has also been documented in primates (Montefalcone et al. 1999). The placement of two PAR genes, AMELX and ANT3, at the distal end of BTAX is direct confirmation that the PAR region of cattle resides on the distal q-arm (Ponce de Leon et al. 1996). Comparison of the human, cat (Murphy et al. 1999) and cattle X chromosomes shows almost complete conservation of order with the exception of the inverted p-arm of cattle. However, chromosome-banding studies by Robinson et al. (1998) suggested many rearrangements of X chromosome segments within the bovidae. These observations imply a much larger variation of X chromosome gene order within the bovidae than among more divergent mammalian orders.
The number of cattle genes on each chromosome was found to be nonrandomly distributed. BTA18 and BTA19 had significantly higher numbers of mapped genes from the expected values at a significance level of P < 0.05. The human homologs of these cattle chromosomes, HSA19 and HSA17, respectively, were also found to have a higher gene density than expected (Deloukas et al. 1998). Although deviations from expected values for other cattle chromosomes were not significant, the inability to detect such differences might have been due to the sample size (n = 465). The conservation of differences in gene density on cattle and human homologs has not been reported previously and may represent conserved heterochromatic regions and/or expression patterns necessary for chromosome function and tissue-specific gene regulation.
The relatively high frequency of novel ESTs identified in the ovary and spleen libraries raises compelling questions as to their origin and function. The majority of the 48 novels appear to represent the 3′ end of coding sequences because they all had poly(A) tracts at their 3′ ends and many had 5′ open reading frames (ORFs) (data not shown). These sequences are of enormous functional interest as they might represent rapidly diverging orthologs that impart species-specific functions. A classic example of such genes is the novel multigene family encoding the pregnancy-associated glycoproteins, aspartyl proteinases that are expressed in the outer epithelial layer of the placenta of ruminants (Xie et al. 1997). With the map information we have obtained it will be of great interest to explore human genome sequence at the homologous chromosome positions to see if the ESTs represent previously undetected orthologs or divergent orthologs that are not discerned by DNA sequence similarity. The functional characterization of these genes might contribute to a better understanding of the genetic basis of phenotypic differences among mammals.
The COMPASS tool for in silico mapping proved to be exceptionally accurate on the set of 465 randomly chosen sequences, and very useful for closing gaps in the comparative map. The comparative mapping table used to make the predictions was created almost exclusively from data derived from synteny mapping of > 500 genes (Bovine Genome Database; http://bos.cvm.tamu.edu/bovgbase.html/). The accuracy of chromosome predictions, and the relatively small number of new conserved segments defined, clearly demonstrates the importance and fidelity of this base knowledge to the COMPASS process. Our findings suggest that COMPASS will also be useful for in silico mapping in a number of agriculturally important species that already have synteny maps, such as the pig, sheep, and horse. Moreover, the approach should be generally useful for any pairwise comparison of species for which there is a reference genome available. An important advantage of the COMPASS approach is that from among thousands of ESTs rapidly entering the public domain, markers useful for sealing gaps can be identified, thus greatly reducing the overall cost of generating comprehensive RH and comparative maps. As new comparative mapping data gets incorporated into the relational genome tables, including updates of UniGene, positional information from the human genome sequence, and map locations within comparative bins, COMPASS should improve in both accuracy and precision. When comparative coverage of the human genome is complete after the next phase of COMPASS-guided RH mapping, the in silico mapping approach will greatly facilitate the identification of candidate genes within conserved segments.
The new era in comparative mapping made possible by RH technology, high-throughput DNA sequencing, and bioinformatics, will reveal the evolutionary history of chromosomes. This history should shed light on the karyology of speciation events, and should provide a new context for understanding how organismal form and function relate to positional information of genes on chromosomes. The predictive power of mammalian comparative genomics will be critical for elucidating the fine genetic differences that result in phenotypic changes among closely related species. In particular, the functional characterization of the novel genes identified in this study, and those harvested from newly obtained EST and DNA sequence information, may provide the raw material for understanding adaptive selection in higher vertebrates.
METHODS
Library Construction
Directionally cloned cDNA libraries were created from ovary and spleen tissue collected from a healthy, adult Aberdeen–Angus cow. Tissue samples were ground after freezing in liquid nitrogen, and total RNA was extracted using TRIZOL (GIBCO BRL) reagent followed by chloroform and acid-phenol (pH 4.5; Ambion Inc.) extractions to remove traces of DNA. Poly(A) RNA was isolated using the Oligotex (Qiagen) affinity chromatography reagent according to the manufacturer's instructions. Libraries were constructed in the pBluescript SK(±) phagemid vector using the ZAP-cDNA Synthesis Kit and ZAP-cDNA Gigapack III Gold Cloning Kit (Stratagene) according to the manufacturer's instructions.
Template Isolation and Characterization
Approximately 250 colony-forming units of excised phagemids were combined with 1.6 * 108 SOLR cells (Stratagene), plated, and cultured according to the ZAP-cDNA Gigapack III Gold Cloning Kit protocols in 2 × LB broth. Before harvesting, glycerol stocks were made and stored at −80°C. The cDNA templates were isolated using the QIAprep 96 Turbo Miniprep Kit and the QIAvac 96 (Qiagen) following the manufacturer's instructions. DNA quantity and quality were analyzed by electrophoresis of 5 μl (100–500 ng) of each sample in 1.0% agarose gels, 1 × TAE buffer, and stained with ethidium bromide. Inserts were excised by digestion with XbaI and XhoI and sized in 1% agarose gels. The average insert size was 1.4 kb and 1.8 kb for the ovary and spleen libraries, respectively.
DNA Sequencing, Analysis, and Annotation
Plasmid inserts were sequenced with Dye Terminator or Big Dye Cycle Sequencing Kits (Perkin-Elmer) on ABI 373A or ABI 377 automated DNA sequencers (Applied Biosystems). The T3 primer (5′-AATTAACCCTCACTAAAGGG-3′) was used for 5′ end sequencing and a modified T7 primer (5′-TACGACTCACTATAGGGCGAAT-3′) for 3′-end sequencing. Ovary ESTs were sequenced from both 5′ and 3′ ends, whereas spleen ESTs were sequenced from 3′ ends only. Gel files were tracked manually, and raw sequence data were extracted using the ABI data collection software. Sequence chromatograms were processed manually using SeqEd (Applied Biosystems). Sequences were trimmed of vector and parsed for similarity against dbEST (Boguski et al. 1993) and nonredundant (NR) sequences in GenBank (Benson et al. 1998) using BLASTN (Altschul et al. 1997). Clones containing mitochondrial RNA, ribosomal RNA, or repetitive elements were removed from the data set.
GenBank Sequences
More than 2500 cattle mRNA sequences were collected from GenBank. We distilled these entries to 387 unique cattle genes (mapped sequences listed in supplement Table 2, available online at www.genome.org). Partial mRNA sequences were excluded from further analysis, as were sequences redundant to any previously mapped cattle ovary EST (this study). Cattle sequences with significant similarity to multiple closely related human paralogs were also excluded from further analysis due to the expected difficulty in identifying the correct ortholog.
EST Distribution Analysis
The randomness of chromosomal distribution of cattle ESTs was tested using a χ2 goodness-of-fit test. The significance threshold was set at 0.05 using the Bonferroni correction for the number of comparisons (Bortoluzzi et al. 1998). The observed number of genes expressed on each chromosome was calculated from RH mapping assignments of ovary and database genes. Cytogenetic measurements (Chiriaeva et al. 1989) were used to determine the expected number of genes per chromosome.
Primer Design and RH Typing
Oligonucleotide primers for EST and GenBank sequences were designed using the program Primer Designer 3.0 (Scientific & Educational Software). Primers were generally designed within the 3′ UTR to avoid amplification of intronic sequences. To obtain cattle-specific PCR products using RH DNA template, regions of low homology between bovine and rodent species were targeted for primer design. Primer sequences and annealing temperatures are listed at http://www.cagst.animal.uiuc.edu/. Primers for 319 microsatellite markers were obtained from published sources (supplement Table 2, available online at www.genome.org). All primer sets were optimized using cattle genomic DNA and a 1:3 mix of cattle genomic DNA and A23 hamster cell-line DNA. A23 DNA and water were included as controls. Annealing temperature was varied to obtain a strong, cattle-specific product. All primer pairs were typed in duplicate against a cattle 5000 rad RH panel (Womack et al. 1997) in 15 μl reactions as described by Band and coworkers (1998). PCR products were electrophoresed in 1.5% agarose gels. Markers were scored as present (1), absent (0), or ambiguous (2).
Mapping Strategy
We employed a two-stage, integrated mapping strategy to develop a whole-genome RH map and a whole-genome human–cattle comparative map. Our primary goal was to build a comparative map so we emphasized heavily the mapping of genes over anonymous markers to achieve maximum cost efficiency. To accomplish this, we first typed ovary ESTs and database genes (∼ 500) on the RH panel and then generated draft maps of each chromosome using existing comparative mapping information as a guide. The second stage involved adding microsatellite markers and spleen ESTs to the chromosome maps. Microsatellites were selected to target regions of the genome with few mapped genes, to define chromosome ends, and to facilitate comparison with published linkage data. The ESTs were selected from the spleen library using the COMPASS tool (see below), which enabled selection of markers to fill gaps in the comparative map and to increase resolution of the comparative map by targeting intervals with low statistical support for gene order.
Map Construction
Two-point linkage was computed using the mapping program RHMAPPER (Slonim et al. 1997). To avoid spurious linkage, a threshold LOD score of 12 was used to assign markers to established linkage groups for genes with no COMPASS predictions, or LOD 8 for genes with predicted assignments confirmed by the RH data. Initial framework maps were created using the RHMAXLIK program of RHMAP 3.0 (Boehnke 1992) with a LOD threshold of three. This order was further expanded using the grow frameworks option of RHMAPPER, and finally, a placement map was created incorporating all remaining markers in the most likely framework intervals. Markers that were assigned to a chromosome by two-point linkage but not linked to at least one framework marker with LOD > 5 were not placed on the map (linked but ambiguous placement), according to the default parameters of RHMAPPER. Two-point linkage was used to confirm the position of markers mapping outside terminal framework markers. Those without significant linkage to terminal framework markers were removed. Microsatellite markers incorporated in the maps were used to orient multiple linkage groups within chromosomes according to maps published previously. Because of the low retention frequency for BTA3 and BTA6, initial frameworks for these chromosomes were constructed by choosing consensus microsatellites with orders conserved between two or more of the cattle linkage maps. All computations were carried out on a SUN SPARC 20 workstation. Data files were converted from RHMAPPER to RHMAP format with the RHScoresFormat Applet available at http://corba.ebi.ac.uk/RHdb/Clients.
COMPASS
The COMPASS strategy (Ma et al. 1998; Ozawa et al. 2000) permits the prediction of map location on the basis of sequence similarity of orthologous genes if comparative map information is available between two species. A PERL script was written to create and update COMPASS predictions for large sets of cattle sequences generated. The program executes a similarity search for FASTA formatted EST or mRNA sequences against the human UniGene database using the BLAST algorithm (Altschul et al. 1997). A threshold expected value of e−5 is accepted as a significant hit. The UniGene cluster containing the sequence with the best hit is identified and the name, gene symbol, accession number of the cluster, and GenBank accession number for the specific sequence recognized is stored in memory. The first five GB4 and first three G3 map locations (GeneMap '98; http://www.ncbi.nlm.nih.gov/genemap/) are used in conjunction with the cattle-on-human comparative maps (Bovine Genome Database; http://bos.cvm.tamu.edu/bovgbase.html/) to predict cattle chromosome assignment. When comparative mapping data cannot be used to unambiguously predict an EST or gene sequence to one chromosome, (i.e., the sequence fell in a gap in the comparative map) the EST is assigned tentatively to the two most likely chromosomes. An output containing all of the above parameters is then imported into a database spreadsheet. When using GB4 map data, several rules were applied to deal with multiple GeneMap '98 chromosome assignments associated with a single UniGene cluster. When GB4 chromosomal assignments for a UniGene cluster were in conflict, separate COMPASS predictions were made for each human chromosome reported. For multiple assignments on the same chromosome the assignment resulting in the smallest conserved segment was chosen.
Acknowledgments
This work was made possible in part by a grant to H.A.L. and J.E.W. from the United States Department of Agriculture, National Research Initiative, Project No. 98-35205-6644, and a grant from the Japanese Ministry of Agriculture Fisheries and Forestry. M.R.B. is a Binational Agricultural Research and Development Fund (BARD) Postdoctoral Fellow (BARD Fellowship FI-263-97)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL h-lewin@ux1.cso.uiuc.edu; FAX (217) 244-5617.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.145900.
REFERENCES
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band M, Larson JH, Womack JE, Lewin HA. A radiation hybrid map of BTA23: Identification of a chromosomal rearrangement leading to separation of the cattle MHC class II subregions. Genomics. 1998;53:269–275. doi: 10.1006/geno.1998.5507. [DOI] [PubMed] [Google Scholar]
- Barendse W, Vaiman D, Kemp SJ, Sugimoto Y, Armitage SM, Williams JL, Sun HS, Eggen A, Agaba M, Aleyasin SA, et al. A medium-density genetic linkage map of the bovine genome. 1997. Mamm Genome. 1997;8:21–28. doi: 10.1007/s003359900340. [DOI] [PubMed] [Google Scholar]
- Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke M. Multipoint analysis for radiation hybrid mapping. Ann Med. 1992;24:383–386. doi: 10.3109/07853899209147842. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for “expressed sequence tags”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Bortoluzzi S, Rampoldi L, Simionati B, Zimbello R, Barbon A, d'Alessi F, Tiso N, Pallavicini A, Toppo S, Cannata N, et al. A comprehensive, high-resolution genomic transcript map of human skeletal muscle. Genome Res. 1998;8:817–825. doi: 10.1101/gr.8.8.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzat, J.L., McNeil, L.K., Robertson, H.M., Solter, L.G., Nixon, J., Beever, J.E., Gaskins, H.R., Olsen G., Subramaniam, S., Sogin M.K., and Lewin, H.A. 2000. Phylogenomic analysis of the alpha proteasome gene family from early diverging eukaryotes. J. Mol. Evol. (in press). [DOI] [PubMed]
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: Genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1127. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Chiriaeva OG, Amosova AV, Efimov AM, Smirnov AF, Kaminir LB, Lakovlev AF, Zelenin AV. Cytogenetic mapping of cattle (Bos taurus). Quantitative analysis of the RBA-map of prometaphase chromosomes. Genetika. 1989;25:1436–1448. [PubMed] [Google Scholar]
- Chowdhary BP, Frønicke L, Gustavsson I, Scherthan H. Comparative analysis of the cattle and human genomes: Detection of ZOO-FISH and gene mapping-based chromosomal homologies. Mamm Genome. 1996;7:297–302. doi: 10.1007/s003359900086. [DOI] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: A somatic cell genetic method for constructing high-resolution maps of mammaliam chromosmes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Eggen A, Fries R. An integrated cytogenetic and meiotic map of the bovine genome. Anim Genet. 1995;4:215–236. doi: 10.1111/j.1365-2052.1995.tb03249.x. [DOI] [PubMed] [Google Scholar]
- Eisen JA. Phylogenomics: Improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Feldman HW. Linkage of albino allelomorphs in rats and mice. Genetics. 1924;9:487–482. doi: 10.1093/genetics/9.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Womack JE, Kirkpatrick BW. A radiation hybrid map of bovine chromosome 7 and comparative mapping with human chromosome 19 p arm. Mamm Genome. 1999;11:1112–1114. doi: 10.1007/s003359901173. [DOI] [PubMed] [Google Scholar]
- Hawken RJ, Murtaugh J, Flickinger GH, Yerle M, Robic A, Milan D, Gellin J, Beattie CW, Schook LB, Alexander LJ. A first-generation porcine whole-genome radiation hybrid map. Mamm Genome. 1999;10:824–830. doi: 10.1007/s003359901097. [DOI] [PubMed] [Google Scholar]
- Hayes H. Chromosome painting with human chromosome-specific DNA libraries reveals the extent and distribution of conserved segments in bovine chromosomes. Cytogenet Cell Genet. 1995;71:168–174. doi: 10.1159/000134100. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes SM, Keele JW, Stone RT, McGraw RA, Sonstegard TS, Smith TPL, Lopez-Corrales NL, Beattie CW. A second-generation linkage map of the bovine genome. Genome Res. 1997;7:235–249. doi: 10.1101/gr.7.3.235. [DOI] [PubMed] [Google Scholar]
- Lyons LA, Laughlin TF, Copeland NG, Jenkins NA, Womack JE, O'Brien SJ. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat Genet. 1997;15:47–56. doi: 10.1038/ng0197-47. [DOI] [PubMed] [Google Scholar]
- Ma RZ, Beever JE, Da Y, Green CA, Russ I, Park C, Heyen DW, Everts RE, Fisher SR, Overton KM, et al. A male linkage map of the cattle (Bos taurus) genome. J Hered. 1996;87:261–271. doi: 10.1093/oxfordjournals.jhered.a022999. [DOI] [PubMed] [Google Scholar]
- Ma RZ, van Eijk MJ, Beever JE, Guérin G, Mummery CL, Lewin HA. Comparative analysis of 82 expressed sequence tags from a cattle ovary cDNA library. Mamm Genome. 1998;9:545–549. doi: 10.1007/s003359900816. [DOI] [PubMed] [Google Scholar]
- Montefalcone G, Tempesta S, Rocchi M, Archidiacono N. Centromere repositioning. Genome Res. 1999;9:1184–1188. doi: 10.1101/gr.9.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Sun S, Chen ZQ, Pecon-Slattery J, O'Brien SJ. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 1999;9:1223–1230. doi: 10.1101/gr.9.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. The lengths of undiscovered conserved segments in comparative maps. Mamm Genome. 1998;9:491–495. doi: 10.1007/s003359900806. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, Stanyon R, Copeland NG, Jenkins NA, Womack JE, Graves JAM. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- Ohno S. Ancient linkage groups and frozen accidents. Nature. 1973;244:259–262. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- Ozawa A, Band MR, Larson JH, Donovan J, Green CA, Womack JE, Lewin HA. Comparative organization of cattle chromosome 5 revealed by COMPASS and radiation hybrid mapping. Proc Natl Acad Sci. 2000;97:4150–4155. doi: 10.1073/pnas.050007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce de Leon FA, Ambady S, Hawkins GA, Kappes SM, Bishop MD, Robl JM, Beattie CW. Development of a bovine X chromosome linkage group and painting probes to assess cattle, sheep, and goat X chromosome segment homologies. Proc Natl Acad Sci. 1996;93:3450–3454. doi: 10.1073/pnas.93.8.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priat C, Hitte C, Vignaux F, Renier C, Jiang Z, Jouquand S, Cheron A, Andre C, Galibert F. A whole-genome radiation hybrid map of the dog genome. Genomics. 1998;54:361–378. doi: 10.1006/geno.1998.5602. [DOI] [PubMed] [Google Scholar]
- Rexroad CE, III, Schläpfer JS, Yang Y, Harlizius B, Womack JE. A radiation hybrid map of bovine chromosome one. Anim Genet. 1999;30:325–332. doi: 10.1046/j.1365-2052.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- Rexroad III, C.E., Owens, E.K., Johnson, J.S., and Womack, J.E. 2000. A 12,000 rad whole genome radiation hybrid panel for high resolution mapping in cattle. Anim. Genetics, in press. [DOI] [PubMed]
- Robinson TJ, Harrison WR, Ponce de Leon FA, Davis SK, Elder FF. A molecular cytogenetic analysis of X chromosome repatterning in the Bovidae: Transpositions, inversions, and phylogenetic inference. Cytogenet Cell Genet. 1998;80:179–184. doi: 10.1159/000014976. [DOI] [PubMed] [Google Scholar]
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP. Comparative gene mapping: A fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res. 1998;8:901–915. doi: 10.1101/gr.8.9.901. [DOI] [PubMed] [Google Scholar]
- Slonim D, Kruglyak L, Stein L, Lander E. Building human genome maps with radiation hybrids. J Comput Biol. 1997;4:487–504. doi: 10.1089/cmb.1997.4.487. [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Lengauer C, Fries R. Comparative genome map of human and cattle. Genomics. 1995;27:489–496. doi: 10.1006/geno.1995.1081. [DOI] [PubMed] [Google Scholar]
- Van Etten WJ, Steen RG, Nguyen H, Castle AB, Slonim DK, Ge B, Nusbaum C, Schuler GD, Lander ES, Hudson TJ. Radiation hybrid map of the mouse genome. Nat Genet. 1999;4:384–387. doi: 10.1038/11962. [DOI] [PubMed] [Google Scholar]
- Wakefield MJ, Graves JAM. Comparative maps of vertebrates. Mamm Genome. 1996;10:715–716. doi: 10.1007/s003359900221. [DOI] [PubMed] [Google Scholar]
- Watanabe TK, Bihoreau MT, McCarthy LC, Kiguwa SL, Hishigaki H, Tsuji A, Browne J, Yamasaki Y, Mizoguchi-Miyakita A, Oga K, et al. A radiation hybrid map of the rat genome containing 5,255 markers. Nat Genet. 1999;22:27–36. doi: 10.1038/8737. [DOI] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack JE, Kata S. Bovine genome mapping: Evolutionary inference and the power of comparative genomics. Curr Opin Genet Dev. 1995;5:725–733. doi: 10.1016/0959-437x(95)80004-o. [DOI] [PubMed] [Google Scholar]
- Womack JE, Johnson JS, Owens EK, Rexroad CE, III, Schlapfer J, Yang YP. A whole-genome radiation hybrid panel for bovine gene mapping. Mamm Genome. 1997;8:854–856. doi: 10.1007/s003359900593. [DOI] [PubMed] [Google Scholar]
- Xie S, Green J, Bao B, Beckers JF, Valdez KE, Hakami L, Roberts RM. The diversity and evolutionary relationships of the pregnancy-associated glycoproteins, an aspartic proteinase subfamily consisting of many trophoblast-expressed genes. Proc Natl Acad Sci. 1997;94:12809–12816. doi: 10.1073/pnas.94.24.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YP, Womack JE. Parallel radiation hybrid mapping: A powerful tool for high-resolution genomic comparison. Genome Res. 1998;8:731–736. doi: 10.1101/gr.8.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]