Abstract
After screening the Berkeley Drosophila Genome Project database with sequences from a recently characterized Leu-rich repeats-containing G protein-coupled receptor (LGR) from Drosophila (DLGR-1), we identified a second gene for a different LGR (DLGR-2) and cloned its cDNA. DLGR-2 is 1360 amino acid residues long and shows a striking structural homology with members of the glycoprotein hormone [thyroid-stimulating hormone (TSH); follicle-stimulating hormone (FSH); luteinizing hormone/choriogonadotropin (LH/CG)] receptor family from mammals and with two additional, recently identified mammalian orphan LGRs (LGR-4 and LGR-5). This homology includes the seven transmembrane region (e.g., 49% amino acid identity with the human TSH receptor) and the very large extracellular amino terminus. This amino terminus contains 18 Leu-rich repeats—in contrast with the 3 mammalian glycoprotein hormone receptors and DLGR-1 that contain 9 Leu-rich repeats, but resembling the mammalian LGR-4 and LGR-5 that each have 17 Leu-rich repeats in their amino termini. The DLGR-2 gene is >18.6 kb pairs long and contains 15 exons and 14 introns. Four intron positions coincide with the intron positions of the three mammalian glycoprotein hormone receptors and have the same intron phasing, showing that DLGR-2 is evolutionarily related to these mammalian receptors. The DLGR-2 gene is located at position 34E-F on the left arm of the second chromosome and is expressed in embryos and pupae but not in larvae and adult flies. Homozygous knock-out mutants, where the DLGR-2 gene is interrupted by a P element insertion, die around the time of hatching. This finding, together with the expression data, strongly suggests that DLGR-2 is exclusively involved in development.
[The nucleotide sequence(s) reported in this paper has been submitted to the GenBank/EMBL database with accession no. AF142343.]
Insects, which comprise 75% of all animal species, are of extreme ecological and economical importance, because 70% of all flowering plants depend on insects for their pollination and because insects can be severe pests, destroying about 30% of our potential annual harvest and transmitting major diseases such as malaria.
Despite the importance of insects, however, the molecular mechanisms controlling their reproduction are largely unknown. This is in contrast to the situation in mammals, where reproduction is known to be controlled by the hypothalamic neuropeptide GnRH (gonadotropin-releasing hormone), the pituitary glycoprotein hormones LH (luteinizing hormone) and FSH (follicle-stimulating hormone), and the gonadal steroid hormones.
Recently, using homology screening, we found that Drosophila produces a receptor that was structurally and evolutionarily related to the LH/FSH receptors from mammals (Hauser et al. 1997). This was an exciting finding, because it opened the possibility that insects use the same hormonal mechanisms as mammals to steer their reproduction. Subsequently, we cloned a receptor from Drosophila that was structurally and evolutionarily related to the GnRH receptors from mammals (Hauser et al. 1998), making the finding of a glycoprotein hormone receptor in Drosophila even more interesting.
Mammals have at least four glycoprotein hormones [LH, FSH, choriogonadotropin (CG), and thyroid-stimulating hormone (TSH)] and at least three glycoprotein hormone receptors (the LH/CG, FSH, and TSH receptors) that are all closely related, forming a subfamily of the large family of G protein-coupled (seven transmembrane) receptors. A characteristic of these glycoprotein hormone receptors is the presence of a very large, extracellular amino terminus that constitutes about half of the receptor protein and that contains 9 Leu-rich repeats, each measuring about 24 amino acid residues. These nine Leu-rich repeats probably form a horseshoe-like structure to which the glycoprotein hormone ligand binds (Jiang et al. 1995; Kajava et al. 1995). Similar Leu-rich repeats have been found in the Drosophila receptor (which is called Drosophila Leu-rich repeats-containing G protein-coupled receptor, or DLGR-1) (Hauser et al. 1997).
In addition to the three mammalian glycoprotein hormone receptors and DLGR-1, other LGRs have been cloned from sea anemones (Nothacker and Grimmelikhuijzen 1993), nematodes, and snails (Tensen et al. 1994). Based on the conserved sequences between the sea anemone and the Drosophila LGRs, two other LGRs (LGR-4 and LGR-5) were recently cloned from rat and man (Hsu et al. 1998). Similar receptors have been found by other groups (McDonald et al. 1998; Hermey et al. 1999). All of these novel invertebrate and mammalian LGRs are orphan receptors, that is, receptors for which the ligands are unknown.
In this paper we describe a second Drosophila receptor (DLGR-2) that is closely related to the three mammalian glycoprotein hormone receptors. Instead of 9 Leu-rich repeats, however, its amino terminus contains 18 Leu-rich sequences. We also describe the developmental expression of the DLGR-2 gene and the isolation of a knock-out mutant. These data suggest that DLGR-2 is exclusively involved in development.
RESULTS
Identification of the Drosophila Receptor Protein
In an attempt to find a second Drosophila LGR, we screened (Spring 1997) the database of the Berkeley Drosophila Genome Project with the sequences of various transmembrane helices of the first Drosophila LGR, using the tblastn program provided by the National Center for Biotechnology Information (NCBI). This resulted in the identification of two overlapping genomic clones, P1 DS00180 (GenBank accession no. AC001660) and P1 DS01514 (GenBank accession no. AC002515), containing open reading frames resembling the genes of the three mammalian glycoprotein hormone receptors (McFarland et al. 1989; Sprengel et al. 1990; Heckert et al. 1992; Koo et al. 1991; Tsai-Morris et al. 1991). Using GENESCAN software, we predicted the exons of this presumed Drosophila receptor gene and designed oligonucleotide probes that could be used in PCR to amplify the Drosophila receptor cDNA. After PCR and 5′- and 3′-RACE, we obtained the composite cDNA coding for the Drosophila receptor, which we named DLGR-2 (Fig. 1A,B). Subsequently, the presence of the actual full-length transcript was confirmed with several long-range PCR experiments (Fig. 1A).
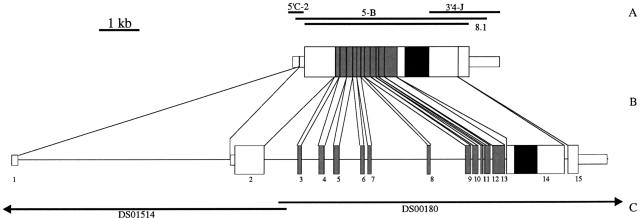
Figure 1.
Schematic representation of the DLGR-2 cDNA and genomic clones and the organization of the DLGR-2 gene. (A) Positions of the PCR clones. (B) Schematic drawing of the composite cDNA (top) and the organization of the receptor gene (bottom). The exons are given as bars and numbered 1–15. We named the introns after the preceeding exons(e.g., intron 1 follows exon 1). The narrow and broad bars represent noncoding and coding regions, respectively. The DNA region coding for the transmembrane domain is black and that coding for the Leu-rich repeats are gray. (C) The positions of the genomic P1 clones, DS00180 and DS01514, from the Berkeley Drosophila Genome Project.
The receptor cDNA is 5399 bp long and is shown in Figure 2. Its transcription start site was determined by 5′-RACE to be at nucleotide position −628 relative to the start codon, although some 5′-RACE products indicated an additional transcription start site at position −624. Similarly, the 3′-RACE experiments indicated various transcription termination points. The longest identified transcript extended 689 bp beyond the stop codon and the transcription stop at this point indicated the use of an imperfect polyadenylylation signal (Proudfoot 1991), AATTAA, at nucleotide positions 4735–4740 of Figure 2, that is, 32 bp upstream of the poly(A)+ tail. Other 3′-RACE experiments indicated additional transcription termination points at nucleotide positions 4753 and 4623 of Figure 2 and, hence, twice the use of another imperfect polyadenylylation signal, CATAAA. All putative polyadenylylation signals are underlined in Figure 2.
Figure 2.
cDNA and deduced amino acid sequence of DLGR-2. This figure is compiled from the sequences of the overlapping cDNA clones 5′C-2, 5B, and 3′4-J (Fig. 1A). Nucleotides are numbered from 5′ to 3′ end, and the amino acid residues are numbered starting with the first ATG in the open reading frame. Introns are indicated by arrows and numbered 1–14. The seven membrane-spanning domains are boxed and labeled TM I—VII. The proposed signal sequence is shaded. Spades indicate potential amino-glycosylation sites. The inframe stop codon in the 5′ region, upstream of the assigned start codon is underlined twice; the translation termination codon is indicated by an asterisk (*). Putative polyadenylylation sites at the 3′ end are underlined.
Although the flanking regions of the ATG codon at nucleotide positions 1–3 of Figure 2 do not fully match the Drosophila consensus sequence for a translation start site (Cavener 1987), it is probably the start codon, because there is an in-frame stop codon shortly upstream of this codon (at position −42 to −40 of Fig. 2). Furthermore, this potential start codon is followed by a series of nucleotides that code for the signal sequence, which probably is cleaved-off between amino acid positions 33 and 34 of Figure 2 (von Heijne 1986, 1990).
The cDNA of Figure 2 codes for a protein of 1360 amino acid residues, which corresponds to a molecular mass of 150.8 kD. Several potential amino glycosylation sites on the extracellular portion of the protein (seven on the amino terminus, but none on the extracellular loops; Fig. 2) suggest that the true mass might be considerably higher, although removal of the signal sequence will, of course, somewhat reduce the molecular mass of the mature protein.
The receptor protein has seven putative transmembrane domains (Fig. 2) characteristic for G protein-coupled receptors. The intracellular loops in between these transmembrane domains and the carboxy-terminal intracellular part of the protein, contain multiple serine and threonine residues, which are potential phosphorylation sites (e.g., Ser-1204 is part of the consensus sequence RRHS for A-kinase phosphorylation; see Kemp and Pearson 1990).
Comparison of DLGR-2 with Other Related Receptors
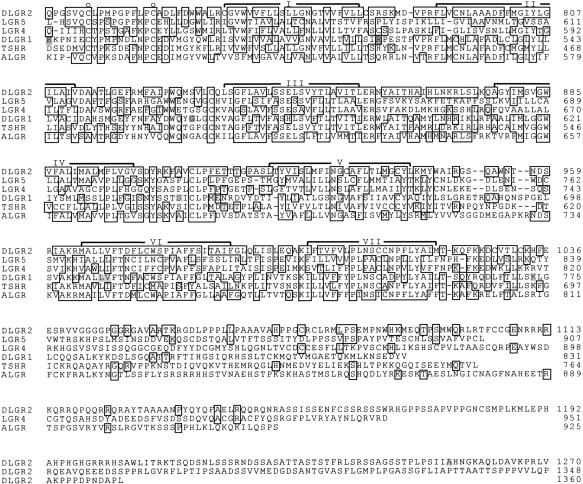
Figure 3 shows a comparison of the amino acid sequence of DLGR-2 with that of DLGR-1, the sea anemone LGR, the human TSH receptor, and the two newly discovered mammalian orphan receptors, LGR-4 and LGR-5. The transmembrane region of DLGR-2 (amino acid positions 758–1021 of Fig. 2) shows a high sequence identity with the human TSH receptor (49%), the sea anemone LGR (47%), and DLGR-1 (45%). Less sequence identity is found with LGR-4 and LGR-5 (30%–31%). When the region of the amino terminus of DLGR-2, containing the Leu-rich repeats (amino acid positions 205–617 of Fig. 2) is compared, however, most sequence identity is found with LGR-4 and LGR-5 (36%–37%), which is higher than when the transmembrane regions are compared. Less sequence identity is found between the Leu-rich repeats of DLGR-2 (amino acid positions 393–617 of Fig. 2) and the human TSH receptor (27%), DLGR-1 (25%), and the sea anemone LGR (22%). Furthermore, a long Gln-rich region of about 100 amino acid residues that is located shortly after the signal sequence is only found in DLGR-2 (Fig. 2).
Figure 3.
Amino acid sequence comparison between DLGR-2, LGR-5, LGR-4, DLGR-1, the human TSH receptor (TSHR), and the LGR from the sea anemone Anthopleura elegantissima (ALGR). Broken lines represent spaces introduced to optimize alignment. The Gly-rich repeats of ALGR (Nothacker and Grimmelikhuijzen 1993) were omitted to facilitate alignment. Amino acid residues that are identical between DLGR-2 and at least one of the other receptors are boxed. Known intron–exon transitions in the genes coding for the receptors are shaded at the corresponding amino acid residues. The positons of the aliphatic and aromatic residues characteristic for the Leu-rich repeats in DGLR-2 are marked by asterisks (*) or solid circles (●). The solid circles also mark intron–exon transitions in the DGLR-2 gene that occur at the same positions and have the same intron phasing as in the DLGR-1, TSHR, and ALGR genes. The seven membrane-spanning domains are indicated by I–VII. The open circles (○) mark conserved cystein residues, flanking the region containing the Leu-rich repeats. The amino acid residue positions are given at right. The amino acid sequences of DLGR-1 and the mammalian and sea anemone receptors as well as the intron–exon positions in their genes are from Misrahi et al. (1990); Gross et al. (1991); Nothacker and Grimmelikhuijzen (1993); Hauser et al. (1997); Hsu et al. (1998); and Vibede et al. (1998).
The amino terminus of DLGR-2 contains a region with 18 Leu (or Ile/Val/Ala/Phe)-rich repeats (marked by solid circles and asterisks in Fig. 3). This region is flanked by clusters of cysteine residues, which is a typical combination also found in other Leu-rich repeats-containing proteins (Kobe and Deisenhofer 1994, 1995). The aliphatic and aromatic residues of the Leu-rich repeats of DLGR-2 form a pattern that strongly resembles that of the other LGRs (Figs 3 and 4). Compared with the other LGRs, however, DLGR-2 has the largest number of these repeats, although it resembles LGR-4 and LGR-5, which have 17 repeats (Hsu et al. 1998).
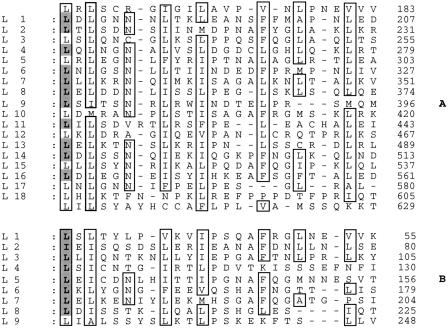
Figure 4.
Leu (Ile/Val/Ala/Phe)-rich repeats in the amino-terminal region of DLGR-2 and the rat LH receptor. (A) Consecutive segments (L1–L18) within the amino terminus of DLGR-2 were aligned and small gaps (–) were introduced to show that many of the aliphatic and aromatic residues in one segment occur at a similar position in the other segments. These residues are boxed. In addition, Asn residues at positions typical for Leu-rich repeats (Kobe and Deisenhofer 1994, 1995) are also marked. The shaded aliphatic residues correspond to intron–exon transitions in the receptor gene and are given at the start of the repeating segments. Most of the repeating segments, therefore, are coded for by distinct exons. Only complete segments, lying within the cluster of Cys residues bordering the Leu-rich repeats of Leu-rich repeats-containing proteins (Kobe and Deisenhofer 1994, 1995), are taken into account. However, 0.8 repeat flanking L1 and 0.3 repeat flanking L18 might contribute to an additional repeat (altogether 19 Leu-rich repeats) in DLGR-2. (B) Similar alignment of the rat LH receptor. Data from Bhowmick et al. (1996).
DLGR-2 is considerably larger than the other known LGRs. This is not only due to the large amino terminus, but also to the intracellular carboxyl terminus that is 340 amino acid residues in length, which is 2–4 times longer than the carboxyl termini of the other receptors of Figure 3. Homology screening with the receptor carboxyl terminus did not reveal the existence of other known proteins with resembling sequences.
Genomic Organization of the Drosophila Receptor Gene
The genomic organization of the gene coding for DLGR-2 is summarized in Figure 1B. Figure 1C shows the position of the gene on the genomic P1 clones DS00180 and DS01514 from the Berkeley Drosophila Genome Project. The total length of the transcribed portion of the gene is 18.6 kb. No conventional TATA box could be recognized on the P1 clones within a region of 1 kb upstream of the identified transcription start site.
The gene contains 14 introns and 15 exons. Four introns in the gene occur at exactly the same positions and have the same intron phasing (intron phase 2) as four introns in the rat TSH, FSH, and LH/CG receptor genes; the DLGR-1 gene; and the sea anemone LGR gene (indicated by solid circles in Fig. 3), strongly suggesting that these genes are evolutionarily related. The introns have a length ranging from 55 bp to more than 7 kb (Table 1), and most of them are located in the region coding for the receptor amino terminus, in such a way that each of the resulting exons codes for one or more Leu-rich repeats. There are no introns in the gene region coding for the seven-transmembrane domain, but there is one intron in the gene region coding for the long intracellular carboxyl terminus (Fig. 1B).
Table 1.
Intron/Exon Boundaries of the DGLR-2 Gene
| Intron | 5′ Donor | Intron size (bp) | 3′ Acceptor | Intron phase |
|---|---|---|---|---|
| 1 (variant1) | TTT gtaaggt.... | ∼7 kb | ....cccttag ACA | – |
| 1 (variant2) | AAG gtgccac.... | ∼7 kb | ....cccttag ACA | – |
| 1 (variant3) | TTG gtgccac.... | ∼7 kb | ....cccttag ACA | – |
| 1 (variant4) | TTT gtaaggt.... | ∼7 kb | ....taaatag ATA | – |
| 2 | CT gtgagta.... | 1223 bp | ....ccaacag A | 2 |
| Leu | Leu | |||
| 3 | CT gtgagta.... | 583 bp | ....tttgcag A | 2 |
| Leu | Leu | |||
| 4 | CT gtaagta.... | 325 bp | ....cacacag A | 2 |
| Leu | Leu | |||
| 5 | CT gtgagcc.... | 591 bp | ....tcgacag C | 2 |
| Leu | Leu | |||
| 6 | CT gtaagta.... | 74 bp | ....tttgcag T | 2 |
| Leu | Leu | |||
| 7 | TT gtaagta.... | 1747 bp | ....tttgcag A | 2 |
| Leu | Leu | |||
| 8 | CT gtaagta.... | 1083 bp | ....ccttcag T | 2 |
| Leu | Leu | |||
| 9 | CT gtgagta.... | 55 bp | ....ttttcag G | 2 |
| Leu | Leu | |||
| 10 | TT gtgagtt.... | 65 bp | ....actgcag A | 2 |
| Leu | Leu | |||
| 11 | TT gtaagtt.... | 57 bp | ....cacccag A | 2 |
| Leu | Leu | |||
| 12 | CT gtgcgtt.... | 61 bp | ....ttcacag T | 2 |
| Leu | Leu | |||
| 13 | C gtaagtg.... | 56 bp | ....tccccag AT | 1 |
| His | His | |||
| 14 | G gtaagtg.... | 57 bp | ....tgtacag CG | 1 |
| Ala | Ala |
The sequence of each of the intron-exon boundaries is shown, as well as the codons for the amino acid residues. Uppercase and lowercase letters represent nucleotides in the exons and introns, respectively. For intron 1, the four splicing variants that we have isolated are given. These variants differ from each other, depending on which 5′ donor and 3′ acceptor sites are used. The sequences of the introns are given in our GenBank/EMBL Database submission. The overall positions of the introns are shown in Figs. 1 and 2. Introns 9-12 occur at the same positions within the gene and have the same intron phasing as four introns in the genes of the mammalian glycoprotein hormone receptors (• in Fig. 3).
Alternative Splicing
During the analysis of several clones, containing the 5′ noncoding region of the receptor cDNA, four splicing variants were identified that all concerned exons 1 and 2 and intron 1. Three donor and two acceptor sites were found to exist (Fig. 5), which means that there are six possible transcripts in which the contributions from exon 1 can be either 45, 50, or 217 nucleotides and from exon 2 either 894 or 1037 nucleotides. Four of these transcripts have been identified (see legend to Fig. 5).
Figure 5.
Partial nucleotide sequence of the genomic DNA around intron 1. The numbers in this figure refer to the nucleotide positions of the cDNA of Fig. 2. Uppercase and lowercase letters represent the nucleotides in exons 1 and 2 and in intron 1, respectively. The three gt splicing donor sites and the two ag acceptor sites are underlined and printed in boldface type. The three donor and two acceptor sites give six possible mRNAs, of which four have been identified. The four identified mRNAs have the combinations d1/a1 (donor 1/acceptor 1), d1/a2, d2/a2, and d3/a2. Fig. 2 corresponds to d3/a2.
Sequence Deviations Between the Cloned Receptor cDNA and the Genomic Database DNA
The coding region of our cDNA (Fig. 2) is identical to the corresponding region in the genomic sequence from the Drosophila Genome Project, except for 36 nucleotides. Most of these substitutions do not lead to changes in the amino acid residues, but three of them do, two of them being conservative (Glu207 → Asp207 and Met446 → Leu446), whereas one is not (Pro409 → Ala409). The nucleotide substitutions within the coding region are given in Table 2. In the noncoding region of the cDNA, 16 nucleotide differences were identified. Most of these differences were replacements, but in the 3′ noncoding region, one nucleotide insertion (one within the stretch of Gs at nucleotide positions 4200–4205 of Fig. 2) and eight nucleotide deletions occurred (four TA sequences were deleted with in the repetitive TA sequence at nucleotide positions 4622–4636 of Fig. 2, resulting in the TA motif only being repeated 7 times compared with 11 times in the genomic sequence). In the 5′ noncoding region, the cDNA sequence contains one A less within the stretch of As starting at nucleotide position −573 of Figure 2. All above-mentioned sequence deviations are probably due to a small genetic difference (∼1%) between our own laboratory D. melanogaster Canton S. population and the one used in the Drosophila Genome Project. The differences are probably not due to PCR artefacts, because they were found in several independent cDNA clones.
Table 2.
Codon Differences Between the Coding Region of our Cloned cDNA and that of the Genomic Sequences from the Berkeley Drosophila Genome Project
| Position of the different nucleotides | Codon in the gene | Codon in the cDNA residue | Change in amino acid |
|---|---|---|---|
| 498 | CGG | CGA | – |
| 507 | GGT | GGG | – |
| 585 | GAG | GAA | – |
| 621 | GAG | GAT | Glu → Asp |
| 750 | CTT | CTC | – |
| 1017 | TCT | TCC | – |
| 1086 | CTG | CTA | – |
| 1209 | CCA | CCT | – |
| 1225 | CCA | GCA | Pro → Ala |
| 1336 | ATG | CTG | Met → Leu |
| 1806 | AGA | AGG | – |
| 1827 | TCG | TCT | – |
| 1854 | CTA | CTC | – |
| 2313 | AAC | AAT | – |
| 2337 | CTC | CTT | – |
| 2349 | TCA | TCT | – |
| 2418 | CTG | CTC | – |
| 2424 | ATT | ATC | – |
| 2451 | TTG | TTA | – |
| 3120 | GTC | GTT | – |
| 3198 | CCA | CCC | – |
| 3285 | ACA | ACG | – |
| 3399 | GCC | GCA | – |
| 3474 | AAT | AAC | – |
| 3534 | CCA | CCG | – |
| 3687 | GCC | GCG | – |
| 3711 | TTC | TTT | – |
| 3771 | AAC | AAT | – |
| 3855 | CGC | CGG | – |
| 3864 | GTA | GTC | – |
| 3867 | CGC | CGT | – |
| 3873 | CTA | CTG | – |
| 3981 | AGT | AGC | – |
| 4053 | CCT | CCA | – |
| 4059 | CCC | CCT | – |
| 4071 | GAT | GAC |
The position of the changed nucleotide (Fig. 2) is given in the first column, the affected codon in the genomic sequence in the second column, and the cDNA in the third column. Most nucleotide differences do not lead to a difference in amino acid residue (fourth column).
Chromosomal Localization
The chromosomal localization of the two P1 clones (Fig. 1C) have been determined by the Berkeley Drosophila Genome Project to be on chromosome 2L, position 34E-F (http://www.fruitfly.org/).
Southern Blot Analysis
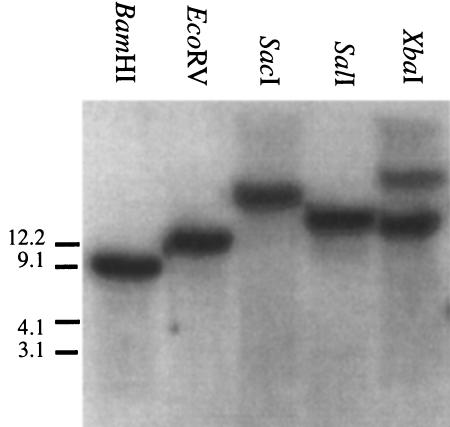
A Southern blot analysis, using a cDNA probe representing most of the coding region of the receptor (including the Leu-rich repeats 10–18 and the seven-transmembrane domain) and BamHI-, EcoRV-, SacI-, or SalI-digested fragments of Drosophila genomic DNA, showed single hybridizing bands (Fig. 6). The sizes of these single bands fully agreed with the genomic restriction maps of the P1 clones DS01514 and DS00180, suggesting that a single gene codes for DGLR-2. Digestion with XbaI yielded, in addition to the expected band of ∼11 kb, a slightly weaker hybridizing band of ∼30 kb (Fig. 6). This extra band might be due to partial digestion of the genomic DNA, or to genotypic variations, with the result that one of the XbaI sites in the DLGR-2 gene region is only present in part of our Drosophila population.
Figure 6.
Southern blot analysis. Genomic DNA from D. melanogaster Canton S. was digested with one of five restrictions enzymes (BamHI, EcoRV, SacI, SalI, and XbaI). After electrophoresis and blotting, the genomic fragments were hybridized with a cDNA fragment coding for the Leu-rich repeats 10–18 and the transmembrane domain of DLGR-2. The size of the markers (left) is in kb. All lanes show a single hybridization band with exception of the lane containing the XbaI fragment.
Developmental Regulation of the Drosophila Receptor
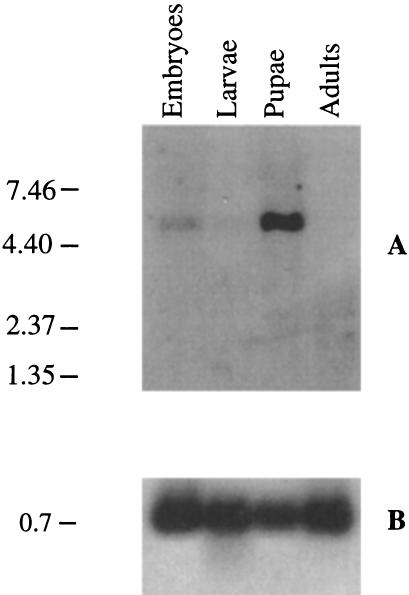
mRNA isolated from different developmental stages was analyzed in a Northern blot for the presence of DLGR-2 mRNA, using a cDNA probe coding for the seven-transmembrane domain. This showed that the receptor gene was only expressed in embryos and pupae, but not in larvae or adult flies (Fig. 7). The transcript size of ∼5.5 kb corresponded well with the size of the cloned receptor cDNA (Fig. 2). The blot was also hybridized with a probe coding for the ribosomal protein RP49 to check for uniform mRNA loading (Fig. 7).
Figure 7.
Northern blot analysis of the expression of the DLGR-2 gene at several developmental stages. Marker size (left) is given in kb. (A) Poly(A)+ RNA from each developmental stage was hybridized with a cDNA fragment coding for the seven-transmembrane domain of DLGR-2. This Northern blot shows that mRNA is only present in embryos and pupae, not in larvae and adult (mixed male and female) flies. (B) Hybridization of the same blot as in A with a cDNA probe coding for RP49. The RP49 gene is regarded to be expressed in all developmental stages (O'Connell and Rosbash 1984; Kerrebrock et al. 1995).
Isolation of a Knock-Out Mutant
We screened several Drosophila mutants from the Bloomington Stock Center (University of Indiana, Bloomington) that were known to have a P element insertion close to the chromosomal region of the DLGR-2 gene. This screening was performed using a P element-specific primer together with one of several gene-specific primers, covering the complete receptor sequence. One mutant (P919) had a P element inserted in exon 1 of the DLGR-2 gene (between nucleotide positions −584 and −583 of Fig. 2). In a cross between flies that were heterozygous for this mutation, about one-quarter of the offspring died around the time of hatching, suggesting that the mutation is homozygous lethal and that it roughly follows mendelian genetics (Table 3). This was confirmed by PCR, showing that all animals that were homozygous for the mutation indeed died around the time of hatching. On the other hand, all larvae that survived were either heterozygous or did not carry the mutation.
Table 3.
Distribution of Genotypes Among the Offspring from a Cross Between Male and Female Flies, Being Heterozygous for the P Element Insertion in Exon 1 of the DLGR-2 Gene
| Dead embryos | Third instar larvae | Total | |
|---|---|---|---|
| P/P | 23 | 0 | 23 |
| P/+ | 2 | 42 | 44 |
| +/+ | 1 | 20 | 21 |
Of 100 eggs collected from a cross, 26 animals died during embryonic development (including both unhatched eggs and larvae that died within 1 hr after hatching), 69 developed into third instar larvae. Five animals disappeared during the experiments. All animals were investigated by PCR to reveal their genotype. For seven of the third instar larvae, the PCR gave no products, so the genotype of these animals is unknown. The dead embryos of the genotype P/+ and +/+ could easily be distinguished from the P/P animals by their less-developed appearance. Dead P/P animals had the gross anatomy of normal, wild-type first-instar larvae. The crossing experiment shows that the mutation is homozygous lethal and that it roughly follows mendelian genetics.
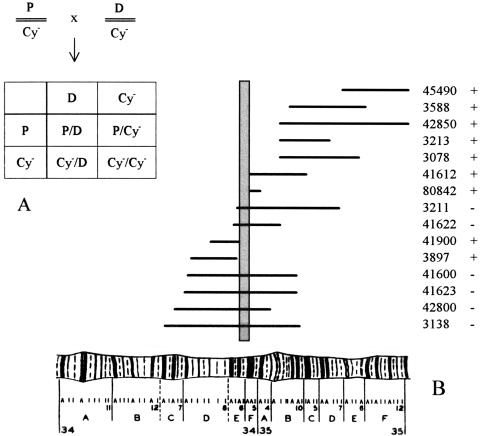
We also investigated whether the observed lethality was caused by the mutated DLGR-2 gene, or by an eventual second mutation located on the same chromosome, which, therefore, would always accompany the DLGR-gene during our crossing experiments. To establish this, we carried out classical complementation experiments (Fig. 8), where heterozygous DLGR-2 mutant flies (containing one chromosome 2 with the P element inserted into the DLGR-2 gene and the other chromosome 2 with the dominant CyO mutation, which gives the heterozygous flies a “curly wing” phenotype) were crossed with 15 different heterozygous mutants, carrying single, well-defined deletions close to or including the DLGR-2 gene region (these flies contained one chromosome 2 with a well-defined deletion and the other chromosome 2 without a deletion, but with the dominant CyO mutation). We found that the various deletion mutants could not be rescued by our P element insertion mutant P919 if their deletions included the chromosome 2L map coordinates 34E5-F1, which is exactly the region where the DLGR-2 gene is located. In fact, this independent genetic localization of the defect in mutant P919 is even more precise than the chromosomal localization carried out by the Berkeley Drosophila Genome Project of the two P1 clones, containing the DLGR-2 gene, which was 34E-F. These results, therefore, strongly indicate that the lethality found in our homozygous DLGR-2 gene mutants (Table 3) is indeed caused by a defect in the DLGR-2 gene itself.
Figure 8.
Results of crossing 15 different deficiency stains (carrying a well-defined deletion in an area of chromosome 2 close to or including the DLGR-2 gene, see Table 4) with mutant P919 that carries a P element insertion in the DLGR-2 gene. (A) Schematic description of the crossings. (P) The DLGR-2 gene, carrying the P element insertion; (Cy-) curly wing mutation; (D) deletion mutation. The Cy- mutation is dominant and homozygous lethal. Therefore, if the offspring only consists of curly wing flies (P/Cy- or Cy-/D), the P/D combination is nonviable and the deletion can not be rescued by the P element-inserted DLGR-2 region. On the other hand, if the offspring contains flies with normal flat wings (P/D), the deletion mutant can be rescued. (B) Map of the deletion strains used in the rescue experiments (horizontal lines). The abscissa shows the region 34A-35F of Drosophila chromosome 2L. The stock numbers of the mutants are given at right (ordinate), together with the information about whether the deletion mutant can be rescued (+) or not (−) in a cross with mutant fly P919. For stock numbers 45490 and 42850 only the left ends of the deletions are shown, as these deletions are very large. The vertical shaded bar indicates the borders of the chormosomal region determined by the inability of mutant P919 to rescue the deletion mutants. This region is 34E5-F1, which is exactly the region where the DLGR-2 gene has been located. This is an independent and strong indication that the P element that we have earlier shown to be inserted in the DLGR-2 gene, is the only cause of lethality in mutant P919.
When we investigated the gross anatomy of the homozygous DLGR-2 mutants around hatching, we found that the animals looked like fully developed first instar larvae, having normal segmentation, tracheal trees, mouth hooks, denticle belts, and no obvious defects in the gastrointestinal tract.
DISCUSSION
Mammals have at least three glycoprotein hormone receptors (the TSH, FSH, and LH/CG receptors) and two other LGRs for which the ligands yet are unknown (Hsu et al. 1998; McDonald et al. 1998; Hermey et al. 1999). It was, therefore, interesting to find that Drosophila also contains at least two LGRs. The first Drosophila LGR (DLGR-1) is both structurally and evolutionarily closely related to the mammalian glycoprotein hormone receptors (Hauser et al. 1997). The same is true of DLGR-2 for a number of reasons: (1) There is 49% amino acid residue identity between its seven-transmembrane domain and that of the human TSH receptor (Fig. 3). This identity is high, if one takes into account that the sequence identity between the seven-transmembrane domains of the mammalian TSH, FSH, and LH/CG receptors is only 67%–70% (Salesse et al. 1991); (2) the amino terminus has Leu-rich repeats, similar to those of the mammalian glycoprotein hormone receptors (Fig. 4); and (3) four introns in its gene occur at the same positions and have the same intron phasing as four introns in the mammalian glycoprotein hormone receptors.
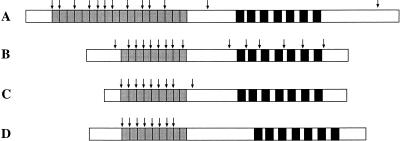
DLGR-2 is not more related to DLGR-1 than to the mammalian glycoprotein hormone receptors. This is evident from both the sequence of its transmembrane domain (45% amino acid residue identity between the two Drosophila receptors), the unusual large number of Leu-rich repeats in its extracellular amino terminus (18 in DLGR-2 and 9 in DLGR-1; Fig. 9), and its genomic organization (no introns present in the gene region coding for the seven-transmembrane domain of DLGR-2 and four introns in the corresponding region of DLGR-1; Fig. 9).
Figure 9.
Schematic representation of four LGR cDNAs. The regions coding for the seven-transmembrane loops are given as black bars, those for the Leu-rich repeats as gray bars. Only complete Leu-rich repeats (see Fig. 4) are taken into account. Intron positions in the four LGR genes are indicated by arrows. (A) The cDNA coding for DGLR-2. (B) The cDNA coding for DLGR-1 (Hauser et al. 1997). (C) The cDNA coding for the rat FSH receptor (Heckert et al. 1992). (D) The cDNA coding for the LGR from the sea anemone A. elegantissima (Nothacker and Grimmelikhuijzen 1993; Vibede et al. 1998).
The genomic organization of the two Drosophila receptor genes gives some interesting information. Previously, it has been assumed that the glycoprotein hormone receptor genes originated from exon shuffling of an intronless gene region coding for the seven-transmembrane domain and an intron-rich region coding for the Leu-rich repeats and the rest of receptor amino terminus (Gross et al. 1991; Tsai-Morris et al. 1991; Heckert et al. 1992). The gene structures of all mammalian glycoprotein hormone receptors (Fig. 9C) and the gene structure of the sea anemone LGR, which should closely resemble that of the ancestral LGR gene (Vibede et al. 1998; Fig 9D) are in accordance with this idea. Also, the gene structure of DLGR-2 fits the original idea of the evolution of the LGRs (Fig. 9A). The gene structure of DLGR-1, however, conflicts with this model, because it contains four introns in its region coding for the seven-transmembrane domain (Fig. 9B). If one accepts the hypothesis that the number of introns tends to decrease, instead of increase during eukaryote evolution (Gilbert et al. 1986), then both DLGR-1 and DLGR-2 would have originated from a single gene containing at least four exons in its region coding for the seven-transmembrane domain. The same would be true for the mammalian and the sea anemone receptors (Vibede et al. 1998; Fig. 9). Thus, Drosophila contains at least two classes of LGR genes, one class (to which the DLGR-1 gene belongs) would be more closely related to the ancestral gene, whereas the other class (to which the DLGR-2 gene belongs) would have lost most of its introns in the region coding for the transmembrane domain, thereby resembling the known genes of the other LGRs.
DLGR-2 is considerably larger than the mammalian TSH, FSH, and LH/CG receptors (Fig. 3 and Fig. 9; Salesse et al. 1991). This is mainly due to the presence of extra Leu-rich repeats in the amino terminus and the presence of a large intracellular carboxyl terminus (Fig. 9A). The extra Leu-rich repeats have probably originated by exon shuffling, because they are all (in single or multiple copies) represented by separate exons in the gene (Fig. 1B). The length of the receptor amino terminus and the number and amino acid sequence of its Leu-rich repeats place DLGR-2 structurally closer to the two mammalian orphan receptors LGR-4 and LGR-5 than to the three mammalian glycoprotein hormone receptors. When the transmembrane regions are compared, however, DLGR-2 is more closely related to the mammalian TSH, FSH, and LH/CG receptors (∼50% sequence identity). DLGR-2, therefore, appears to be a naturally occurring chimaera, with its amino terminus more closely resembling LGR-4 and LGR-5 and its transmembrane region more closely resembling the three mammalian glycoprotein hormone receptors.
The Leu-rich repeats of the mammalian glycoprotein hormone receptors probably form a horseshoe-like structure to which the glycoprotein hormone binds at the inner, concave side (Jiang et al. 1995; Kajava et al. 1995). We assume that the Leu-rich repeats of DLGR-2 also form such a horseshoe, because these repeats very closely resemble those of the mammalian receptors (Fig. 4). Because DLGR-2 has twice as many Leu-rich repeats as the mammalian glycoprotein hormone receptors, we assume that the horseshoe is correspondingly larger. This might indicate that the ligand is somewhat different from the mammalian glycoprotein hormones.
Two of the three mammalian glycoprotein hormone receptors are involved in reproduction. Therefore, we monitored the expression of the DLGR-2 gene during several developmental stages of Drosophila to see whether it was expressed in adult male or female flies. We found, however, that the receptor was only expressed in embryos and pupae, that is, in stages where there are active cell division, cell differentiation, and other forms of development but not in the three larval stages of Drosophila or adult male or female flies (Fig. 7). This exclusive expression of DLGR-2 in embryos and pupae already points to an important role for the receptor in development. This idea was confirmed when we investigated the phenotype of the homozygous knock-out mutants of the DLGR-2 gene. These mutant flies have a P element insertion in exon 1, which is the noncoding 5′ region of the DLGR-2 gene, meaning that the homozygous mutants are devoid of DLGR-2 (because the P element contains a gene, including stop codons; Hazelrigg et al. 1984). We observed that all mutants died around the time of hatching. It is interesting that the homozygous mutants had the overall appearence of normal larvae. This means that DLGR-2 does not play a role in morphogenesis but rather is important in a more subtle developmental process. This process must be absent in hatched first-, second-, and third-instar larvae and in adult flies, because the receptor is not expressed in these stages. This exclusive role of an LGR in development is unique and has not been described previously for the other known glycoprotein hormone receptors.
METHODS
Animals
Wild-type Drosophila melanogaster Canton S. were reared under standard conditions (Roberts 1986).
Database Screening
Amino acid residues 703–728 of DLGR-1 (Hauser et al. 1997) were used as a probe for the electronic screening of the Berkeley Drosophila Gene Project database, using the NCBI Search Engine BLAST (Altschul et al. 1990). The same search engine was used for homology screening.
Preparation of Poly(A)+ RNA and cDNA Synthesis
Poly(A)+ RNA from various Drosophila stages was purified with the Oligotex Direct mRNA Kit from Qiagen. Oligo(dT)-primed cDNA was synthesized from 0.4 μg poly(A)+ RNA, as recommended by the manufacturer, using the RT-PCR Kit (Stratagene) and an oligo (dT) primer supplied with the kit.
PCR
The PCR reactions used for cloning of the cDNA were carried out in a 50 μl volume as described in Sambrook et al. (1989), with the exception that a final concentration of 1.75 mm MgCl2 was used. The template was 5 μl of a first-strand cDNA reaction mixture. Various primers were used, based on the predicted exon sequences of the DLGR-2 gene (see Results). The PCR products were separated on 2% agarose gel, and bands of the expected size were isolated (Qiaquick extraction kit, Qiagen), subcloned into PCR 2.1 with the Original TA Cloning Kit (Invitrogen), and sequenced.
The final full-length products were generated by use of the Expand Long Template PCR System (Boehringer Mannheim). Two independent PCR reactions were carried out. In each of these PCRs, the 50-μl reaction mixture consisted of Buffer System 1, 2 mm final concentration of MgCl2, 500 μm of each dNTP, 0.3 μm of each primer (corresponding to positions −560 to −537 and 4661 to 4682 of Fig. 2 for the first reaction, and positions −1 to 21 and 4068 to 4087 for the second reaction), 275 mU of Long Range Polymerase, and cDNA from Drosophila embryos as a template. Cycling parameters were 2 min of initial denaturation at 94°C; 10 cycles of the following step program: 94°C for 15 sec, 60°C for 30 sec, 68°C for 4 min; then 25 cycles of 94°C for 15 sec, 60°C for 30 sec, 68°C for 4 min. This last 4-min period was increased with 20 sec for every new cycle.
3′-RACE
First-strand cDNA synthesis was performed according to the protocol for the 5′/3′-RACE Kit (Boehringer Mannheim), using 2 μg of poly(A)+ RNA from 16–24-hr-old Drosophila embryos as a template, and the oligo d(T) primer from the kit. This cDNA was used directly for PCR amplification with a sense DLGR-2-specific primer (positions 2985 to 3006 from Fig. 2) in combination with a PCR anchor primer supplied with the kit. A second round of PCR using 2 μl of the first PCR reaction as a template and employing the anchor primer and a second DLGR-2-specific sense primer (positions 3013 to 3033 of Fig. 2) further to the 3′ end, was necessary to obtain a specific PCR product. The reaction mixture was in both cases as recommended by the manufacturer. Cycling parameters were as follows 3 min of initial denaturation at 95°C; 10 cycles of 95°C for 30 sec, 62°C for 45 sec, 72°C for 2 min. The reactions were held at 72°C while the anchor primer was added, and then further 25 cycles were run as described above.
5′-RACE
For the first strand cDNA synthesis, 2 μg of poly(A)+ RNA from 16–24-hour-old embryos and a DLGR-2-specific antisense primer (position 1149 to 1170 of Fig. 2) were used. 5′-RACE PCR was carried out using the 5′/3′-RACE Kit (Boehringer Mannheim), following the instructions of the manufacturer, and employing two nested antisense primers (positions 837 to 858 and −318 to −297 of Fig. 2).
DNA Sequencing and Sequence Analysis
DNA sequences were determined by the chain termination method (Sanger et al. 1977), using the T7 Sequenase Version 2.0 DNA Sequencing Kit (Amersham Life Science). GC-rich sequences were determined, using the Thermo Sequenase Radiolabeled Terminator Cycle Sequencing Kit (Amersham Life Science). DNA sequence compilation and nucleotide and amino acid sequence comparisons were performed using the Lasergene DNA Software (DNASTAR Inc.).
Promotor Analysis
The genomic sequence was searched for promotor sequences up to 1 kb upstream of the identified transcription start site, employing the TSSG promoter search engine at http://dot.imgen.bcm.tmc.edu:9331/genefinder/gf.html.
Prediction of Gene Structures in Genomic DNA
To predict exons, introns, and polyadenylylation signals, the GENSCAN web server at http://stanford.edu/∼chis/GENSCAN.html was used.
Radioactive Labeling of DNA Probes
DNA fragments to be labeled were excised from vector DNA by restriction enzymes and purified by agarose gel electrophoresis. Probes were labeled with [α-32P]dCTP (3000 Ci/mmole) from Amersham by use of the Ready-To-Go DNA Labeling Beads (Pharmacia Biotech), according to the manual.
Northern Blot Analysis
Poly(A)+ (2.5 μg) from various stages of Drosophila (obtained from Clontech or purified as described above) were electrophoresed on a gel containing 1% agarose and 0.22 m formaldehyde. The 0.24–9.5-kb RNA Ladder (GIBCO-BRL) was included as a size marker. RNA was capillary transferred onto ZetaProbe membranes (BioRad) and cross-linked as recommended by the manufacturer. Hybridization was carried out as recommended by BioRad for 18 hr at 65°C in the presence of heat-denatured radioactive probe in a final concentration af 1–2 × 106 cpm/ml. This probe corresponded to nucleotide positions 1806 to 3204 of Figure 2. The ribosomal protein 49 (RP49) probe was prepared as described in Hauser et al. (1997).
Southern Blot Analysis
Genomic DNA (10 μg) from D. melanogaster was digested with one of the restriction enzymes, BamHI, EcoRV, SacI, SalI, or XbaI, and separated on a 0.7% agarose gel. DNA was capillary transferred to a Hybond-N nylon membrane, hybridized, and washed as recommended by the supplier (Amersham). The radioactive probe corresponded to nucleotide positions 1149 to 3083 of Figure 2.
Isolation of a Knock-Out Mutant
Various Drosophila mutants from the Bloomington Stock Center were screened by PCR for a P element insertion in the DLGR-2 gene. This screen was performed using a P element-specific primer, 5′-CGACGGGACCACCTTATGTTATTTCATCATG-3′, along with various DLGR-2 gene-specific primers, covering the complete receptor sequence. For one mutant, P919 with the genotype w[1118]; P{w+[tAR] ry[+t7.2AR]=wA[R]}4-34/CyO, a 1200-bp PCR product was seen, using the exon 1-specific primer 5′-GGCTGTGCCGACAATTGAAC-3′ together with the P element-specific primer. The location of the P element in exon 1 of the receptor gene (between nucleotide positions −584 and −583 of Fig. 2), was confirmed by sequencing the PCR product with the primer 5′-GAAATCTCTGTGCCACTG-3′.
To obtain homozygous mutants, male P919 flies were crossed with female virgin Oregon R. flies to get P{w[+tAR] ry[+t7.2AR]=wA[R]}4-34/+ flies. Mating between flies of this last genotype gave the results of Table 3. To investigate the presence of a P-element insertion in the offspring of Table 3, DNA was extracted from single animals by grinding the animal in 100μl 5% chelex-100 (Bio-Rad Laboratories) in an Eppendorf tube (Sweet et al. 1996). The tube was then incubated for 30 min at 56°C and 8 min at 100°C. After a 3-min centrifugation at 13000 rpm, the supernatant was transferred to a clean Eppendorf tube and stored at −20°C. DNA extract (5–10 μl) was used in a PCR reaction, using the primers 5′-GAGCATAACCCTCTTCTTGT-3′ and 5′-TGATCTGGGAGTTTGGAGTG-3′, that spans a 564-bp region in intron 1 of the receptor gene. For unknown reasons, the P919 strain has an insertion of ∼184 bp in intron 1 (2120 bp downstream of the transcription start) of that allele that bears the P-element insertion, whereas in the allele that does not bear the P-element insertion, this 184-bp insertion is lacking. Because our two primers lie at each side of the 184-bp insertion, an animal that is homozygous for the P-element insertion will give a PCR product of 751 bp, whereas a homozygous wildtype will give a PCR product of 567 bp and a heterozygous animal both PCR products.
Complementation Experiments
The deficiency strains of Drosophila that were used in our complementation experiments of Figure 8 are given in Table 4. The deficiencies in the heterozygous flies were balanced with the CyO chromosome (see also legend to Fig. 8). After crossing, we verified by PCR that the Cy+ offspring (flat wings; P/D, see Fig. 8A) did indeed carry the wild type DLGR-2 gene (D) together with the P element-inserted DLGR-2 gene (P).
Table 4.
Deficiency Strains of Drosophila Used in the Complementation Experiments of Figure 8 to Independently Localize the Deficiency in Mutant P919, which Carries a P Element Insertion in the DLGR-2 Gene
| Stock number | Deficient chromosomal region |
|---|---|
| 45490 | 35E1-2 to 36A6-7 |
| 3588 | 35B4-6 to 35F1-7 |
| 42850 | 35B3 to 38D3-5 |
| 3213 | 35B2-3 to 35D5-7 |
| 3078 | 35B1-3 to 35E6 |
| 41612 | 34E5-F1 to 35C3-9 |
| 80842 | 34F1-2 to 35A2 |
| 3211 | 34E3 to 35D7 |
| 41622 | 34E1-2 to 35B3-5 |
| 41900 | 34D4-6 to 34E5-6 |
| 3897 | 34D2 to 34E3 |
| 41600 | 34D1-2 to 35B9-C1 |
| 41623 | 34C6-7 to 35B9-C1 |
| 42800 | 34C4 to 35A4 |
| 3138 | 34B12-C1 to 35B10-C1 |
Stock numbers with five digits are from the Umeå Drosophila Stock Center (Umeå University, Sweden), those with four digits from the Bloomington Drosophila Stock Center (Indiana University, Bloomington). The deficient chromosomal region refers to the left arm of chromosome 2.
Two deficiency strains from the Umeå Drosophila Stock Center (42450 and 41624) could be rescued by mutant P919 (which carries a P-element insertion in the DLGR-2 gene), despite the fact that their records indicated a deletion in the DLGR-2 gene area. PCR tests of the Cy+ offspring (see Fig. 8A) demonstrated that these flies carried a wild-type DLGR-2 gene together with the P element-inserted DLGR-2 gene, showing that the two deficiency strains either have been mapped incorrectly or have lost their deficient chromosomes. These two strains were, therefore, withdrawn from the complementation experiments.
Acknowledgments
This work was supported by the Danish Biotechnological Research and Development Programme of the Danish Research Agency, Læge- og Naturvidenskabelige Komité of the Novo Nordisk Foundation and Carlsberg Foundation.We thank Lotte Steffensen for typing the manuscript and for help with Fig. 3.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cgrimmelikhuijzen@zi.ku.dk; FAX 0045 35 32 12 00.
REFERENCES
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bhowmick N, Huang JN, Puett D, Isaacs NW, Lapthorn AJ. Determination of residues important in hormone binding to the extracellular domain of the luteinizing hormone/chorionic gonadotropin receptor by site-directed mutagenesis and modeling. Mol Endocrinol. 1996;10:1147–1159. doi: 10.1210/mend.10.9.8885249. [DOI] [PubMed] [Google Scholar]
- Cavener D. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Marchionni M, McKnight G. On the antiquity of introns. Cell. 1986;46:151–154. doi: 10.1016/0092-8674(86)90730-0. [DOI] [PubMed] [Google Scholar]
- Gross B, Misrahi M, Sar S, Milgrom E. Composite structure of the human thyrotropin receptor gene. Biochem Biophys Res Commun. 1991;177:679–687. doi: 10.1016/0006-291x(91)91842-z. [DOI] [PubMed] [Google Scholar]
- Hauser F, Nothacker H-P, Grimmelikhuijzen CJP. Molecular cloning, genomic organization and developmental regulation of a novel receptor from Drosophila melanogaster structurally related to members of the thyroid-stimulating hormone, follicle-stimulating hormone, luteinizing hormone/choriogonadotropin receptor family from mammals. J Biol Chem. 1997;272:1002–1010. doi: 10.1074/jbc.272.2.1002. [DOI] [PubMed] [Google Scholar]
- Hauser F, Søndergaard L, Grimmelikhuijzen CJP. Molecular cloning, genomic organization and developmental regulation of a novel receptor from Drosophila melanogaster structurally related to gonadotropin-releasing hormone receptors from vertebrates. Biochem Biophys Res Commun. 1998;249:822–828. doi: 10.1006/bbrc.1998.9230. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T, Levis R, Rubin GM. Transformation of white locus DNA in Drosophila: Dosage compensation, zeste interaction, and position effects. Cell. 1984;36:469–481. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- Heckert LL, Daley IJ, Griswold MD. Structural organization of the follicle-stimulating hormone receptor gene. Mol Endocrinol. 1992;6:70–80. doi: 10.1210/mend.6.1.1738373. [DOI] [PubMed] [Google Scholar]
- Hermey G, Methner A, Schaller HC, Hermans-Borgmeyer I. Identification of a novel seven-transmembrane receptor with homology to glycoprotein receptors and its expression in the adult and developing mouse. Biochem Biophys Res Commun. 1999;254:273–279. doi: 10.1006/bbrc.1998.9882. [DOI] [PubMed] [Google Scholar]
- Hsu SY, Liang S-G, Hsueh AJW. Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol Endocrinol. 1998;12:1830–1845. doi: 10.1210/mend.12.12.0211. [DOI] [PubMed] [Google Scholar]
- Jiang X, Dreano M, Buckler DR, Cheng S, Ythier A, Wu H, Hendrickson WA, El Tayar N. Structural predictions for the ligand-binding region of glycoprotein hormone receptors and the nature of hormone-receptor interactions. Structure. 1995;3:1341–1353. doi: 10.1016/s0969-2126(01)00272-6. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Vassart G, Wodak SJ. Modeling of the three-dimensional structure of proteins with the typical leucine-rich repeats. Structure. 1995;3:867–877. doi: 10.1016/S0969-2126(01)00222-2. [DOI] [PubMed] [Google Scholar]
- Kemp BE, Pearson RB. Protein-kinase recognition sequence motifs. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat—A versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- ————— Proteins with leucine-rich repeats. Curr Opinion Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- Koo YB, Ji I, Slaughter RG, Ji TH. Structure of the luteinizing hormone receptor gene and multiple exons of the coding sequence. Endocrinology. 1991;128:2297–2308. doi: 10.1210/endo-128-5-2297. [DOI] [PubMed] [Google Scholar]
- McFarland KC, Sprengel R, Phillips HS, Köhler M, Rosemblit N, Nikolics K, Segaloff DL, Seeburg PH. Lutropin-choriogonadotropin receptor: An unusual member of the G protein-coupled receptor family. Science. 1989;245:494–499. doi: 10.1126/science.2502842. [DOI] [PubMed] [Google Scholar]
- McDonald T, Wang R, Bailey W, Xie G, Chen F, Caskey CT, Liu Q. Identification and cloning of an orphan G protein-coupled receptor of the glycoprotein hormone receptor subfamily. Biochem Biophys Res Commun. 1998;247:266–270. doi: 10.1006/bbrc.1998.8774. [DOI] [PubMed] [Google Scholar]
- Misrahi M, Loosfelt H, Atger M, Sar S, Guiochon-Mantel A, Milgrom E. Cloning, sequencing and expression of human TSH receptor. Biochem Biophys Res Commun. 1990;166:394–403. doi: 10.1016/0006-291x(90)91958-u. [DOI] [PubMed] [Google Scholar]
- Nothacker H-P, Grimmelikhuijzen CJP. Molecular cloning of a novel, putative G protein-coupled receptor from sea anemones structurally related to members of the FSH, TSH, LH/CG receptor family from mammals. Biochem Biophys Res Commun. 1993;197:1062–1069. doi: 10.1006/bbrc.1993.2586. [DOI] [PubMed] [Google Scholar]
- O'Connell PO, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A+) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Roberts DB. Drosophila—A practical approach. Oxford, UK: IRL Press; 1986. [Google Scholar]
- Salesse R, Remy JJ, Levin JM, Jallal B, Garnier J. Towards understanding the glycoprotein hormone receptors. Biochimie. 1991;73:109–120. doi: 10.1016/0300-9084(91)90083-d. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-termination inhibitiors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengel R, Braun T, Nikolics K, Segaloff DL, Seeburg PH. The testicular receptor for follicle stimulating hormone: Structure and functional expression of cloned cDNA. Mol Endocrinol. 1990;4:525–530. doi: 10.1210/mend-4-4-525. [DOI] [PubMed] [Google Scholar]
- Sweet D, Lorente M, Valenzuela A, Lorente JA, Alvarez JC. Increasing DNA extraction yield from saliva stains with a modified Chelex method. Forensic Sci Int. 1996;83:167–177. doi: 10.1016/s0379-0738(96)02034-8. [DOI] [PubMed] [Google Scholar]
- Tensen CP, van Kesteren ER, Planta RJ, Cox KJA, Burke JF, van Heerikhuizen H, Vreugdenhil E. A G protein-coupled receptor with low density lipoprotein-binding motifs suggests a role for lipoproteins in G-linked signal transduction. Proc Natl Acad Sci USA. 1994;91:4816–4820. doi: 10.1073/pnas.91.11.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai-Morris CH, Buczko E, Wang W, Xie X-Z, Dufau ML. Structural organization of the rat luteinizing hormone (LH) receptor gene. J Biol Chem. 1991;266:11355–11359. [PubMed] [Google Scholar]
- Vibede N, Hauser F, Williamson M, Grimmelikhuijzen CJP. Genomic organization of a receptor from sea anemones, structurally and evolutionarily related to glycoprotein hormone receptors from mammals. Biochem Biophys Res Commun. 1998;252:497–501. doi: 10.1006/bbrc.1998.9661. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The signal peptide. J Membrane Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]