Abstract
The promyelocytic leukemia (PML) protein is the core component of nuclear substructures that host more than 70 proteins, termed nuclear domains 10 or PML-nuclear bodies. PML was first identified as the gene participating in the translocation responsible for the pathogenesis of acute promyelocytic leukemia (APL). The notion that PML is a tumor suppressor gene was soon extrapolated from leukemia to solid tumors. The last decade has radically changed the view of how this tumor suppressor is regulated, how it can be therapeutically targeted, and how it functions. Notably, one of the most recent and striking features uncovered is how PML regulates cellular homeostasis outside its original niche in the nucleus. These new findings open an exciting new area of research in extra-nuclear PML functions.
The identification of PML and the APL saga
The 90s started with a breakthrough discovery from several groups that would change the research of the deadly acute promyelocytic leukemia (APL): the mapping of the breakpoint of the reciprocal translocation with chromosome 17 [1]. Soon after, the promyelocytic leukemia gene (PML, MYL, RNF71, PP8675, and TRIM19) was identified as the most frequent translocation partner of the retinoic acid receptor alpha (RARα in APL) [2–5] (Figure 1).
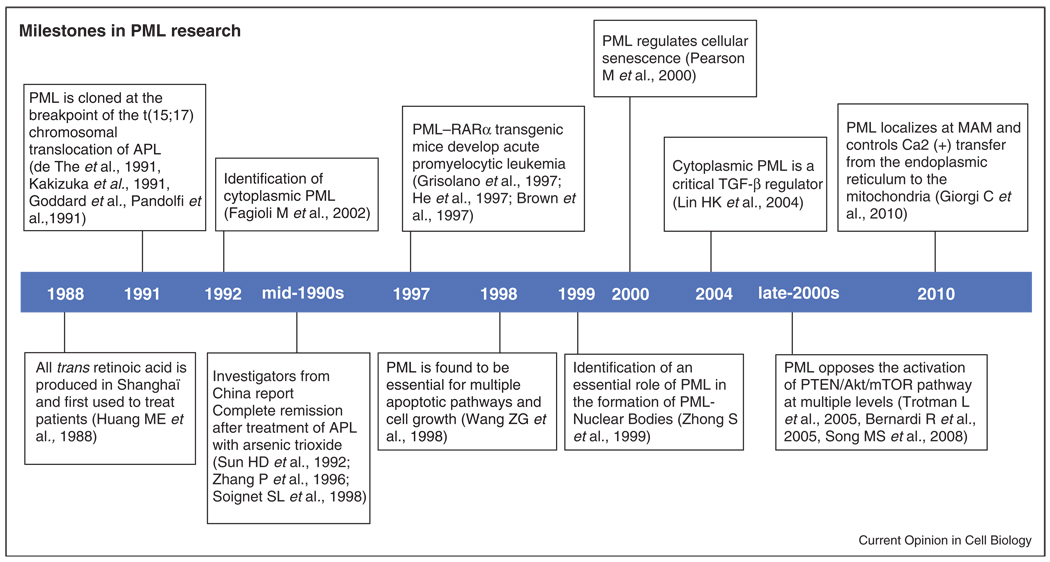
Figure 1.
Milestones in PML research from its discovery in the beginning of the 90s.
Since its discovery and for the next five years, the tumor suppressive activity of PML was restricted to leukemia, but was soon extrapolated to solid tumors [6,7]. This led to the current notion that PML is a tumor suppressor lost in cancers from multiple origins [8].
One of the most relevant breakthroughs in PML research was the discovery of two drugs which would target PML and/or the PML–RARα fusion oncoprotein. On the one hand, All-Trans Retinoic Acid (ATRA), produced in Shangai and used to treat patients with APL [9], was found to promote the rapid degradation of the fusion protein [10]. On the other hand, two groups described that same year the potential mechanism of arsenic trioxide (ATO), an ancient Chinese medicine used for the treatment of APL [11–13], inducing apoptosis with the concomitant regulation of PML localization and stability [14,15]. This phenomenon would be, a decade later, shown to depend on the binding of the arsenic molecule to PML to promote its SUMOylation-dependent ubiquitin- mediated degradation by RNF4 [16••,17••,18••].
The notion that the PML–RARα fusion oncoprotein is sufficient to drive APL was recapitulated in the mouse by several groups [19–21], in turn generating preclinical tools which would later become key in the discovery of an effective therapy for APL. These preclinical efforts in faithful mouse models of APL proved definitive game changer in the treatment of this deadly disease. Genetic mouse models of APL were critical for the formal demonstration of the combined efficacy of ATRA and ATO in APL [22,23]. In turn, these seminal studies would transform a devastating disease in a curable one.
PML nuclear functions
PML is the essential component of a macromolecular nuclear substructure, the PML-nuclear bodies (PML-NB [24]). Indeed, PML functions as the scaffold of this structure allowing other proteins to shuttle in and out, a process which is regulated by SUMO-mediated modifications and interactions [25,26•]. PML has multiple splicing variants, giving rise to a wide variety of isoforms, whose differential expression and function is yet poorly understood [27,28]. These PML isoforms move dynamically between NBs at a different rate, thus suggesting that the PML-NB composition might be heterogeneous and functionally different [29].
Among the recently discovered nuclear functions of PML, we will focus on the control of gene expression and protein modification, which have become of great interest.
PML and transcription
PML-NBs host a wide variety of transcriptional regulators, including transcriptional activators, repressors, and histone modifiers. Moreover, PML-NBs have been shown to localize adjacent to transcriptionally active chromatin regions, to Major Histocompatibility Class 1 and p53 loci [30–33]. Furthermore, PML colocalizes with the histone acetyltransferase Creb Binding Protein (CBP) and with RNA Pol II in a cell cycle-dependent manner [34]. On the other hand, PML can repress transcription through the interaction with histone deacetylases and heterochromatin protein 1 (HP1) [35,36] and through the regulation of heterochromatin recondensation in satellite DNA [37]. The transcriptional regulation and heterochromatin remodeling induced by PML have highlighted the importance of this tumor suppressor in the induction of cellular senescence [38,39], with recent evidence pointing at a role for PML in regulating this process through the Rb-E2F pathway [40].
PML and post-translational modifications
PML-NBs are a factory for protein modifications. It has been shown that PML regulates protein acetylation, phosphorylation, ubiquitination, and SUMOylation among others, through the correct formation of the PML-NBs. These structures host all kinds of protein modifiers, from acetyltransferases to deacetylases, E3 ligases, deubiquitinases, phosphatases, kinases, and more. These regulatory processes have been shown to be of critical importance for cell homeostasis, and in turn, loss of PML results in deregulated modulation of protein function.
Firstly, PML is found to modulate the activity of both protein phosphatases and kinases. On the one hand, PML tunes the function of the protein phosphatases 1A and 2A (PP1A and PP2A). Loss of Pml results in increased proliferation and reduced differentiation of neural progenitors [41•], an event that arises from the delocalization of PP1A and reduced activity towards pRb. On the other hand, PML positively regulates the activity of PP2A towards AKT in the PML-NBs, and, as a result, Pml-loss exacerbates the phenotype of Pten heterozygous mice and leads to more aggressive forms of cancer [42]. Furthermore, PML regulates the activity of several kinases to promote protein phosphorylation. As an example, PML promotes the phosphorylation of p53 by HIPK2, which increases the rate of acetylation of p53 and its transcriptional activation [43].
Secondly, PML regulates protein stability and function by affecting the activity of E3-ligases and deubiquitinases (DUBs). PML inhibits KLHL2 through its recruitment to the PML-NBs and the physical separation from its target, DAPK, which results in apoptosis and autophagy [44]. Also, PML has been shown to recruitMDM2 to a distinct subnuclear structure, the nucleolus, hence preventing p53 ubiquitination and proteosomal degradation [45]. Conversely, PML negatively regulates the DUB HAUSP/USP7, thus opposing PTEN deubiquitination and cytoplasmic translocation. This molecular framework is found altered in APL, where PML–RARα disrupts the PML-NBs, increases PTEN deubiquitination by HAUSP, and leads to PTEN nuclear exclusion [46].
Lastly, the PML nuclear bodies have a predominant role in the regulation of protein acetylation. PML activates p53-dependent gene expression during oncogene-induced senescence by promoting its acetylation by CBP in the PML-NBs [47]. The regulation of p53 by PML is a balance between the acetylation by CBP and the deacetylation by SIRT1, which also resides in the PML-NBs. Indeed, SIRT1 overexpression tilts the equilibrium towards p53 deacetylation and transcriptional repression, which results in the impairment of the cellular senescence program [48]. Interestingly, PML–RARα exerts the opposite activity on p53, by promoting its deacetylation by a different family of deacetylases, HDACs [49].
The numerous functions described for PML in the control of post-translational modifications raise the question of whether the nuclear bodies serve the main function of providing a microenvironment where these reactions can occur. It remains to be clarified whether other activities of PML attributed to a scaffolding function (e.g., regulation of mTOR by PML [50,51•]) are also the result of the modulation of post-translational modification pathways.
PML cytoplasmic functions
As we mentioned above, a nuclear localization signal (NLS) located in exon 6 of PML isoforms restricts the localization of the majority of PML protein to the nucleus. However, nucleo-cytoplasmic fractionation reveals that a fraction of PML resides in the cytoplasm [28,52,53••], a 10% of total PML in asynchronously growing primary mouse cells, and probably a higher percentage in other cell types (A.C. and P.P.P., unpublished observations). Of note, a PML isoform, namely PML-VII, lacks the fraction encoding for the NLS and it is therefore purely cytoplasmic (reviewed in [54]).
Functions of PML-VII in the cytoplasm
The first report to unveil a cytoplasmic function of PML came upon the study of the activity of PML-VII [52]. PML-VII or PMLc (cytoplasmic) was found to be essential for the proper activation of the cytokine transforming growth factor beta (TGFβ) signaling. In the absence of PML (using Pml knockout mouse embryonic fibroblasts) the intracellular signaling elicited by TGFβ receptor activation was diminished, and in line with this notion, these cells were refractory to the induction of growth arrest, senescence, and apoptosis by the cytokine. This effect was shown to be mediated by the interaction of PMLc with SMAD2/3 and Smad anchor for receptor activation (SARA), and the regulation of their cytoplasmic compartmentalization. In addition, the activation of TGFβ signaling by PMLc is negatively regulated by the homeodomain protein TGIF (TG-interacting factor), which disrupts its association with SARA in concert with c-Jun [55]. Further research will be needed in order to determine whether this activity of PMLc is also exerted by other PML isoforms in the cytoplasm (Figure 2).
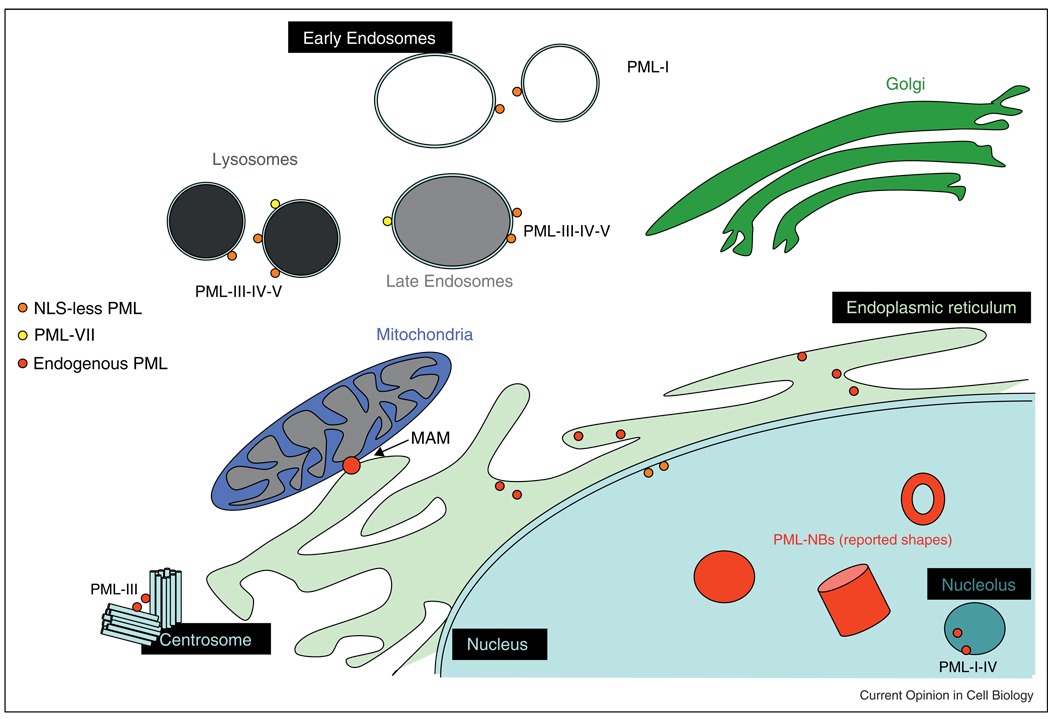
Figure 2.
Current view of PML compartmentalization in the cell. Nuclear PML in PML-NBs (with suggested shapes and forms) and cytoplasmic PML localization.
Although most of the research related to cytoplasmic PML has been linked to the use of PML-VII, in the recent years, more data have been brought up to light regarding the function of other PML isoforms and mutants in this compartment.
Tumor suppressive activities of cytoplasmic PML
As we mentioned before, all PML isoforms can be found in the cytoplasm upon subcellular fractionation analysis. One of the most exciting new roles of this cytosolic PML pool was unveiled when carefully evaluating the precise localization of cytoplasmic PML fraction [53••]. Surprisingly, PML was found to localize to specific membrane structures termed mitochondria-associated membranes, or MAMs for short, which connect the endoplasmic reticulum (ER) and the mitochondria. The MAMs play a central role in apoptosis through the regulation of calcium influx from the ER to the mitochondria. Indeed, Pml-deficient cells exhibited impaired calcium influx to the mitochondria and resistance to ER-stress-induced cell death. In line with the notion that PML in the cytoplasm could form structures that host proteins also found in the PML-NB [56], PML was found to exert this proapoptotic activity through the regulation of a large complex involving PP2A, AKT, and the inositol triphosphate receptor (IP3R), which ultimately controls calcium flux to the mitochondria. To formally demonstrate the relevance of cytoplasmic PML fraction in this activity, the resistance to apoptosis of Pml-deficient cells was reverted upon expression of an ER-targeted PML construct. These results provide an exciting new potential mechanism whereby cytoplasmic PML could regulate apoptosis from MAM in parallel with the modulation of proapoptotic factors by nuclear PML [57].
Oncogenic activities of cytoplasmic PML mutants
The localization of PML to the nucleus requires a functional NLS located within exon 6. In this sense, genetic mutations affecting this domain might result in dysfunctional PML. Indeed, two independent studies described that mutations in PML leading to NLS-less PML mutants have oncogenic consequences. On the one hand, in a plasmacytoma cell linePML was found to be mutated in exon 3, which resulted in the expression of a truncated protein confined to the cytoplasm [58]. Surprisingly, this cytoplasmic PML functions as a dominant negative, oncogenic form of the tumor suppressor. On the other hand, in two aggressive forms of PML–RARα APL, the nontranslocated moiety of PML was found mutated [59]. Both mutations led to the expression of a cytoplasmic PML mutant. Interestingly, these mutants form cytoplasmic PML bodies (PML-CB) and host some of the components found in the PML-NB. Indeed, PML-CB was able to bind PML–RARα, sequester it in the cytoplasm, further inhibit RA-dependent transcription, and potentiate the block in differentiation in APL [56]. These reports raise the interesting possibility that an aberrant PML in the cytoplasm could function as an oncogenic cue. However, it remains to be determined whether an aberrant cytoplasmic PML is oncogenic through its activity on nuclear or cytoplasmic PML physiological functions.
Cytoplasmic PML: what next?
The view of how this multitasking protein operates has dramatically changed with the most recent developments, attributing to PML extra-nuclear oncogenic and tumor suppressive activities. However, the field is just in its infancy. As an example, a recent interesting study analyzed systematically the sites of PML cytoplasmic localization by expressing all PML isoforms harboring mutations in the NLS. Surprisingly, many of PML mutant isoforms localized to endosomes and lysosomes [60••]. This study, along with the various mentioned in this review raises new and exciting questions in need of answers: Where else does PML localize in the cytoplasm and what does it do there? The notion that PML could reside or shuttle between these compartments opens the possibility that, as in the MAMs and the nucleus, this protein could be regulating the localization, post-translational modification, and function of residents of ‘PML-related bodies’. How is PML nucleocytoplasmic shuttling regulated? PML presents multiple phosphorylation and acetylation sites surrounding the NLS [61,62] and it is therefore plausible that these modifications alter the accessibility to the NLS and in turn the compartmentalization of PML. Two decades of PML research has therefore identified multiple new unexpected lines of research to be pursued. On the basis of this premise, the next decade of research will certainly change further our view of this mysterious and exciting protein.
Acknowledgements
We thank all members of the Pandolfi laboratory for comments and discussion. We apologize to those whose publication related to the discussed issues could not be cited due to space limitations. A.C. was supported by the Ramón y Cajal award from the Spanish Ministry of Education. K.I. was supported by a K99 NIH grant.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Longo L, Donti E, Mencarelli A, Avanzi G, Pegoraro L, Alimena G, Tabilio A, Venti G, Grignani F, Pelicci PG. Mapping of chromosome 17 breakpoints in acute myeloid leukemias. Oncogene. 1990;5:1557–1563. [PubMed] [Google Scholar]
- 2.Pandolfi PP, Grignani F, Alcalay M, Mencarelli A, Biondi A, LoCoco F, Pelicci PG. Structure and origin of the acute promyelocytic leukemia myl/RAR alpha cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- 3.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML–RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 4.Kakizuka A, Miller WH, Jr, Umesono K, Warrell RP, Jr, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 5.Goddard AD, Borrow J, Freemont PS, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 6.Koken MH, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thepot J, Juhlin L, Degos L, Calvo F, de The H. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- 7.Liu JH, Mu ZM, Chang KS. PML suppresses oncogenic transformation of NIH/3T3 cells by activated neu. J Exp Med. 1995;181:1965–1973. doi: 10.1084/jem.181.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- 9.Huang ME. Treatment of acute promyelocytic leukemia with all-trans retinoic acid. Zhonghua Yi Xue Za Zhi. 1988;68:131–133. [PubMed] [Google Scholar]
- 10.Raelson JV, Nervi C, Rosenauer A, Benedetti L, Monczak Y, Pearson M, Pelicci PG, Miller WH., Jr The PML/RAR alpha oncoprotein is a direct molecular target of retinoic acid in acute promyelocytic leukemia cells. Blood. 1996;88:2826–2832. [PubMed] [Google Scholar]
- 11.Sun HD, Ma L, Hu XC, Zhang TD. Ai-Lin I treated 32 cases of acute promyelocytic leukemia. Chin J Integrat Chin West Med. 1992:12. [Google Scholar]
- 12.Zhang P, Wang SY, Hu LH, Shi FD, Qiu FQ, Hong GJ, Han XY, Yang HF, Sun YZ, Liu YP, et al. Arsenic trioxide treated 72 cases of acute promyelocytic leukemia. Chin J Hematol. 1996:2. [Google Scholar]
- 13.Soignet SL, Maslak P, Wang ZG, Jhanwar S, Calleja E, Dardashti LJ, Corso D, DeBlasio A, Gabrilove J, Scheinberg DA, et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med. 1998;339:1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 14.Chen GQ, Zhu J, Shi XG, Ni JH, Zhong HJ, Si GY, Jin XL, Tang W, Li XS, Xong SM, et al. In vitro studies on cellular and molecular mechanisms of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia: As2O3 induces NB4 cell apoptosis with downregulation of Bcl-2 expression and modulation of PML-RAR alpha/PML proteins. Blood. 1996;88:1052–1061. [PubMed] [Google Scholar]
- 15.Andre C, Guillemin MC, Zhu J, Koken MH, Quignon F, Herve L, Chelbi-Alix MK, Dhumeaux D, Wang ZY, Degos L, et al. The PML and PML/RARalpha domains: from autoimmunity to molecular oncology and from retinoic acid to arsenic. Exp Cell Res. 1996;229:253–260. doi: 10.1006/excr.1996.0368. [DOI] [PubMed] [Google Scholar]
- 16. Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. The studies by Tatham et al. and Lallemand-Breitenbach et al. [17••] define the biological mechanism leading to the degradation of PML and PML-RARa by the action of RNF4, a SUMO-dependent E3 ligase. RNF4 deficiency results in the accumulation of PML with mixed SUMO and poliubiquitinated chains, along with resistance to Arsenic trioxide-mediated PML degradation (see also [17••]).
- 17. Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML–RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. The studies by Lallemand-Breintebach et al. and Tatham et al. [16••] define the biological mechanism leading to the degradation of PML and PML–RARα by the action of a SUMO-dependent E3 ligase, RNF4. Upon arsenic trioxide treatment, RNF4 localizes to the PML nuclear bodies, together with ubiquitin and proteasome components. RNF4 promotes SUMO-mediated PML ubiquitination and degradation. Importantly, a SUMOylation-deficient PML is resistant to the effect of arsenic trioxide, and APL cells expressing a nondegradable SUMO-deficient mutant PML–RARα are resistant to the prodifferentiation activity of arsenic trioxide.
- 18. Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, et al. Arsenic trioxide controls the fate of the PML–RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. The authors deconstruct the molecular mechanism of PML and PML–RARα regulation by arsenic trioxide. They show that the compound binds to cysteines within the RBCC domain of PML, in turn promoting oligomerization, enhanced SUMOylation and degradation.
- 19.Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML–RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 20.He LZ, Tribioli C, Rivi R, Peruzzi D, Pelicci PG, Soares V, Cattoretti G, Pandolfi PP. Acute leukemia with promyelocytic features in PML/RARalpha transgenic mice. Proc Natl Acad Sci U S A. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci PG, Atwater S, Bishop JM. A PMLRARalpha transgene initiates murine acute promyelocytic leukemia. Proc Natl Acad Sci U S A. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lallemand-Breitenbach V, Guillemin MC, Janin A, Daniel MT, Degos L, Kogan SC, Bishop JM, de The H. Retinoic acid and arsenic synergize to eradicate leukemic cells in a mouse model of acute promyelocytic leukemia. J Exp Med. 1999;189:1043–1052. doi: 10.1084/jem.189.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rego EM, He LZ, Warrell RP, Jr, Wang ZG, Pandolfi PP. Retinoic acid (RA) and As2O3 treatment in transgenic models of acute promyelocytic leukemia (APL) unravel the distinct nature of the leukemogenic process induced by the PML–RARalpha and PLZF–RARalpha oncoproteins. Proc Natl Acad Sci U S A. 2000;97:10173–10178. doi: 10.1073/pnas.180290497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- 25.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lang M, Jegou T, Chung I, Richter K, Munch S, Udvarhelyi A, Cremer C, Hemmerich P, Engelhardt J, Hell SW, et al. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci. 2010;123:392–400. doi: 10.1242/jcs.053496. Lang et al. utilize 4Pi fluorescence laser-scanning microscopy analysis to ascertain the three-dimensional structure of the PML nuclear bodies. They define these structures as subcompartments that allow for the exchange of components with the nucleoplasm, therefore explaining the critical function of the nuclear bodies as modulators and centers for the concentration of biochemical activities and biological modules.
- 27.Fagioli M, Alcalay M, Pandolfi PP, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci PG. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene. 1992;7:1083–1091. [PubMed] [Google Scholar]
- 28.Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, de The H. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 2006;66:6192–6198. doi: 10.1158/0008-5472.CAN-05-3792. [DOI] [PubMed] [Google Scholar]
- 29.Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. 2008;121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiels C, Islam SA, Vatcheva R, Sasieni P, Sternberg MJ, Freemont PS, Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- 32.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Durrin LK, Krontiris TG. Specific interaction of PML bodies with the TP53 locus in Jurkat interphase nuclei. Genomics. 2003;82:250–252. doi: 10.1016/s0888-7543(03)00075-2. [DOI] [PubMed] [Google Scholar]
- 34.Kiesslich A, von Mikecz A, Hemmerich P. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J Struct Biol. 2002;140:167–179. doi: 10.1016/s1047-8477(02)00571-3. [DOI] [PubMed] [Google Scholar]
- 35.Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci U S A. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu WS, Vallian S, Seto E, Yang WM, Edmondson D, Roth S, Chang KS. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol Cell Biol. 2001;21:2259–2268. doi: 10.1128/MCB.21.7.2259-2268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luciani JJ, Depetris D, Usson Y, Metzler-Guillemain C, Mignon-Ravix C, Mitchell MJ, Megarbane A, Sarda P, Sirma H, Moncla A, et al. PML nuclear bodies are highly organised DNA–protein structures with a function in heterochromatin remodelling at the G2 phase. J Cell Sci. 2006;119:2518–2531. doi: 10.1242/jcs.02965. [DOI] [PubMed] [Google Scholar]
- 38.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 39.Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD. Definition of pRB- and p53-dependent and - independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol. 2007;27:2452–2465. doi: 10.1128/MCB.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, Mes-Masson AM, Ferbeyre G. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011;25:41–50. doi: 10.1101/gad.1975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Regad T, Bellodi C, Nicotera P, Salomoni P. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat Neurosci. 2009;12:132–140. doi: 10.1038/nn.2251. The authors unravel a function for PML in the regulation of neural progenitor function. They identify PML as a regulator of Retinoblastoma protein (pRb) activity, an event that depends on PML-mediated dephosphorylation of pRb by PP1. In turn, Pml-deficient mice exhibit reduced thickening of the cortex wall and impared transition of neural progenitors from radial glia to basal progenitors.
- 42.Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441:523–527. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 44.Lee YR, Yuan WC, Ho HC, Chen CH, Shih HM, Chen RH. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 2010;29:1748–1761. doi: 10.1038/emboj.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 46.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 48.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Insinga A, Monestiroli S, Ronzoni S, Carbone R, Pearson M, Pruneri G, Viale G, Appella E, Pelicci P, Minucci S. Impairment of p53 acetylation, stability and function by an oncogenic transcription factor. EMBO J. 2004;23:1144–1154. doi: 10.1038/sj.emboj.7600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Simon MC, Rafii S, Pandolfi PP. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- 51. Bernardi R, Papa A, Egia A, Coltella N, Teruya-Feldstein J, Signoretti S, Pandolfi PP. Pml represses tumour progression through inhibition of mTOR. EMBO Mol Med. 2011 doi: 10.1002/emmm.201100130. Bernardi et al. define the contribution of PML to tumor initiation and progression in the kidney, along with the crosstalk between PML and mTOR regulatory cues. Through the analysis of Pml-Tsc2 compound mutants, the authors show that Pml-loss results in exacerbated mTOR signaling in the kidney, and that this effect impacts in tumor progression but not in initiation.
- 52.Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 53. Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M, Bononi A, Bonora M, Duszynski J, Bernardi R, et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 2010;330:1247–1251. doi: 10.1126/science.1189157. This important study defines a proapoptotic activity for the tumor suppressor PML which is exerted from the endoplasmic reticulum–mitochondria contact regions (mitochondria-associated membranes or MAMs). PML in the MAMs regulates calcium influx to the mitochondria through the modulation of a complex comprising PP2A, AKT, and the IP3 receptor.
- 54.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 55.Seo SR, Ferrand N, Faresse N, Prunier C, Abecassis L, Pessah M, Bourgeade MF, Atfi A. Nuclear retention of the tumor suppressor cPML by the homeodomain protein TGIF restricts TGF-beta signaling. Mol Cell. 2006;23:547–559. doi: 10.1016/j.molcel.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 56.Bellodi C, Kindle K, Bernassola F, Dinsdale D, Cossarizza A, Melino G, Heery D, Salomoni P. Cytoplasmic function of mutant promyelocytic leukemia (PML) and PML–retinoic acid receptor-alpha. J Biol Chem. 2006;281:14465–14473. doi: 10.1074/jbc.M600457200. [DOI] [PubMed] [Google Scholar]
- 57.Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 58.Zheng P, Guo Y, Niu Q, Levy DE, Dyck JA, Lu S, Sheiman LA, Liu Y. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]
- 59.Gurrieri C, Nafa K, Merghoub T, Bernardi R, Capodieci P, Biondi A, Nimer S, Douer D, Cordon-Cardo C, Gallagher R, et al. Mutations of the PML tumor suppressor gene in acute promyelocytic leukemia. Blood. 2004;103:2358–2362. doi: 10.1182/blood-2003-07-2200. [DOI] [PubMed] [Google Scholar]
- 60. Jul-Larsen A, Grudic A, Bjerkvig R, Boe SO. Subcellular distribution of nuclear import-defective isoforms of the promyelocytic leukemia protein. BMC Mol Biol. 2010;11:89. doi: 10.1186/1471-2199-11-89. This elegant study evaluates the cytoplasmic localization of PML isoforms through the expression of nuclear localization signal-mutant PML constructs. The authors define the endosome–lysosome organelles as potential targets for the activity of PML in the cytoplasm.
- 61.Hayakawa F, Abe A, Kitabayashi I, Pandolfi PP, Naoe T. Acetylation of PML is involved in histone deacetylase inhibitor-mediated apoptosis. J Biol Chem. 2008;283:24420–24425. doi: 10.1074/jbc.M802217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reineke EL, Lam M, Liu Q, Liu Y, Stanya KJ, Chang KS, Means AR, Kao HY. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol Cell Biol. 2008;28:997–1006. doi: 10.1128/MCB.01848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]