Abstract
Technical advances in recent years, such as laser microirradiation and chromatin immunoprecipitation, have led to further understanding of DNA damage responses and repair processes as they happen in vivo and have allowed us to better evaluate the activities of new factors at damage sites. Facilitated by these tools, recent studies identified the unexpected roles of heterochromatin factors in DNA damage recognition and repair, which also involves poly(ADP-ribose) polymerases (PARPs). The results suggest that chromatin at damage sites may be quite structurally dynamic during the repair process, with transient intervals of “closed” configurations prior to a more “open” arrangement that allows the repair machinery to access damaged DNA.
Introduction
Genome integrity is continually threatened by endogenous metabolic products generated during normal cellular respiration, by errors that arise during DNA replication and recombination, and by exogenous exposure to DNA damaging agents. The resulting DNA lesions, if not faithfully repaired, can accumulate as mutations ranging from single nucleotide changes to chromosomal rearrangements and loss that can lead to cancer, developmental abnormalities, and cell death. Various types of DNA damage are constantly occurring in the cell. Different insults to DNA are recognized by lesion-specific repair factors, which invoke distinct repair pathways including nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), and double-strand break (DSB) repair. The factors most critical for DNA repair, as well as the major players in the DNA damage response (DDR), have been largely identified through genetics-based studies using model organisms or through investigation of human diseases (see the recent comprehensive review on DDR and repair [1••]). However, how their activities are coordinated in the cell nucleus is still not well understood. In vivo, DNA is organized in the form of chromatin by interacting with histones and other factors, and it is widely acknowledged that regulation of chromatin structure is of paramount importance for DNA repair. Recent studies, taking advantage of the experimental tools that allow in vivo analysis of the cellular response at damage sites, revealed the previously unrecognized roles of chromatin factors in DNA repair. In this review, we briefly discuss these tools and summarize recent unexpected findings on chromatin regulation in DNA damage signaling and repair in vivo, with particular emphasis on the damage-induced recruitment of heterochromatin factors and the expanding role of poly(ADP-ribose) polymerase (PARP) activity.
Biochemical and cytological analyses of cellular responses at in vivo damage sites
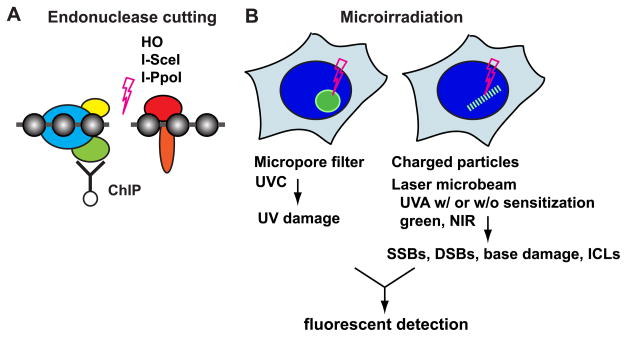
Chromatin immunoprecipitation (ChIP) analysis at endonuclease-induced DSB sites and cytological analysis of microirradiation-induced damage constitute two powerful tools to probe protein recruitment and/or modifications at locally induced DNA lesions in the cell nucleus (Fig. 1). These methods are particularly useful for those factors that do not form irradiation-induced foci (IRIF) [2,3•]. DSB sites for ChIP [4] can be introduced at specific sites in the genome by the HO mating-type switch endonuclease in yeast and the I-SceI or I-PpoI endonucleases in mammalian cells (Fig. 1A) [5–8]. The cytological methods include the partial exposure of cells to ultraviolet C (UVC) irradiation using a micropore filter to study UV damage as well as microirradiation of the cell nucleus using charged particles or highly focused optical lasers (such as UVA, green, and near-infrared (NIR)) to study primarily DSB repair (Fig. 1B) [2,3•,9–12]. In addition, interstrand crosslinking damage (ICLs) can be induced at specific subnuclear regions using photo-reactive psoralen derivatives combined with microirradiation [13••]. These cytological methods are particularly useful for the spatio-temporal kinetic studies of DDRs by quantitative fluorescence microscopy analyses, including fluorescence resonance energy transfer (FRET), fluorescent loss in photobleaching (FLIP), and fluorescent recovery after photobleaching (FRAP) (for example, [14•,15•,16•,17••]).
Figure 1. In vivo tools to study DNA damage recognition and response pathways.
A. ChIP analysis of endonuclease-induced DSBs. The HO endonuclease in yeast, and the I-SceI and I-PpoI homing endonucleases in mammalian cells, have been used to induce DSBs. While HO and I-PpoI cut endogenous recognition sequences, the I-SceI recognition sequence is introduced into the genome by stable transfection. Factor binding or modification in the vicinity of DSB sites can be analyzed by ChIP using specific PCR primers.
B. Fluorescent microscopy analysis following nuclear microirradiation. Selective sub-nuclear exposure of UVC using micropore filters has been used to study the NER pathway. Focused laser microbeams (of various wavelengths, energies, etc.) or charged particles, delivered to sub-micron regions in the cell nucleus (with or without DNA sensitization), have been used mainly to study DSBs. However, other types of damage are also induced, including CPD, SSBs, and in some cases, base damage. In addition, photo-activation of psoralen derivatives by microirradiation induces interstrand crosslinking damage (ICLs).
While the endonucleases induce only DSBs, microirradiation induces complex DNA damage, including crosslinking damage (e.g., cyclobutane pyrimidine dimers (CPD)), base damage, and both single strand breaks (SSBs) and DSBs, which may result in different DDRs. DSBs induced by ionizing radiation (IR), which is known to induce complex DNA damage, and those induced by endonucleases are indeed processed differently [18]. Furthermore, individual optical laser systems, depending on system parameters, can induce different mixtures of DNA damage, thus leading to different DDRs. For example, TRF2 recruitment to damage sites was observed only with certain laser systems but not others [19–21]. This is possibly due to distinct features of the particular induced DSBs and/or the co-induction of other types of damage that may dictate the cellular response. Thus, the methods used to induce DSBs may affect the DDR and repair pathway choice. Although this obviously requires further investigation, these systems have proven nonetheless to be powerful tools for demonstrating the damage site-specific molecular events in vivo, and have greatly facilitated discoveries of the chromatin organization events at damage sites discussed below.
Chromatin relaxation at DNA damage sites
It is widely accepted that chromatin near the damage site should be in an “opened” state for efficient repair factor recruitment and subsequent repair. The damage-site recruitment of ATP-dependent chromatin remodeling complexes, such as INO80 and Trrap/Tip60, leads to chromatin relaxation important for efficient DDR and/or repair [22]. It was shown that Trrap/Tip60 increases acetylation of histone H4 in the vicinity of the damage sites [6]. Indeed, DNA damage regions induced by UVA laser microirradiation with Hoechst 33342 were found to expand and decrease chromatin density, consistent with chromatin opening [23]. Depletion of the heterochromatin-binding transcription repressor proteins HP1 and KAP1 (also known as TIF1β KRIP-1, or TRIM28) as well as the Class I histone deacetylases 1 and 2 (HDAC1/2) alleviates the requirement for ATM in DNA repair, indicating that damage sites within heterochromatin require special processing for proper repair [24•]. Both HP1 and KAP1 are phosphorylated in response to DNA damage. HP1 phosphorylation weakens its affinity for the heterochromatic histone modification (i.e., the methylated lysine 9 residue of histone H3 (H3K9me)), and KAP1 phosphorylation results in increased micrococcal nuclease accessibility [14•,25]. All of these studies support a model in which chromatin relaxation is an essential early step in DNA damage repair. However, several recent studies have identified PARP-regulated damage site recruitment of multiple factors that are well-described players in transcriptional repression/heterochromatin formation, providing a different view on chromatin structural alterations at DNA lesions.
The role of PARP1 in chromatin relaxation at DNA damage sites
PARP1 is a DNA nick sensor activated by DNA damage that functions in BER and DSB repair [26–28]. Activated PARP1 not only (ADP-ribosyl)ates its targets but also itself, resulting in its dissociation from DNA [26,29]. Laser-induced damage enabled visualization of poly(ADP-ribose) (PAR) at, and in some cases the rapid and transient recruitment of PARP1 itself to, damage sites [29–31]. Histone H1, as well as all of the core histones, was shown to be (ADP-ribosyl)ated in vivo [32]. However, the identity of the major target(s) of PARP1, other than itself, at the damage sites is unclear.
PARP activity in the presence of NAD+ leads to decondensation of chromatin in vitro [33,34], and is required for gene activation-associated puffing of Drosophila polytene chromosomes in vivo [35]. This was thought to be mediated by (ADP-ribosyl)ation of histones. Thus, activated PARP1 recruitment to DNA damage sites was thought to induce chromatin opening [26,28]. Several recent studies have shown, however, that PAR at the damage sites also serves as an important binding site for the recruitment of various factors. A number of DNA repair/checkpoint proteins were found to contain a novel PAR-binding zing finger (PBZ) motif, which appears to direct their damage site recruitment [36]. Macrodomain-containing ALC1 (Amplified in Liver Cancer 1, or CHD1L), a member of the SNF2 superfamily of ATPases, is recruited to the damage sites via its interaction with PAR. In addition, its chromatin remodeling activity is stimulated by activated PARP1, likely facilitating chromatin relaxation at the damage sites [37•,38•].
PARP-dependent recruitment of heterochromatin components to DNA damage sites
Interestingly, contrary to PARP1’s function in chromatin opening described above, several new studies uncovered the role of PARP1 (and possibly PARP2) in the recruitment of multiple gene silencing/heterochromatin factors, which appear to be also important for DNA repair.
The chromatin remodeling factor chromodomain helicase DNA-binding protein 4 (CHD4; or Mi2β) is an integral component of the NuRD complex. The NuRD complex contains chromatin remodeling and histone deacetylase and demethylase activities and functions in transcriptional repression [39]. Four recent reports showed that the NuRD complex is recruited to DNA damage sites [40••,41•,42,43]. Elledge’s and Lukas’s groups both used stable isotope labeling with amino acids in cell culture (SILAC)-mass spectrometry analyses to screen for proteins that are induced to associate with chromatin in response to different types of DNA damage (UV and IR damage, respectively) and identified multiple NuRD subunits [40••,41•]. Both Jackson’s and Elledge’s groups showed that CHD4 directly interacts with PAR and that the NuRD complex is recruited in a PAR-dependent manner to the damage sites [40••,42]. In these studies, no clear functional distinction was made between PARP1 and PARP2. The related but less characterized PARP2 interacts with PARP1 and is also involved in DNA repair, though the kinetics of its localization to DNA damage sites appear to differ from PARP1 [29,44•]., Interestingly, van Attikum’s and Lukas’s groups reported the impairment of RNF168-dependent ubiquitylation and BRCA1 assembly at damage sites following CHD4 depletion, while Jackson’s group reported that CHD4 is important for DNA repair and G1/S cell cycle control through p53 deacetylation but has no effect on BRCA1 assembly [41•–43]. Thus, the exact role of the NuRD complex in DDR and repair requires further investigation.
Interestingly, Elledge’s group also found that polycomb (PcG) proteins known to be involved in epigenetic gene silencing are also recruited to damage sites in PARP-dependent manner [40••]. PcG proteins form two major complexes: polycomb repressive complex 1 (PRC1) with H2A ubiquitylation activity and PRC2 with H3K27 di- and tri-methylation activity. In gene silencing, PRC2 is first recruited to target genes and mediates local H3K27 methylation. This mark in turn recruits PRC1, which monoubiquitylates lysine 119 of H2A (H2AUb1) in the vicinity [45–47]. Elledge’s group found both PRC1 and PRC2, as well as H3K27me3, at the damage sites and showed the essential role of PARP1/PARP2 in recruiting PRCs to the DNA damage sites [40••]. Based on the lack of the m7G cap of mRNA in the laser damage area, they suggested that the recruitment of these repressive chromatin remodeling complexes is important for local silencing of transcription near DNA lesions, proposing that PRCs indeed form transcriptionally repressive heterochromatin in this region [40••]. Interestingly, Hendzel’s group also reported the PRC1 recruitment to damage sites, but observed that its recruitment is Nbs1-dependent and PRC2-independent, raising the possibility that the role of PRC1 at damage sites is distinct from gene silencing [48•]. Indeed, PRC1 appears to play an important role in monoubiquitylation of phosphorylated histone H2AX (γH2AX), which contributes to the efficient recruitment of 53BP1 and BRCA1 [48•]. Thus, the involvement of PARPs, the relationship of PRC1 and PRC2, and their roles at the damage sites need to be further evaluated. Nevertheless, both studies demonstrate the presence at damage sites of factors that are capable of establishing repressive chromatin environments.
The macrodomain-containing histone H2A variant, macroH2A1.1, was also found to be recruited to laser-induced damage sites in a PARP1 activation-dependent manner [44•]. Only weak localization of PARP2 to damage sites was observed in this study, and PARP1 depletion and complementation was sufficient to dictate the macroH2A1.1 recruitment to damage sites. MacroH2A has been associated with gene silencing found, for example, at the inactive X chromosome in mammalian cells [49]. This recruitment of macroH2A1.1 appears to induce transient localized chromatin compaction based on the change of DAPI staining density surrounding the UVA-induced damage site [44•]. The recruitment of Ku70, a critical factor for initiating the non-homologous endjoining (NHEJ) pathway of DSB repair, appeared to be decreased in response to this chromatin structural change, suggesting a direct impact on the DNA damage response [44•]. It would be interesting to confirm these findings using other damage induction systems.
Other heterochromatin factors recruited to the damage sites
We referred earlier to work indicating that particular heterochromatic factors (i.e. KAP-1, HP1, and HDAC1/2) are essentially antagonistic to DNA repair, necessitating the actions of ATM to effect repair in heterochromatic regions [24•]. However, recent reports demonstrate that HP1 and HDACs are actually recruited to damage sites [15•,16•,50••]. HP1 was originally identified in Drosophila as a non-histone chromosomal protein functioning in heterochromatin-mediated gene silencing [51]. The recruitment mechanism mediating HP1 localization to lesions, which is H3K9me-independent, and HP1’s specific function at the damage sites, are currently undefined. Curiously, HP1 accumulation occurs at different types of damage (both strand breaks and UV crosslinks) [15•]. Whether HP1 is there to mediate some aspect of heterochromatinization or to carry out completely distinct functions at the damage sites remains to be determined.
Recruitment of HDAC1/2 to DNA damage sites was also reported. These HDACs were found to effect deacetylation of H3K56Ac and H4K16Ac and were necessary for normal NHEJ [50••]. HDAC1/2 are also found in the NuRD complex, but whether these HDACs are recruited as part of the NuRD complex was not directly addressed. How does this reconcile with histone H4 hyperacetylation by Trrap/Tip60 at the damage sites described earlier [6]? Using a UVA laser with BrdU presensitization, Miller et al observed a bi-phasic response at the damage sites, a rapid deacetylation (at ~5 min) followed by subsequent hyperacetylation (at 2 hr) [50••]. Since Trrap/Tip60 was shown to be important for homologous recombination (HR), the second major pathway of DSB repair [6], and HDACs for NHEJ, the authors suggest that the rapid initial deacetylation is important for NHEJ (perhaps in a more “repressed” chromatin structure) while subsequent hyperacetylation (favoring chromatin “opening”) facilitates HR. This is consistent with the sequential, rather than parallel, nature of NHEJ and HR factor assembly and function at the damage sites [52–54]. Interestingly, HDAC inhibition resulted in NHEJ factor persistence at the damage sites [50••]. How HDAC1/2 impact NHEJ remains unresolved, though the authors suggest possible transcriptional repression, chromatin structural change favoring NHEJ, or direct modification of NHEJ factors. It should be noted that HDAC1/2 depletion had no obvious effect on DSB repair (as judged by γH2AX focus formation) in ATM-proficient human fibroblasts, though it restored wild type DSB repair activity when ATM was inhibited [24•]. These different findings may be due to the types of cells used in each study (i.e., U2OS human osteosarcoma cancer cell line [50••] versus human primary fibroblasts [24•]) and/or the distinct effects of HDAC1/2 depletion on DNA repair in euchromatic versus heterochromatic regions. Regardless, additional study is necessary to gain better understanding of HDAC1/2 functions at damage sites.
Conclusions
There has been an explosion of important discoveries of new factors and posttranslational modifications (e.g., poly(ADP-ribosyl)ation and, though not discussed in this review, recent exciting findings on SUMO modifications at damage sites [17••,55••]) that are involved in DDR and DNA repair. Identification of these factors/modifications and analysis of their precise temporal and/or spatial interaction with damage sites were facilitated by the use of various laser microirradiation systems and chromatin analysis by ChIP.
One of the highlights of the recent discoveries as described in this review is the damage site recruitment and repair function of “heterochromatin factors” (Fig. 2) in addition to the previously identified chromatin “opening” factors. These results suggest an unexpectedly fluid chromatin structure at DNA lesions, providing a new perspective on the chromatin response at DNA damage sites. Furthermore, these studies redefined the roles of PARPs and HDACs, which is also very exciting from a clinical perspective. There is intense interest in exploiting defective repair mechanisms often seen in tumor cells, and PARPs and HDACs are both promising targets for anti-cancer therapy [56,57]. PAR and γH2AX are also being investigated as possible biomarkers to both plan and evaluate the efficacy of cancer therapeutic regimens for individual patients [58]. Therefore, the new findings concerning the roles of PARPs and HDACs in DDR/DNA repair provide important new insight into their relevance as therapeutic targets.
Figure 2. Recruitment of heterochromatic factors to DNA damage sites.
PARP is activated and rapidly recruited to the DNA damage sites resulting in the accumulation of PAR in the vicinity of the damage sites. PAR serves as a landing pad for macroH2A, polycomb repressor complex(es), and NuRD. The PRC recruitment is accompanied by histone H3K27me3. In addition, both HDAC1 and HDAC2, as well as HP1, are also recruited to the damage sites, although the recruitment mechanism(s) is unclear. These heterochromatin/gene silencing factors may be important for local transcriptional silencing at the damage sites, dictating DNA repair pathway choice, and/or recruiting DNA repair factors, such as 53BP1 and BRCA1, to promote efficient DNA repair.
Though the DNA repair field has made remarkable advances, significant challenges remain. In fact, the successful identification of so many new factors recruited to damage sites has made it particularly difficult to synthesize an all-inclusive picture of what occurs at a DNA lesion. Are all of the factors recruited to a damage site or is there a selection process determining which pathway and factor assembly should be dominant at a given lesion? What dictates this preference? In some cases, the choice may be cell cycle stage-specific, cell type-specific, cell state-specific (cancer cells, primary cells, stem cells, or ageing cells), species-specific (mouse versus human cells), and, of course, be dependent on the nature of the DNA damage. Furthermore, as many chromatin factors found to play a role in DNA repair also affect transcription, one must be cautious about the interpretation of their depletion/mutation phenotype. These phenotypes may be induced indirectly by gene expression changes, or by their effect on genome-wide chromatin regulation rather than by their specific roles at the damage sites. Additional detailed studies are essential to confirm these initial fascinating discoveries discussed here and to create a cohesive and coherent model for how mammalian cells recognize and address DNA damage.
Acknowledgments
Research in the Yokomori laboratory is supported by the National Institutes of Health (CA100710) to KY. The authors apologize for not being able to cite and discuss all other relevant papers for this review due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ••1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. Comprehensive review of the current knowledge of DNA damage response/repair pathways and their association with human diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J-S, Heale JT, Zeng W, Kong X, Ball JAR, Yokomori K. In situ analysis of DNA damage response and repair using laser microirradiation. Methods Cell Biol. 2007;82 :377–407. doi: 10.1016/S0091-679X(06)82013-3. [DOI] [PubMed] [Google Scholar]
- •3.Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ, Kim JS, Yokomori K, Berns MW. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nuc Acids Res. 2009 doi: 10.1093/nar/gkp221. The first systematic comparison of different laser systems, including UVA, green, and NIR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- 5.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 6.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 7.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkovich E, Monnat RJJ, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 9.Lukas C, Bartek J, Lukas J. Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma. 2005;114:146–154. doi: 10.1007/s00412-005-0011-y. [DOI] [PubMed] [Google Scholar]
- 10.Tobias F, Durante M, Taucher-Scholz G, Jakob B. Spatiotemporal analysis of DNA repair using charged particle radiation. Mutat Res. 2010;704:54–60. doi: 10.1016/j.mrrev.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 12.Botchway SW, Reynolds P, Parker AW, O’Neill P. Use of near infrared femtosecond lasers as sub-micron radiation microbeam for cell DNA damage and repair studies. Mutat Res. 2010;704:38–44. doi: 10.1016/j.mrrev.2010.01.003. [DOI] [PubMed] [Google Scholar]
- ••13.Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J Biol Chem. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. The authors developed a strategy to specifically induce inter-strand crosslinking damage using localized photoactivation of psoralen and related agents with UVA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •14.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. The authors found displacement of HP1 from heterochromatin upon damage, and identified specific phosphorylation of HP1 in response to DNA damage that decreases the affinity of HP1 for H3K9me. [DOI] [PubMed] [Google Scholar]
- •15.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. The authors demonstrated the H3K9me-independent recruitment of HP1 to DNA damage induced by various modalities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •16.Zarebski M, Wiernasz E, Dobrucki JW. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry. 2009;75:619–625. doi: 10.1002/cyto.a.20734. [DOI] [PubMed] [Google Scholar]
- ••17.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. Similar to the paper by Galanty et al. (Ref.55), this group showed the PIAS-dependent SUMO responses at damage sites induced by hydroxyurea. In addition to the accrual of DNA repair proteins and efficient repair, this group showed that PIAS-dependent SUMOylation of BRCA1 is required for activation of its Ub ligase activity. [DOI] [PubMed] [Google Scholar]
- 18.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradshaw PS, Stavropoulos DJ, Meyn MS. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- 20.Williams ES, Stap J, Essers J, Ponnaiya B, Luijsterburg MS, Krawczyk PM, Ullrich RL, Aten JA, Bailey SM. DNA double-strand breaks are not sufficient to initiate recruitment of TRF2. Nat Genet. 2007;39:696–698. doi: 10.1038/ng0607-696. [DOI] [PubMed] [Google Scholar]
- 21.Splinter J, Jakob B, Lang M, Yano K, Engelhardt J, Hell SW, Chen DJ, Durante M, Taucher-Scholz G. Biological dose estimation of UVA laser microirradiation utilizing charged particle-induced protein foci. Mutagen. 2010;25:289–297. doi: 10.1093/mutage/geq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 23.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. This paper presents important evidence for the role of ATM in DNA repair in heterochromatin regions. [DOI] [PubMed] [Google Scholar]
- 25.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8 :870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 26.Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: Functional synergy in mouse development. DNA Repair. 2004;3 :1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Iliakis G. Backup pathways of NHEJ in cells of higher eukaryotes: cell cycle dependence. Radiother Oncol. 2009;92:310–315. doi: 10.1016/j.radonc.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortusewicz O, Amé JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nuc Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J-S, Krasieva TB, LaMorte VJ, Taylor AMR, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- 31.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda K, Omachi A, Kawaichi M, Hayaishi O. Natural occurrence of poly(ADP-ribosyl) histones in rat liver. Proc Natl Acad Sci. 1975;72:205–209. doi: 10.1073/pnas.72.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirier G, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 36.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. This paper introduces the poly(ADP-ribose)-binding zinc finger (PBZ) motif as the third described example of a PAR-binding motif. The authors systematically analyze the PBZ domains in the antephase checkpoint protein CHFR and the DNA damage response factor APLF, demonstrating that an intact PBZ domain is essential for CHFR’s checkpoint function. [DOI] [PubMed] [Google Scholar]
- •37.Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. The data in this study shows that ALC1, an oncogene linked to primary hepatocellular carcinomas and a chromatin remodeler, is a DNA damage response protein that is rapidly recruited to sites of DNA damage in a PAR-dependent manner. See also Gottschalk et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •38.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. See also Ahel et al. (09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramírez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenet. 2009;4:532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- ••40.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiácovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. The authors utilized a SILAC/mass spectrometry approach to identify proteins that are induced to interact with chromatin after DNA damage, and found several components of polycomb repressor complexes (PRC1 and PRC2) and NuRD. Consistent with PRC2 recruitment, H3K27me3 was enriched at laser-induced damage sites. Activated PARP was necessary for the recruitment of both PRC and NuRD complexes to damage sites. The authors proposed the idea of a “repressive” chromatin environment that inhibits transcription as part of DNA repair. See also papers by Larsen et al, Smeenk et al, and Polo et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Danielsen JM, Menard P, Sand JC, Stucki M, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •44.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struc Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. The authors used laser-induced DNA damage to show that the histone macroH2A1.1 is recruited to sites of PARP activation. This recruitment was dependent on interaction with PAR through the macrodomain. They went on to address how macroH2A1.1 localization to damage sites leads to chromatin structural alterations. [DOI] [PubMed] [Google Scholar]
- 45.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 47.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- •48.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. This paper presents evidence of the role of BMI1, a component of the polycomb complex PRC1, in the DNA damage response. A particularly intriguing aspect of this work is that BMI1 (and thus PRC1) is recruited to damage sites in a PRC2-independent, NBS1-dependent manner. It is useful to compare this study to that of Chou et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ladurner AG. Inactivating chromosomes: a macro domain that minimizes transcription. Mol Cell. 2003;12:1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- •50.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struc Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. The authors used multiple methods to introduce DSB damage and found that HDAC1 and HDAC2 are recruited to damage sites where they deacetylate H3K56Ac and H4K16Ac. Loss of these HDACs resulted in compromised NHEJ. Importantly, this paper showed an initial decrease and subsequent enrichment of H4K16Ac at the damage sites, proposing a model that involves a fluid change from a “closed” chromatin signature to one that is more “open” as the damage response/repair process proceeds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SCR. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophiola melanogaster. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J-S, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 54.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An XRCC4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nuc Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••55.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. This paper together with the paper by Morris et al. (ref.17) presented the first cytological demonstration of SUMO response at DNA damage sites (by laser and IRIF) mediated by PIAS1/4 E3 SUMO ligases, and its connection with damage-induced ubiquitin (Ub) response. They found that 53BP1 and BRCA1 are targets of SUMO modification. PIAS1/4 are important for the recruitment of 53BP1, Ub ligases RNF8, RNF168 and BRCA1, and the Ub response at damage sites, affecting both NHEJ and HR repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan J, Cang S, Ma Y, Petrillo RL, Liu D. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J Hematol Oncol. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Redon CE, Nakamura AJ, Zhang YW, Ji JJ, Bonner WM, Kinders RJ, Parchment RE, Doroshow JH, Pommier Y. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res. 2010;16:4532–4542. doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]