Abstract
Frizzled/planar cell polarity (PCP) signaling regulates cell motility in several tissues, including ommatidial rotation in Drosophila melanogaster. The Nemo kinase has also been linked to cell motility regulation and ommatidial rotation. The mechanistic role(s) of Nemo during rotation remain however obscure. We demonstrate that nemo functions throughout the entire rotation movement promoting rate of rotation. Genetic and molecular studies indicate that Nemo binds both the core PCP factor complex of Strabismus–Prickle, and the E-cadherin–β-catenin (Armadillo) complex, which colocalize and like Nemo also promote rotation. Strabismus/Vang binds and stabilizes Nemo asymmetrically within the ommatidial precluster. Nemo and β-catenin then act synergistically promoting rotation, which is mediated in vivo through Nemo phosphorylation of β-catenin. Our data suggest that Nemo serves as a conserved molecular link between core PCP factors and E-cad/β-catenin complexes, promoting ommatidial rotation and cell motility in general.
The retina of Drosophila melanogaster is a highly organized structure composed of ~800 units, called ommatidia, each containing 8 photoreceptor (R-) cells forming a cluster. Clusters are precisely arranged with respect to each other and the axes of the eye field1,2. 5-cell preclusters are first organized in the anterior–posterior (A/P) axis, and as R-cells get specified, preclusters start to undergo a 90o rotation towards the dorso–ventral midline. Frizzled planar cell polarity (Fz/PCP) signaling is associated with cell fate specification of the R3/R4 photoreceptor pair3,4. In response to Fz/PCP signaling ommatidial preclusters rotate a precise 90° in opposite directions in either half of the eye, creating a mirror image symmetry across the dorso-ventral midline1. Ommatidial rotation follows R3/R4 cell fate specification1,2. During rotation, R-cell precursors of each cluster are held together tightly by increased E-cadherin (E-cad) localization to membranes between precluster cells, but the process also requires the presence of E-cad on the outside membranes of the preclusters to facilitate rotation (as in shotgun/E-cad hypomorphic mutants rotation is strongly reduced)5. Besides Drosophila E-cad, N-cadherins have also been implicated in the process, with E-cad/shg again promoting rotation and the N-cadherins (N-cad1 and 2) restricting movement5. The combination of motility of the whole cluster relative to surrounding interommatidial epithelial cells and tight association of precluster cells to each other suggests a complex regulation of cell adhesive behavior. Ommatidial rotation is not the only cell motility process regulated by PCP signaling as convergent extension associated cell movements in vertebrates, for example, are also affected by Fz–PCP signaling3,6. Molecular links between the PCP factors and the mechanisms underlying cell motility have remained elusive.
Only a few factors that affect rotation have been identified, but the mechanism of action for any of them remain unclear. Nemo (Nmo) is the founding member of the Nlk subfamily of MAPKs, and the first rotation-specific gene described7,8. Other factors/pathways required in the process include Rho kinase (dROK; ref9), Egfr-signaling10–12, scabrous13 and Zipper–Myosin II14. It remains unclear how the activity of E-cad (and associated proteins) is regulated during rotation, as the molecular links between the PCP complexes and classical Cadherins have not been described, although a link has been suggested15.
Cadherins are homophilic transmembrane adhesion proteins, comprising a major intercellular adhesion system in epithelia16,17. Their cytoplasmic domains interact with the cytoskeleton through an association with Catenins: α-catenin, β-catenin (armadillo/arm in Drosophila), and p120ctn. Differential association and activity of Catenins can help to determine different states of Cadherin-mediated adhesion18. β-catenin directly binds the Cadherin cytoplasmic domain, promoting the stabilization and cell surface transport of cadherins19,20. α-catenin binds β-catenin through its N-terminal region, and can bundle F-actin as well as other actin-associated proteins through its C-terminal region (e.g. refs21–23). Several mechanisms regulate adhesion strength and dynamics to maintain tissue integrity, and allow morphogenetic movements. Adhesion regulation may occur via several broad mechanisms: (1) regulation of cadherin cell surface delivery/stabilization20, (2) phospho-modulation of β-catenin binding to cadherin (refs24,25) or α-catenin binding to β-catenin26, (3) the relationship of α-catenin to the actin cytoskeleton21,27, and/or (4) cis-clustering of Cadherins28.
Although the Nmo kinase was identified as a rotation specific factor7, subsequent work has revealed multiple mechanistic roles for Nmo and Nlks in other contexts: Nlk family members can phosphorylate TCF and/or β-catenin (depending on the organism), preventing the complex from binding to DNA29,30, accordingly, in Drosophila, Nmo can antagonize Wg-signaling and affect peak levels of the pathway31. In addition, it can interact with Notch and Dpp-signaling in Drosophila and vertebrates32–34.
Using a new null allele of nmo (nmoDB), we demonstrate here that it is required throughout ommatidial rotation for the rate of rotation. Genetic and molecular experiments indicate that Nmo binds the core PCP factors Strabismus (Stbm; a.k.a. Van Gogh/Vang) and Prickle, and also interacts genetically and physically with the E-cad/β-catenin complex. Nmo binds and phosphorylates Arm/β-catenin and also E-cad, which are both required for rotation5. Stbm is required for Nmo localization in R4, which then synergizes with β-catenin/Arm in vivo through its phosphorylation of Arm to promote rotation. Our data indicate that Nmo serves as a molecular link between core PCP factors (Stbm/Vang) and E-cad/β-catenin complexes, regulating the function of such complexes through β-catenin phosphorylation during ommatidial rotation.
Results
nmo is required autonomously during ommatidial precluster rotation
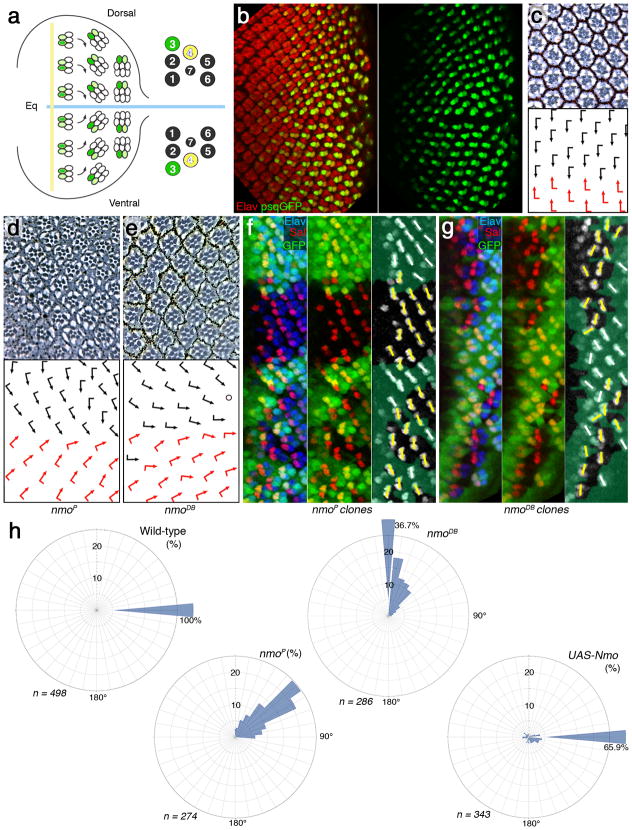
To define the role of nmo in rotation, we have analyzed a new null allele, nmoDB (Supplementary Fig. 1). In comparison to the hypomorphic nmoP allele, nmoDB showed a much more severe underrotation, with many clusters unable to initiate rotation and remaining parallel to the equator (Fig. 1). The defects in nmoDB mutant ommatidia were largely restricted to rotation (Fig. 1e). Strong underrotation was evident from early stages of precluster rotation in eye discs (Fig. 1f,g). Mutant preclusters (in both alleles, nmoDB and nmoP) displayed rotation defects as early as in row 5 (the earliest stage when rotation is detectable with molecular markers; e.g. anti-Sal staining marks R3/R4 and indicates degree of rotation of respective clusters just after the onset of rotation (Fig. 1f,g). In nmoDB eyes, many clusters do not rotate at all and remain in the orientation parallel to the equator at 0°. Similarly, at later stages of rotation (evident for example with the psq-GFP R3/R4 marker which is first detected from about a 45° angle to the end of rotation, where clusters rotated 90° relative to their original position35) many mutant clusters displayed severe under-rotation or no rotation at all (Supplementary Fig. 2; this marker confirmed that R3/R4 cell fate decisions were not affected in nmoDB mutants). A quantification of rotation angles in adult retinas (Fig. 1h) shows that in wildtype, all ommatidia are at 90° (100%), in nmoP the average is around 50°, whereas in nmoDB the majority (36.7%) is at 0° angle. Note also that in sevGal4, UAS-Nmo 34% of clusters are overrotated to random degrees (only 65.9% remain at 90°). Taken together with the disc analyses, this indicates that nmo is required throughout the rotation process.
Figure 1. nmo is required throughout ommatidial rotation.
(a) Schematic presentation of ommatidial rotation in 3rd instar larval eye disc, posterior to the morphogenetic furrow (MF, vertical yellow line; equator, horizontal blue line). Cells acquiring R3 fate are labeled in green. Left panel shows organization of individual photoreceptors within an ommatidium in adult eye. (b) 3rd instar eye disc stained with antibody against a pan-neuronal marker, Elav (red) and psq>GFP reporter (green; strong in R3 and weaker in R4; single channel in right panel). (c) Tangential section of adult wild-type eye with ommatidia arranged around the equator (having completed the 90° rotation). Bottom panel: schematic with dorsal and ventral chiral forms indicated by black and red arrows, respectively.
(d–e) Eye sections of the hypomorphic allele nmoP (d) and the null allele nmoDB (e). Arrows indicate degree of rotation (see quantifications of rotation angle distribution in panel h).
(f–g) 3rd instar eye discs (just posterior to MF) stained with anti-Sal (red; marking R3/R4 precursors in rows 2–5), anti-Elav (blue; all R-cell precursors) and anti-GFP (green; mutant tissue marked by GFP absence). Mutant clones of nmoP (f) and nmoDB (g) are shown. Right panels: semi-schematic versions of middle panels of f and g, white bars indicating orientation of Wt clusters and yellow bars indicate orientation of mutant clusters. (h) Rose diagrams displaying the angle distribution of ommatidia (in interval of 10°) of the genotypes indicated. The radial axis displays % (up to 25%), percentages above 25% are written next to sector-bar.
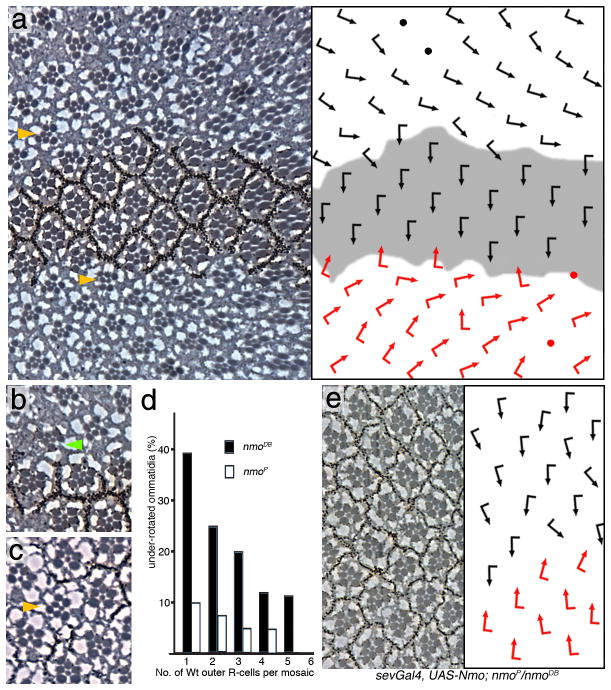
Earlier mosaic analysis of nmoP had suggested that single nmo+ R-cells can rescue the nmo requirement for the whole cluster7. A mosaic analysis with the null nmoDB allele (Fig. 2) revealed the following: (1) nmo is not required in any particular R-cell subtype (in agreement with ref7) (2) nmo is required in a cluster autonomous manner in R-cells only as the mutant phenotype was not influenced by neighboring wildtype or mutant inter-ommatidial cells (mutant ommatidia adjacent to wildtype cells had an equal frequency and degree of under-rotation as clusters surrounded by mutant cells; Fig. 2a–c); (3) a single nmo+ R-cell was not sufficient to rescue a nmoDB mosaic cluster; and (4) normal rotation did correlate with the number of wildtype R-cells in a mosaic ommatidium (Fig. 2d, see cluster marked by green arrow in Fig. 2b with a single mutant R-cell). Specifically, the more wildtype outer photoreceptors a mosaic ommatidium contained the higher was the probability that the cluster rotated correctly (Fig. 2d). This conclusion is supported by the full rescue of nmoP/nmoDB by a tubulin-NmoGFP transgene (expressed in all cells; not shown), whereas other genotypes where Nmo is expressed in a subset of R-cells like sevenless (sev) or m δ-Gal4 displayed an incomplete rescue (Fig. 2e and not shown).
Figure 2. nmo is required in a cluster autonomous manner for rotation.
Panels a–c, and e show tangential sections of adult eyes, (d) shows a quantification; dorsal is up and anterior left. The respective schematic presentations of ommatidial orientation (arrows as in Fig. 1) are shown as right panels in a and e. (a–c) Mutant clones of nmoDB, marked by absence of pigment (Wt area in schematic in a is shaded gray), In a: examples of non-rotated ommatidia adjacent to wt tissue are highlighted by yellow arrowheads. (b) Green arrowhead marks an under-rotated mosaic ommatidium. (c) A mutant ommatidium (yellow arrowhead) surrounded by wild-type clusters.
(d) Quantification of rotation in mosaic ommatidia: (nmoDB is shown with black bars and nmoP in white bars). (e) sev>Nmo; nmoP/nmoDB. Expression of Nmo protein in a subset of R-cells in flies homozygous mutant for nmo.
Increased Nemo levels cause ommatidial overrotation
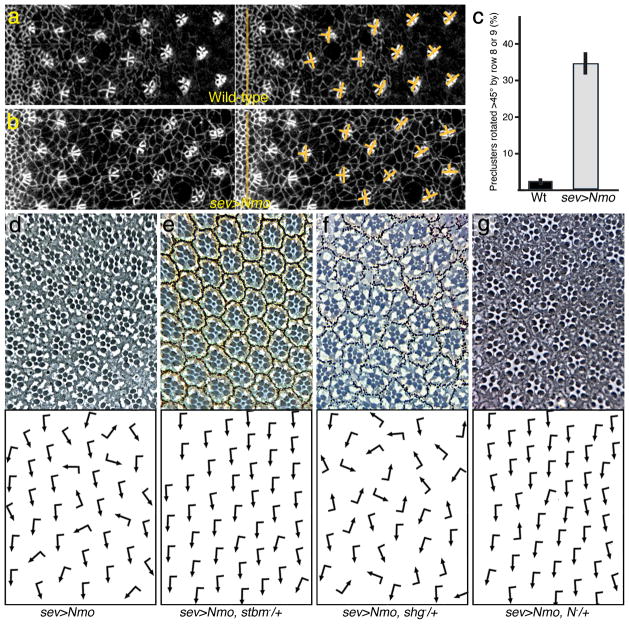
Overexpression of Nmo (e.g. under sev-enhancer control, which is expressed in R3/R4 in rows 2–7 and in R1/R6 and R7 a few rows later36) in otherwise wildtype eye discs caused a large number of ommatidia to rotate faster and often over-rotate (rotate >90°) (Fig. 3a-d; also13). A similar phenotype was seen in clones overexpressing Nmo using the MARCM technique (not shown). Importantly, rotation was faster than wild-type from earliest stages on. In contrast to wildtype, many clusters rotated beyond 45° by rows 9–10 (Fig. 3b–c). This accelerated rotation feature resulted in many ommatidia having rotated over 90° in adult eyes (Fig. 3d; quantified in Fig. 1h). Expression of a kinase-dead Nmo did not cause rotation defects (not shown), indicating that kinase activity is critical. Taken together with the loss of function analyses, these data suggest that Nmo levels/activity directly correlate with the rate of rotation.
Figure 3. Increased Nmo levels cause an increased rotation rate.
(a–b) Confocal microscopy images of 3rd instar eye imaginal discs, anterior is left and dorsal is up; (a) wild-type and (b) sevGal4, UAS-Nmo (sev>Nmo). Discs are stained with anti-Arm, labeling all cell membranes. Arm is enriched on membranes within the forming cluster. Yellow bars delineate rotation angles of preclusters. (c) Quantification of rotation angles in rows up to row 9 in % +/− s.d. . (d–g) Adult eyes of sev>Nemo genotypes as indicated, dorsal area of eye is shown, anterior is left (see Table 1 for quantification). (d) sev>Nmo (in w1118 background). (e) sev>Nemo, stbm−/+. (f) sev>Nemo, shg−/+. (g) sev>Nemo, N−/+. Other genotypes: see Table 1.
Nemo genetically and physically interacts with Stbm–Vang and Pk
As an entry point to potential mechanistic links, we used the sevGal4, UAS-Nemo (sev>Nmo) overrotation phenotype to screen for genetic interactors. sev>Nmo is dosage sensitive as it is suppressed when one copy of endogenous nmo is removed (Table 1). We tested a large set of candidate genes, including all core PCP components, selected factors of most signaling pathways, and many cytoskeletal or cell adhesion factors (Fig. 3). We identified the PCP factors stbm/Vang and prickle (pk) as dominant suppressors of sev>Nmo (Fig. 3e; Table 1). Strikingly, other core PCP factors did not interact with sev>Nmo (Table 1 and not shown). In addition, Notch (N) suppressed sev>Nmo (Fig. 3g; Table 1), suggesting that Nmo regulates or is regulated by input from several distinct pathways (see Discussion).
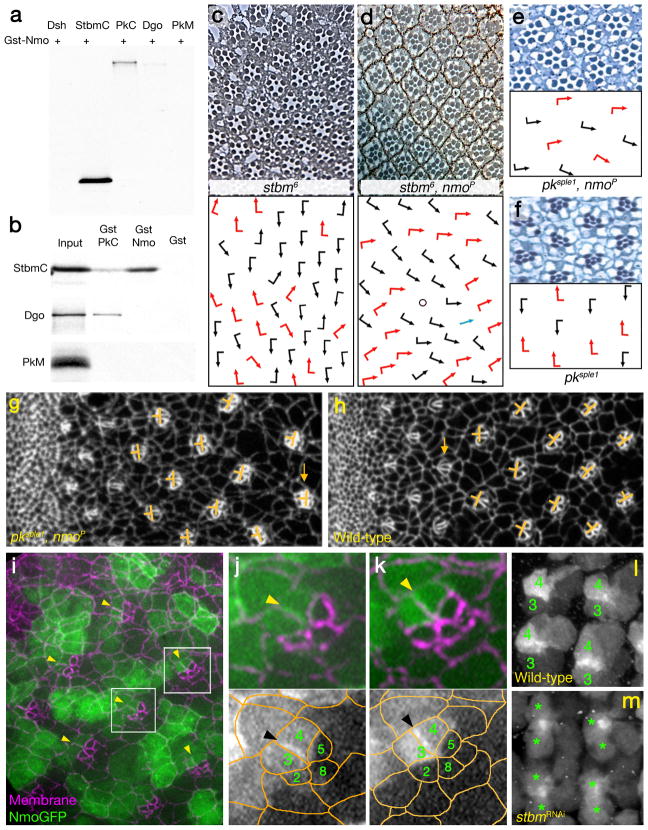
To corroborate the hypothesis of a functional relationship between Nmo and the PCP factors Stbm and Pk, we examined defects in double mutant stocks of the hypomorphic nmoP allele and stbm/Vang and pk alleles (Fig. 4). In stbm/Vang mutants rotation angles and direction become randomized (Fig. 4), and it has been suggested that rotation is slightly delayed37. Whereas ommatidia rotate in nmoP to average of 45° (Fig. 1d, i–j), in the double mutants many ommatidia did not rotate at all and remained in a parallel orientation to the equator (Fig. 4d; chirality was randomized as is common to stbm/Vang− alleles, compare distribution of red and black arrows in Fig. 4c,d). The rotation defect in the double mutant with the hypomorphic nmoP was very similar to that observed in the nmoDB null, suggesting that the partial Nemo activity in nmoP is further impaired in the absence of Stbm/Vang. A very similar effect was observed in pksple1, nmoP double mutants. The enhancement is particularly evident here, as in pksple1 single mutant, rotation is normal despite randomized chirality (Fig. 4f). Note that in double mutants rotation is often not initiated and clusters remain in the original position by row 10 and later (Fig. 4e). In discs, CanoeGFP labels all cells at the adherens junctions (Fig. 4g–h) and failure of rotation at larval stages reflects defects seen in adult eyes. As these defects were detectable from early stages of precluster rotation (Fig. 4g–h), the phenotype is likely due to a failure to properly initiate rotation.
Figure 4. Nemo interacts physically and genetically with Stbm and Pk.
(a) Gst-pulldown assays using Gst-Nmo full length and in vitro translated StbmC, the part common to all Pk isoforms (PkC), Dsh, Dgo and PkM (b) Specificity assays for the Gst-Nmo/StbmC interaction, using Gst-PkC: ref54. The “input” lane is 10%. (c–f) Tangential eye sections of stbm6 (panel c), stbm6; nmoP (d), pksple1, nmoP (e) and pksple1 (f). Schematic presentation of cluster orientation is shown below each micrograph; arrows are is in Fig. 1.
(g–h) Precluster rotation defects in pksple1, nmoP double mutant discs (CnoGFP is used to mark all cells at the adherens junctions). Orientation of clusters is highlighted by orange bars; orange arrows highlight one cluster each in g and h that have not yet initiated rotation.
(i–k) NmoGFP is localized to the membrane at the R4 side of the R3/R4 border. (i) Area of eye imaginal disc posterior to furrow (anterior is left, dorsal up) showing NmoGFP localization (green) and membrane staining (anti-Arm; magenta). Boxes indicate areas shown at high magnification in j and k, respectively. (j–k) High magnifications of preclusters that initiated rotation (j) and one that is approximately at a 45° (k). R4 side of the R3/R4 border in both clusters is highlighted by yellow arrowheads, also in panel i; the 5 R-cells of the precluster are labeled by their numbers; bottom panels: monochromes showing NmoGFP only with a semi-schematic overlay of cell outlines (orange). R4-side of membrane is indicated by black arrowheads.
(l–m) NmoGFP localization and stability depends on the presence of Stbm. (l) NmoGFP localization in wild-type background. R3 and R4 precursors are labeled with 3 and 4, respectively, within each cluster. (m) In GMR>NmoGFP; >stbmRNAi the respective mutant R3/R4 precursors are marked with stars. Anterior is left and dorsal up in both panels.
We next tested for physical interaction(s) between Nmo and core PCP factors using GST pull-down assays. Nmo bound to the Stbm/Vang C-tail (StbmC) and the common part of Pk (PkC; Fig. 4a). Other PCP factors such as Dishevelled (Dsh) or the PkM peptide did not bind Nmo (Fig. 4a,b and not shown). These interactions were confirmed in yeast 2-hybrid assays (not shown). Since Nmo physically binds Stbm, we also asked whether it can phosphorylate Stbm. In in vitro kinase assays with purified, active Nmo kinase and Stbm as a substrate (see Methods), Nmo did not phosphorylate Stbm, whereas it did phosphorylate itself and several other targets (see below).
As there are no reliable antibodies to Nmo, we generated NmoGFP transgenes (expressed under tubulin-Gal4 or sev-Gal4-control). NmoGFP is generally localized in a ubiquitous manner throughout each cell, including association with the membrane and nuclear localization (Fig. 4i, Supplementary Fig. 3, and not shown). A low level of expression under ubiquitous promoter revealed NemoGFP was enriched asymmetrically on the R4 side of the R3/R4 border membranes (Fig. 4i–k, examples marked by arrowheads; note that this is the case throughout the process). Stbm/Vang is asymmetrically localized in R4 in a very similar manner38. At later rotation stages, NmoGFP is higher in R4 (as compared to R3) like the core PCP genes (the likely result of post-transcriptional stabilization as it seen with expression from ubiquitous promoters; Supplementary Fig. 3e–f and not shown).
As nmo genetically and physically interacts with Stbm and is enriched in R4, we next analyzed whether they are required for each other’s localization and/or enrichment. While, nmo loss-of-function or Nmo overexpression did not affect Stbm/Vang localization (or that of other core PCP factors; not shown), localization and enrichment of NmoGFP was affected in stbm/Vang backgrounds. In a stbm mutant background (GMR>stbmRNAi) both aspects of NmoGFP expression are markedly reduced (Fig. 4l–m). In particular, the levels of Nmo in R3 and R4 are indistinguishable in the mutant, in contrast to the high R4 levels in wildtype (cf. Fig. 4l with 4m, see also Supplementary Fig. 3e–f). The membrane enrichment at the R4 side of the R3/R4 border is also lost (Fig. 4l–m). These data suggest that Stbm is required for proper localization or stability of Nmo in R4, and thus Nmo is functionally linked to the R4-specific PCP factors, Stbm/Vang and Pk.
Nemo directly binds and phosphorylates β-catenin and E-cadherin
Among the cell adhesion and cytoskeletal/cell architecture factors, E-cadherin/shotgun and β-catenin/Armadillo (arm) were strong modifiers of the sev>Nmo phenotype (Fig. 3f; Table 1). Of note, only arm null alleles displayed an interaction, whereas Wg-signaling defective arm alleles that retain the cell adhesion function did not interact (Table 1; consistent with observations of a lack of interactions between E-cad mediated rotation and Wg-signaling5). These genetic data suggested that nmo function requires components of the cell adhesion E-cad/β-cat complex.
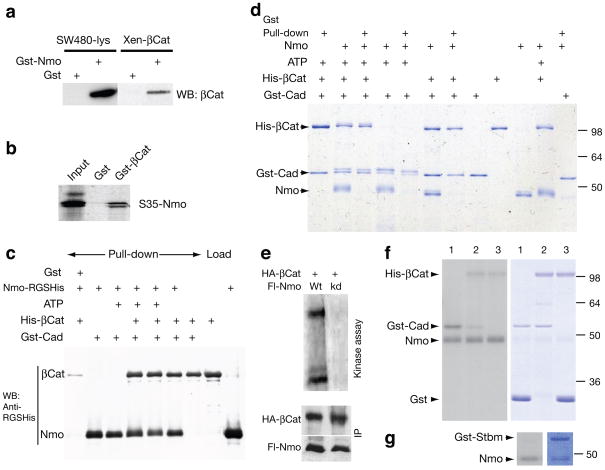
We thus tested whether Nmo can physically interact with and/or phosphorylate E-cad or β-catenin (Fig. 5). Gst–Nmo bound β-catenin from SW480 cell lysates, similar to purified Xenopusβ-catenin (Fig. 5a). The double band in S35Nmo likely represents an autophosphorylated form of Nmo. Gst-β–catenin also bound in vitro translated Nmo (Fig. 5b). Similarly, Gst-Cadherin, containing only the cytoplasmic portion (Gst-Cad), pulled down both Nmo and β-catenin, individually or together (Fig. 5c). Using purified proteins for all three components and testing phosphorylation of each factor (and binding to Gst-Cad; Fig. 5d) we observed that Nmo phosphorylated both β-catenin and Gst-Cad (and itself) in the presence of ATP (Fig. 5d; note slower migrating phosphorylated bands in presence of Nmo and ATP). However, whereas β-catenin bound in a stable, stoichiometric manner to Gst-Cad, only traces of Nmo were detected bound to Gst–Cad, suggesting a transient interaction (Fig. 5d). Nmo can bind Cadherin as detected in a more sensitive assay (Western blot, Fig. 5c; note that anti-RGS-His antibody specific to the epitope of Nmo-His recognizes this with a much higher sensitivity than the common His-epitope present in His-β-cat). Presence of ATP did not affect binding of the kinase.
Figure 5. Nemo binds and phosphorylates β-catenin and Cadherin.
(a) Extracts from SW40 cell lysates incubated with Gst-Nemo. β-catenin is detected with antiβcat (left panel). Recombinant Xenopus β-cat incubated with Gst-Nmo (right panel). (b) Gst–βcat (Arm) incubated with in vitro translated Nmo. (c) Western blot with RGS-His antibody detection of the respective Gst-Cad pulldowns as indicated. (d) Coomassie stained gel of Gst-Cad (C-cadherin) pull-down assays and kinase reactions with Nmo-His and His-β-cat (in the presence or absence of ATP). (e–g) 32P kinase reactions with Nmo. (e) Nmo purified from cell lysates: full length Nemo (Nmo-FL) wild-type protein (lane 1; Wt) or kinase inactive (lane 2; kd) and β-cat (Arm; upper band). (f–g) In vitro kinase reactions with bacculovirus expressed, purified Nmo. In f, lanes 1: Nmo, Gst-Cad, and Gst; lanes 2: Nmo, Gst-Cad, and His-β–catenin: and lanes 3: Nmo, His-β–catenin, and Gst. In g, Nmo and Gst-Stbm. Left panels show radioactive exposures, revealing kinase events of respective Coomassie stained proteins shown on right in panels (f) and (g).
Similarly, immunoprecipitates of Nmo efficiently phosphorylated β-catenin, but those containing a kinase-dead (kd) Nmo isoform did not (Fig. 5e). These phosphorylation events were specific: whereas Nmo phosphorylated β-catenin, Cadherin and itself (Fig. 5f; right panel showing protein staining and left panel showing 32P signal), it did not phosphorylate the PCP factor Stbm/Vang (or control Gst; Fig. 5f–g). In summary, our molecular data indicate that Nmo can associate with Cadherin/β-catenin complexes and specifically phosphorylate them.
Arm/β-catenin and Nmo act synergistically in rotation
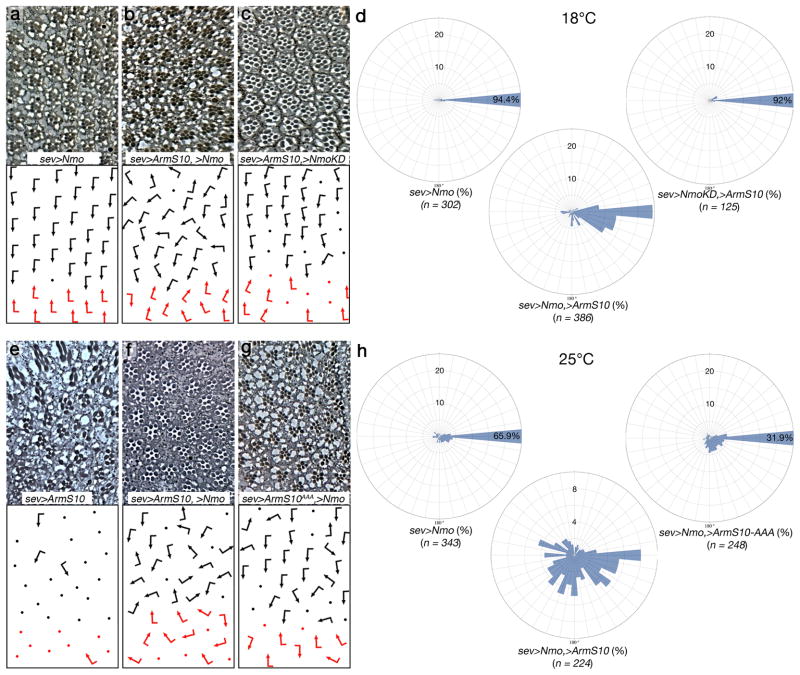
Phosphorylation of E-cad and β-catenin was proposed to impact their association18,39. To determine whether Nmo phosphorylation can affect E-cad/β-catenin association, we performed titration assays, comparing the binding of β-catenin to cadherin in the presence or absence of phosphorylation by Nmo. We did not, however, detect an effect of Nmo on the formation of β-catenin/cadherin complexes in vitro (Supplementary Fig. 4). Since phosphorylation can impact AJ structure and dynamics in several ways we next tested whether Arm/β-catenin and Nmo act together to promote rotation in vivo. Stable Arm/β-catenin isoform ArmS10 (a truncated isoform that is not subjected to degradation40), which displays both nuclear Wg/Wnt-signaling and cell adhesion gain-of-function (GOF) phenotypes, was expressed alone or co-expressed with Nmo (under sev-Gal4 control)(Fig. 6). At 25°C in sev>ArmS10 almost all ommatidia are unscorable due to nuclear Wg-signaling function. Remarkably, in sev>ArmS10, UAS-Nmo most ommatidia are scorable and the majority of these (>90%) are overrotated. The overrotation in the sev>ArmS10, UAS-Nmo genotype is massively stronger than any other genotypes tested and less than 10% stay at a 90° angle (Fig. 6e–f; Supplementary Fig. 5). At 25°C the ArmS10 gain-of-function effects in nuclear Wg-signaling were too severe to accurately allow comparison of ArmS10 co-expressed with Nmo in rotation. At 18°C (with reduced expression due to the temperature sensitivity of Gal4) the effect of Nmo on ArmS10 in rotation was testable. Strikingly, while neither Nmo nor ArmS10 alone showed marked rotation defects at 18°C, co-expression produced many overrotated ommatidia (Fig. 6a–d). ArmS10 co-expressed with the kinase-dead Nmo isoform behaved like ArmS10 alone, confirming the requirement of kinase activity in rotation (Fig. 6c). These data suggest that Nmo and Arm/β-catenin cooperate to promote rotation and that the kinase activity of Nmo is required.
Figure 6. The activity of Arm/β-catenin is modulated by Nmo in vivo.
(a–c) and (e–g) Tangential eye sections of genotypes indicated, anterior is left and dorsal up. Ommatidial orientation is presented schematically in lower panels (arrows are as in Fig. 1, dots represent ommatidia that cannot be scored for orientation).
(a–c) Expression of indicated transgenes at 18°C under sevGal4 control; (a) UAS-Nmo, (b) UAS-Nmo, UAS-ArmS10 (a stabilized form of Arm/β-cat) (c) UAS-ArmS10, UAS-NmoKD (a kinase inactive isoform). (d) Quantification of rotational enhancements: rose diagrams in 10° intervals of the genotypes indicated expressed at 18°C (corresponding to the genotypes shown in panels a–c). The radial axis displays % (up to 25%), and percentages above 25% are written into sector-bar.
(e–g) Expression of indicated transgenes at 25°C under sevGal4 control; (e) UAS-ArmS10, (f) UAS-ArmS10, Uas-Nmo; (g) UAS-ArmS10AAA, UAS-Nmo. (h) Rose diagrams displaying the angle distribution of ommatidia (in intervals of 10°) of the genotypes indicated. The radial axis displays % (up to 25% or 10%, as indicated), and percentages above 25% are written into sector-bar.
To confirm that this effect is due to direct phosphorylation of ArmS10 by Nmo we identified Nmo target phosphorylation sites on Arm in vitro. This revealed three Ser/Thr phosphorylation-sites in the C-terminal region of Arm [S764, S802, T827] as preferential target sites of Nmo (Supplementary Fig. 6). To determine whether these sites are physiologically relevant, we established transgenic flies expressing stable ArmS10 with the three sites mutated to Ala (UAS-ArmS10-AAA, Supplementary Fig. 6). Strikingly, in contrast to normal ArmS10, when ArmS10-AAA was co-expressed with Nmo (under sev-Gal4) the distribution of the rotation angles (overrotation) resembled that of UAS-Nmo alone. It was markedly suppressed as compared to UAS-Nmo, UAS-ArmS10 (Figs. 6f-g, quantified in Fig. 6h and also Supplementary Fig. 7). Expression of ArmS10-AAA at 25°C alone showed typical Wg-signaling gain of function defects with R-cell death similar to ArmS10 (Supplementary Fig. 5 and not shown), precluding an analysis of rotation, and has no rotation phenotype at 18°C. The subcellular localization of Arm or ArmS10 was not affected by the AAA mutations (Supplementary Fig. 6). In addition to data presented, we analyzed two independent insertion lines for sev>ArmS10, UAS-Nmo and sev>ArmS10-AAA, UAS-Nmo and found they displayed very similar behavior (Supplementary Figs. 6c and 7, and not shown). Taken together, these data suggest that Nmo mediated phosphorylation of the Arm C-terminal sites is critical for the regulation of ommatidial rotation in vivo but has no effect on other functions of Arm/β-catenin.
These in vivo data confirm that Nmo binding to E-cad–β-catenin complexes and phosphorylation of Arm/β-catenin on the respective C-terminal sites is physiologically relevant and as such the Nmo kinase promotes rotation via Arm/β-catenin.
Discussion
Ommatidial rotation is an example of Fz–PCP signaling-regulated cell motility. Our data indicate that Nmo serves a regulatory function for the activity of E-cad–β-catenin complexes and a link to the core PCP factor Stbm/Vang. It promotes Arm/β-catenin function in rotation by its direct phosphorylation, while integrating signaling input from PCP (Stbm/Vang–Pk) and (possibly) other signaling pathways including Notch (see below).
Our analyses of a nmo null allele indicate that nmo is required throughout the process and many clusters do not rotate at all. Accordingly, increased Nmo levels (sev>Nmo) cause a faster rotation and overrotated ommatidia, suggesting that Nmo regulates the rate of rotation. Mosaic studies reveal that nmo is required in all outer R-cells, as only fully (wildtype) nmo+ ommatidia have a 100% certainty to rotate normally. A recent report has suggested that nmo is required mainly in R1/R6 cells8. The difference to our data is likely explained with the fact that Fiehler and Wolff (2008)8 used a hypomorphic allele, where a portion of gene function is maintained. Consistent with our study Choi and Benzer (1994)7 also concluded that nmo can function in any R-cell.
Genetic, physical and functional interactions indicate that Nmo regulates the activity of E-cad–β-catenin complexes during rotation. Stable β-catenin/Arm (ArmS10) affects both nuclear Wg-signaling (resulting in photoreceptor loss) and junctional E-cad–β-catenin complexes (resulting in aberrant cell adhesion). Co-expression of Nmo with ArmS10 not only blocks increased nuclear Arm signaling (as expected29,30), but it synergizes with ArmS10 in causing an increase in the rate of rotation (as compared to either alone). This synergy is dependent on the presence of the Nmo phosphorylation sites in Arm and the α-catenin binding region of Arm/β-catenin (Fig. 6 and not shown). Our data indicate that (1) Nmo directly phosphorylates Arm/β-catenin, (2) this phosphorylation is physiologically important, and (3) Nmo phosphorylation of Arm is necessary for them to act synergistically.
Our data suggest that Nmo connects the core PCP Stbm/Vang–Pk complex to the activity of E-cad–β-catenin. Consistently, mutations in stbm/Vang and pk enhance not only the nmoP rotation defects but also rotation defects of hypomorphic shg/E-cad backgrounds41. As the Stbm/Vang–Pk complex appears to enrich Nmo at R4 membranes/junctional complexes, we hypothesized that an increase in Stbm/Vang levels would increase the capability of sev>Nmo to cause an overrotation phenotype. This is indeed the case (Supplementary Fig. 8). These data indicate that Nmo serves as a link from PCP factors to the E-cad–catenin complexes and are consistent with a model suggesting that the Stbm–Pk complex helps to recruit and/or stabilize Nemo at membrane regions (where the PCP factors partially overlap with E-cad/β-cat complexes42–44).
The effect of Nmo on E-cad–β-catenin complexes could be mediated either through the dynamics of lateral clustering (e.g. formation or disassembly of higher order E-cad–β-catenin complexes) or by affecting the interaction of β-catenin with other associated proteins18. An Ecad::α-catenin fusion protein (which bypasses a β-catenin requirement and provides “stable adhesion”45,46) is not influenced by Nmo, suggesting that once α-catenin is part of the E-cad–catenin complex Nmo cannot influence their activity. It is thus possible that phosphorylation by Nmo affects the E-cad–β-catenin complex “activity” via Arm/β-catenin (as an ArmS10-AAA isoform with the Nmo target sites mutated no longer synergizes with Nmo) and possibly modulates its interaction(s) with other binding partners like α-catenin.
The interactions of adhesion and planar polarity during the early “convergence-extension” rearrangements in the fly embryo suggest a mechanism by which a polarized pattern of junction remodeling drives cell intercalation47–49. Polarized activity of RhoA/myosinII (zipper) regulates adherens junction disassembly along the A/P axis, primarily by regulating lateral cadherin clustering without affecting their surface levels50. The specific effect of RhoA on rotation5, and interaction of nmo with myosinII/zipper14 supports the idea for actin–myosin contractility downstream of Nmo. Loss of maternal contribution or Nmo overexpression in the embryonic epidermis phenocopies shg/E-cad alleles or ArmS10 cuticle defects, respectively40,41. Thus, Nmo may be generally required in epithelia undergoing morphogenetic movements, where it modulates polarized remodeling of adherens junctions in response to local asymmetries created by, for example, the activity of PCP signaling complexes.
In conclusion, this study defines a framework for Nmo serving as a link between PCP (Vang/Stbm) and the regulation of adhesive cell behavior at the level of adherens junction complexes. Although Nmo is recruited and/or maintained apically by the Stbm/Vang–Pk complex, other factors must affect Nmo activity or localization as well, because the cellular requirements of nmo (all outer R-cells) are broader than stbm/Vang (R4). First, the rate of rotation could be regulated independently of the PCP complexes through Notch (N) (and/or Egfr) signaling also via Nmo, as suggested by genetic data (Table 1): N− alleles strongly suppress sev>Nmo, and N functions in all R-cells. Second, an asymmetric input/localization of Nmo by Stbm/Vang would provide a direction to rotation. Thus, a Notch–Nmo interaction in all cells and an asymmetric Stbm/Vang effect in R4 could combine to regulate both rate and direction of rotation. The observation that zebrafish Nlk enhances (the PCP dedicated) Wnt11 cell migration defects in prechordal plates51,52, which also requires E-cad complexes53, supports a general Nmo-mediated mechanism in PCP associated cell movements.
Methods
Fly strains and genetics
UAS-Nmo32 and UAS-NmoGFP8 transgenes were as described and driven with the UAS/Gal4 system55. Flip-out GOF clones of UAS-Nemo were generated with MARCM using hsFLP, actin>CD2> and marked with UAS-wRNAi. Interaction crosses were grown at 25°C and eyes from female flies examined. The transgenes that modified sev>Nemo (UAS-EcadWT, UAS-Stbm, UAS-RhoARNAi showed no phenotype with sevGal4 by themselves.
For LOF clonal analyses, nmo alleles were recombined onto w−; FRT80 chromosome and clones induced with eyFLP (marked with ubiGFP). Minute (M) clones (giving mutant tissue a growth advantage) were induced with hsFLP. Rescue experiments were performed with a tub-NemoGFP construct (NemoII cDNA was used), or with UAS-Nemo expressed under sevGal4 or mδGal4 control.
Other mutant strains and UAS lines were as described in Flybase (http://flybase.bio.Indiana.edu). The following alleles were obtained from: shgP34-1, shgIg29 (U. Tepass), DE-cadWT (H. Oda), UAS-DN-cad56, UAS-RhoAIR (ref57), Ncad Δ14 (from T. Clandinin).
Immunohistochemistry and histology
Primary antibodies were: rat anti-DE-cad (DCAD2; gift from H. Oda); rabbit anti-Bar (gift from K. Saigo); rat anti-Spalt (B. Mollereau); rabbit anti-Dlg (gift from Z.H. Chen); rhodamine-phalloidin (Molecular Probes); anti-β-Gal (Cappel, Promega), rat anti-Elav, mouse anti–Arm (N27A1), rat anti-DN-cad (Dn-Ex#8) and mouse anti-Fmi from the DSHB. Secondary antibodies were from Jackson Labs. Eye sections and eye discs were prepared, stained, and analyzed as described 5. For genetic interactions, eyes were sectioned near equatorial region and ommatidia with correct photoreceptor number were scored; a rotation defect was defined as being at least 20° less or more than the 90° in wt.
Biochemical assays
The respective proteins, either GST or His-tagged, were expressed and purified following standard protocols. The associated details and the specifics of kinase and binding assays are described in Supplemental Information.
Supplementary Material
Acknowledgments
We are grateful to Z. Chen (Baylor College of Medicine), T. Clandinin (Stanford School of Medicine), H. Oda and S. Tsukita (Japan Science and Technology Corporation, Kyoto), P. Rørth (Institute of Molecular and Cell Biology, Singapore), K. Saigo (University of Tokyo), U. Tepass (University of Toronto), N. Tolwinski (Sloan-Kettering Institute), T. Uemura (Kyoto University), W. Weis (Stanford University), T. Wolff (HHMI/Janelia Farm), B. Mollereau (Rockefeller University) and the Bloomington stock center for flies and reagents. We thank S. Okello and Y.A. Zeng for technical assistance, N. Maj for help in generating the rose diagrams, J. Delaney and U. Weber for comments on the manuscript, and all members of the Mlodzik lab for helpful discussions. This work was supported by NIH/NEI grant RO1 EY14597 to MM, a NSERC RGPIN/203545 and CIHR MOP 62895 to EV, and NIH GM076561 to CG.
Footnotes
AUTHOR CONTRIBUTIONS
I.M. and M.M. designed experiments; I.M., W.J.G., M.R., A.J., K.G. conducted experiments and analyzed data; A.J., W.J.G. and C.J.G. edited the manuscript; D.B. generated the nmoDB allele; C.J.G. conducted biochemical experiments with β–catenin and Nmo; I.M., E.M.V., M.M. analyzed data and prepared the manuscript.
References
- 1.Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Martinez-Arias MBA, editor. The development of Drosophila melanogaster. Cold Spring Harbor Press; Cold Spring Harbor: 1993. pp. 1277–1326. [Google Scholar]
- 2.Mlodzik M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seifert JR, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–38. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Nathans J. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development. 2007;134:647–58. doi: 10.1242/dev.02772. [DOI] [PubMed] [Google Scholar]
- 5.Mirkovic I, Mlodzik M. Cooperative activities of drosophila DE-cadherin and DN-cadherin regulate the cell motility process of ommatidial rotation. Development. 2006;133:3283–93. doi: 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- 6.Solnica-Krezel L. Conserved patterns of cell movements during vertebrate gastrulation. Curr Biol. 2005;15:R213–28. doi: 10.1016/j.cub.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- 8.Fiehler RW, Wolff T. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev Biol. 2008;313:533–44. doi: 10.1016/j.ydbio.2007.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winter CG, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 10.Brown KE, Freeman M. Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development. 2003;130:5401–12. doi: 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- 11.Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–23. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- 12.Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–7. doi: 10.1016/s0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- 13.Chou YH, Chien CT. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev Cell. 2002;3:839–50. doi: 10.1016/s1534-5807(02)00362-3. [DOI] [PubMed] [Google Scholar]
- 14.Fiehler RW, Wolff T. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev Biol. 2007;310:348–62. doi: 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Classen AK, Anderson KI, Marois E, Eaton S. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev Cell. 2005;9:805–17. doi: 10.1016/j.devcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Takeichi M. Cadherins: a molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237–52. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- 17.Tepass U, et al. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–85. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- 18.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–34. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 19.Chen YT, Stewart DB, Nelson WJ. Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J Cell Biol. 1999;144:687–99. doi: 10.1083/jcb.144.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber AH, Stewart DB, Laurents DV, Nelson WJ, Weis WI. The cadherin cytoplasmic domain is unstructured in the absence of beta-catenin. A possible mechanism for regulating cadherin turnover. J Biol Chem. 2001;276:12301–9. doi: 10.1074/jbc.M010377200. [DOI] [PubMed] [Google Scholar]
- 21.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–15. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. Functional domains of alpha-catenin required for the strong state of cadherin-based cell adhesion. J Cell Biol. 1999;144:1311–22. doi: 10.1083/jcb.144.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weis WI, Nelson WJ. Re-solving the cadherin-catenin-actin conundrum. J Biol Chem. 2006;281:35593–7. doi: 10.1074/jbc.R600027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stappert J, Kemler R. A short core region of E-cadherin is essential for catenin binding and is highly phosphorylated. Cell Adhes Commun. 1994;2:319–27. doi: 10.3109/15419069409014207. [DOI] [PubMed] [Google Scholar]
- 25.Daugherty RL, Gottardi CJ. Phospho-regulation of Beta-catenin adhesion and signaling functions. Physiology. 2007 doi: 10.1152/physiol.00020.2007. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda S, et al. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin- mediated cell-cell adhesion. Science. 1998;281:832–5. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 27.Nieset JE, et al. Characterization of the interactions of alpha-catenin with alpha-actinin and beta-catenin/plakoglobin. J Cell Sci. 1997;110 ( Pt 8):1013–22. doi: 10.1242/jcs.110.8.1013. [DOI] [PubMed] [Google Scholar]
- 28.Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol. 1998;141:779–89. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishitani T, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 30.Meneghini MD, et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- 31.Zeng YA, Verheyen EM. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development. Development. 2004;131:2911–20. doi: 10.1242/dev.01177. [DOI] [PubMed] [Google Scholar]
- 32.Verheyen EM, et al. The tissue polarity gene nemo carries out multiple roles in patterning during Drosophila development. Mech Dev. 2001;101:119–32. doi: 10.1016/s0925-4773(00)00574-8. [DOI] [PubMed] [Google Scholar]
- 33.Zeng YA, Rahnama M, Wang S, Sosu-Sedzorme W, Verheyen EM. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development. 2007;134:2061–71. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- 34.Ishitani T, et al. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol. 2010;12:278–85. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- 35.Weber U, Pataki C, Mihaly J, Mlodzik M. Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Dev Biol. 2008;316:110–23. doi: 10.1016/j.ydbio.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basler K, Siegrist P, Hafen E. The spatial and temporal expression pattern of sevenless is exclusively controlled by gene-internal elements. EMBO J. 1989;8:2381–2386. doi: 10.1002/j.1460-2075.1989.tb08367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff T, Rubin GM. strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development. 1998;125:1149–1159. doi: 10.1242/dev.125.6.1149. [DOI] [PubMed] [Google Scholar]
- 38.Strutt D, Johnson R, Cooper K, Bray S. Asymmetric localization of frizzled and the determination of Notch-dependent cell fate in the Drosophila eye. Curr Biol. 2002;12:813–24. doi: 10.1016/s0960-9822(02)00841-2. [DOI] [PubMed] [Google Scholar]
- 39.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development. 1997;124:2255–66. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- 41.Mirkovic I, Charish K, Gorski SM, McKnight K, Verheyen EM. Drosophila nemo is an essential gene involved in the regulation of programmed cell death. Mech Dev. 2002;119:9–20. doi: 10.1016/s0925-4773(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 42.Djiane A, Yogev S, Mlodzik M. The apical determinants aPKC and dPatj regulate Frizzled-dependent planar cell polarity in the Drosophila eye. Cell. 2005;121:621–31. doi: 10.1016/j.cell.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Klein TJ, Mlodzik M. Subcellular localization of frizzled receptors, mediated by their cytoplasmic tails, regulates signaling pathway specificity. PLoS Biol. 2004;2:1004–1014. doi: 10.1371/journal.pbio.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Courbard JR, Djiane A, Wu J, Mlodzik M. The apical/basal-polarity determinant Scribble cooperates with the PCP core factor Stbm/Vang and functions as one of its effectors. Dev Biol. 2009;333:67–77. doi: 10.1016/j.ydbio.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagafuchi A, Ishihara S, Tsukita S. The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin-alpha catenin fusion molecules. J Cell Biol. 1994;127:235–45. doi: 10.1083/jcb.127.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dumstrei K, Wang F, Shy D, Tepass U, Hartenstein V. Interaction between EGFR signaling and DE-cadherin during nervous system morphogenesis. Development. 2002;129:3983–94. doi: 10.1242/dev.129.17.3983. [DOI] [PubMed] [Google Scholar]
- 47.Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–71. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez-Gonzalez R, Simoes Sde M, Roper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell. 2009;17:736–43. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simoes Sde M, et al. Rho-Kinase Directs Bazooka/Par-3 Planar Polarity during Drosophila Axis Elongation. Dev Cell. 2010;19:377–88. doi: 10.1016/j.devcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shewan AM, et al. Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell-cell contacts. Mol Biol Cell. 2005;16:4531–42. doi: 10.1091/mbc.E05-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulrich F, et al. Slb/Wnt11 controls hypoblast cell migration and morphogenesis at the onset of zebrafish gastrulation. Development. 2003;130:5375–84. doi: 10.1242/dev.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorpe CJ, Moon RT. nemo-like kinase is an essential co-activator of Wnt signaling during early zebrafish development. Development. 2004;131:2899–909. doi: 10.1242/dev.01171. [DOI] [PubMed] [Google Scholar]
- 53.Ulrich F, et al. Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin. Dev Cell. 2005;9:555–64. doi: 10.1016/j.devcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 54.Jenny A, Reynolds-Kenneally J, Das G, Burnett M, Mlodzik M. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat Cell Biol. 2005;7:691–7. doi: 10.1038/ncb1271. [DOI] [PubMed] [Google Scholar]
- 55.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 56.Iwai Y, et al. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 57.Billuart P, Winter CG, Maresh A, Zhao X, Luo L. Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell. 2001;107:195–207. doi: 10.1016/s0092-8674(01)00522-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.