Abstract
We describe a transfection system that induces terminal deletions at specific chromosome ends in malaria parasites using a linear construct containing telomeric repeats at one end and plasmodial sequences able to drive homologous recombination at the other. A site-specific deletion was generated at one extremity of chromosome 5 of Plasmodium berghei, which was stably maintained in the parasite population selected after transfection. The telomeric repeat array introduced with the construct reached the average length observed in natural telomeres of Plasmodium, indicating that in vivo telomere addition occurred at the newly formed extremity. The expression of a mutant dhfr/ts gene conferring pyrimethamine resistance, used as a selectable marker, was not affected by the proximity to the telomeric sequences, either in the presence or absence of drug pressure. In addition, no transcriptional silencing was observed on insertion of the mutant dhfr/ts gene either in subtelomeric or internal positions that are transcriptionally silent in blood-stage parasites. This suggests that the activity of its promoter is not affected by the chromatin organization of the chromosomal context.
Telosomes, the protein–DNA complexes at the ends of eukaryotic chromosomes, are essential for chromosome stability and replication (for review, see Muniyappa and Kironmai 1998). Adjacent to the terminal repeats, shorter or longer arrays of repetitive noncoding elements (telomere-associated sequences) are present both in lower and higher eukaryotes. These subtelomeric elements are generally highly variable in location and copy number, making the subtelomeric region one of the most polymorphic genomic regions.
Plasmodium parasites, haploid during most of their complex life cycle, harbor 14 linear chromosomes bounded by arrays of telomeric tandem repeats of two types (Ponzi et al. 1985; Vernick and McCutchan 1988). Subtelomeric regions containing families of noncoding repetitive DNA have been characterized both in human Plasmodium falciparum (Oquendo et al. 1986) and in rodent P. berghei (Pace et al. 1987) malarias and have been shown to represent preferential sites of chromosome rearrangements both in vivo and under culture conditions (Corcoran et al. 1988; Ponzi et al. 1990).
Gene families mapping to the terminal portion of different P. falciparum chromosomes include the var family encoding the variant surface antigen PfEMP1 involved in parasite cytoadherence (Smith et al.1995; Chen et al. 1998), the rif and stevor multicopy gene families, also believed to encode variant antigens (Cheng et al. 1998; Kyes et al. 1999). Comparison of the complete sequence of chromosomes 2 and 3 of P. falciparum (Gardner et al. 1998; Bowman et al. 1999) indicates that these genes are clustered and arranged in a specific order (telomere: subtelomeric repeat: var: rif: stevor: → centromere). The arrangement of subtelomeric genes is thought to facilitate recombination and generation of genetic diversity.
In addition to rearrangements leading to enhanced genetic variability (Sinnis and Wellems 1988; Hernandez-Rivas et al. 1996), spontaneous terminal deletions can cause the loss of subtelomeric sequences and genes during mitotic propagation in laboratory animals or in in vitro cultures (Pologe and Ravetch 1986; Cappai et al. 1989; Scherf et al. 1992; Day et al. 1993; van Lin et al. 1997).
We report the development of a transfection system designed for inducing and selecting controlled terminal deletions, potentially at any chromosomal end. Recombination in the Plasmodium genome occurs almost exclusively by mechanisms that rely on high homology between the parasite genome and the introduced DNA. We were able, by this method, to reproducibly induce, for the first time in Plasmodium, a terminal deletion at a specific site of chromosome 5 of P. berghei.
Using this deletion mutant, which bears a selectable drug resistance gene close to the telomere of the truncated chromosome, we investigated the possible influence of telosome proximity on the transcription of this marker gene in the presence or in the absence of selective pressure. Transcription levels for the same gene transfected in different chromosomal contexts were determined for comparison. No telomeric silencing effect similar to that described for yeast (Gotta and Gasser 1996) was observed.
RESULTS
Targeting a Terminal Deletion
Chromosome 5 of P. berghei, characterized in a previous work (van Lin et al. 1997), was chosen as a target for transfection-induced deletion. A long-range restriction map is available, along which genes and sequences specific for internal or subtelomeric regions are positioned. In addition, a natural gametocyteless mutant bearing an 80-kb deletion at one extremity of this chromosome (Janse et al. 1992) provides indirect evidence that the lack of this region would not affect parasite mitotic multiplication, that is, it does not contain essential genes or centromeric sequences.
A map of the 100-kb ApaI terminal fragment affected by the spontaneous deletion is presented in Figure 1a. Two members (orfA and orfB) of a family of three related genes map within this region, whereas the third member (orfC) is located at the other extremity of the same chromosome. The three genes, highly transcribed during the blood stages, share the amino portion and differ at the carboxyl terminus of the predicted translation products (T. Pace, C. Birago, E. Pizzi, L. Pizzi, and M. Ponzi, in prep.). Probe A and Probe B, used in this work, are PCR fragments specific for orfA and orfB.
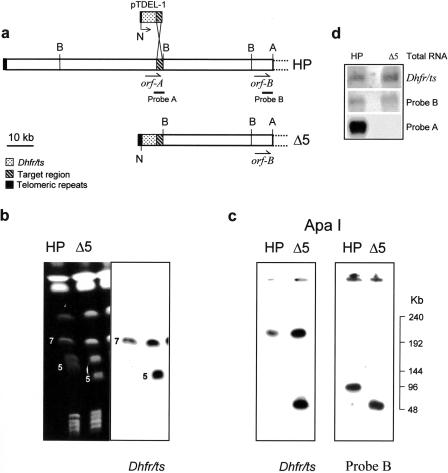
Figure 1.
(a) Site-directed deletion strategy. A schematic map of the subtelomeric region of P. berghei chromosome 5 where the deletion is to occur is shown and the homology region with the transfecting plasmid is indicated. (A) ApaI; (N) NotI; (B) BamHI. (b) CHEF-separated chromosomes (left) of 8417HP and HPΔ5 (250 V, 55-sec pulse, 16 hr; 120 V, 270-sec pulse, 48 hr) blotted and hybridized with dhfr/ts probe (right). (c) 8417HP and HPΔ5 total DNA blocks digested with ApaI, electrophoresed by CHEF (250 V,12-sec pulse, 15 hr), blotted, and hybridized with the indicated probes. (d) Northern blot of total RNA from 8417HP (HP) and HPΔ5-clone 1 (Δ5) hybridized with dhfr/ts, probe A, and probe B. The latter has been used to normalize the blotted RNAs.
The deletion-inducing transfection construct pTDEL-1, described in Methods, contains the P. berghei dihydrofolate reductase–thymidylate synthetase mutant gene (dhfr/tsR), conferring pyrimethamine resistance to transfected parasites (van Dijk et al. 1996), under its own promoter. The selectable marker is flanked upstream by a 550-bp fragment containing exclusively Plasmodium telomeric tandem repeats (Ponzi et al. 1990) and downstream by a 2.4-kb fragment chosen to drive specific recombination. This genomic region includes the unique carboxy-terminal portion of the coding region and the 3′ UTR of orfA, but lacks the promoter and 5′ portion of the open reading frame. This ORF, lost as a consequence of the natural 80-kb deletion, can be considered a “dispensable” gene.
Once this construct is transfected, homologous recombination at the chosen site should result in the concomitant loss of the telomere-proximal portion of the target chromosome (Fig. 1a).
The presence of telomeric sequences at the free extremity of the recombination event should favor in vivo maintenance of the newly formed chromosome end. A NotI site, placed at the 3′ end of the telomeric sequence, makes it possible to observe whether telomere elongation occurs during mitotic multiplication of the transfected parasites.
Analysis of Transfectants
Linear pTDEL-1 construct was used to transfect mature schizonts from P. berghei clone 8417HP as detailed in Methods. Successfully transfected parasites were selected for their resistance to pyrimethamine. Karyotype analysis of the resistant parasite population (HPΔ5) showed a shift of the band corresponding to chromosome 5 to a molecular weight lower than in the parental clone 8417HP (Fig. 1b). Chromosome attribution was confirmed using specific internal probes (data not shown).
When karyotypes from HPΔ5 population and from 8417HP parental clone were probed with the P. berghei dhfr/ts gene, in addition to the resident copy located on chromosome 7, a second copy of the gene could be detected exclusively in the shortened chromosome 5 of HPΔ5 parasites (Fig. 1b).
Identical results were obtained in six sibling clones derived by limiting dilution from the HPΔ5 population after a second round of selection and in two additional independent transfection experiments using the pTDEL-1 construct to promote site specific recombination.
To map the deletion produced by pTDEL-1 plasmid more precisely, hybridization experiments were performed on ApaI-digested total DNA blocks from 8417HP and from the mutant HPΔ5-clone 1. Probe B, which recognizes the target ApaI-terminal fragment both in the intact and in the deleted version of chromosome 5, confirmed the 50-kb shortening of the terminal fragment in HPΔ5 parasites (Fig. 1c) that also contains the dhfr/tsR gene.
BamHI digestion of purified chromosome 5 from HPΔ5 provides further evidence that the deletion occurred at the predicted site.
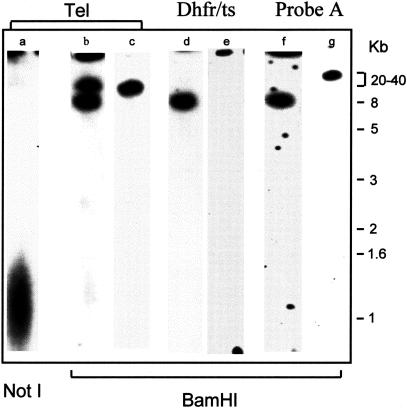
The two ends of the parental chromosome 5 share common sequences and identical restriction pattern within the 20-kb region delimited by the first BamHI site, whereas probe A specifically recognizes the second, 40-kb BamHI fragment of the targeted extremity (see Figs. 1 and 2). Two distinct telomeric bands are instead highlighted in HPΔ5 parasites (Fig. 2): the one at 20 kb, coincident with the size of both parental telomeric fragments, and the other at 8 kb, which is the size expected as the result of the induced deletion. Both the selectable marker (dhfr/tsR gene) and the remaining portion of the target gene (orfA) mapped within this smaller telomeric fragment.
Figure 2.
Hybridization pattern of chromosome 5 purified from HPΔ5 (clone 1) (lanes a,b,d,f) and from 8417HP parental clone (lanes c,e,g) digested and separated in 1% agarose gel.
Taking advantage of a NotI site artificially introduced at the 3′ end of the 550-bp telomeric region in pTDEL-1 construct (and absent in the natural chromosome 5), it was observed that the length of the telomeric region at the newly generated extremity increased in the course of mitotic propagation of the HPΔ5 population, reaching an average length of 1–1.2 kb, the normal telomere length in P. berghei blood stages (Dore et al. 1994), during three mechanical passages in laboratory animals (Fig. 2, lane a).
pTDEL-1 construct contains only part of the coding region of orfA, lacking the promoter and the 5′ portion of the gene. Hence, the recombination event should truncate orfA leaving only the 3′ portion of the gene. As demonstrated by Northern blot hybridization with probe A, the transcription of the target gene was completely abolished in the HPΔ5 mutant. Conversely, an increased amount of dhfr/ts transcript was observed in HpΔ5 when compared with the parental 8417HP clone (Fig. 1d), as expected because of the presence of two copies of dhfr/ts gene in the mutant parasites. Signals of comparable intensity between the two parasite populations were observed using probe B as a control.
Transcription of the dhfr/tsR Gene Transfected in Different Locations of P. berghei Genome
Several transfected clones of P. berghei 8417HP are presently available that contain dhfr/tsR gene integrated in different genomic regions, always under the control of its own promoter. This gave us the opportunity to observe whether chromosomal position effects might influence the transcription level of the reporter gene in parasites grown in the presence or in the absence of drug selective pressure.
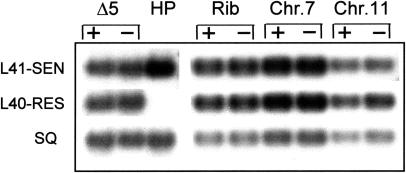
The HPΔ5 1–6 cloned lines, generated by transfection with the pTDEL-1 linear construct and bearing the 60-kb terminal deletion in chromosome 5, contain dhfr/tsR gene with its 2.5-kb, 5′ regulatory region adjacent to the telomere. The oligonucleotides chosen for RT-PCR experiments allow the amplification of a 682-bp fragment within the coding region, which includes the single base substitution (G→A) conferring the pyrimethamine resistance. Therefore, RT-PCR would equally amplify transcripts originating from both the resident dhfr/ts gene and the subtelomeric dhfr/tsR gene. PCR products were blotted and hybridized with oligonucleotides L41-SEN and L40-RES, which are able to discriminate, at high stringency, between the “resistant” and the “sensitive” transcripts (van Dijk et al. 1996), as demonstrated by control PCR experiments using the parental clone 8417HP (Fig. 3) and the dhfr/tsR gene present in the original PMD204 plasmid (not shown). Thus quantitation of the hybridization signals should give an accurate measure of the relative steady state level of the two mRNAs species.
Figure 3.
RT-PCR amplifications of dhfr/ts mRNAs from transfected clones harboring the dhfr/tsR gene in different chromosome locations—near the telomere in HPΔ5 parasites (Δ5); within C-type rRNA locus (Rib); and within the subtelomeric regions of chromosomes 7 and 11. Parasites were grown in the presence (+) or in the absence (−) of pyrimethamine selection. Hybridizations were performed with end-labeled oligonucleotides L41-SEN and L40-RES able to distinguish between the two versions of the dhfr/ts gene, as shown using the parental 8417HP as a control (HP), and probe SQ, which recognizes both transcripts, to normalize blotted DNA.
Results for HPΔ5 clone 1 parasites either in the presence or in the absence of drug selection (Fig. 3) showed that both versions of the dhfr/ts transcript were detected at comparable levels in the RT-PCR product, indicating that neither transcriptional silencing nor up-regulation of the telomeric copy occurred. These results were reproducibly obtained in three independent PCR experiments. The consistency of the results confirmed the reliability of this approach. The presence in the RT-PCR products of both versions of the dhfr/ts transcript was also confirmed by direct sequencing the amplified fragments.
Dhfr/ts transcription pattern was also analyzed in transformants carrying the dhfr/tsR gene integrated within the C-unit rRNA locus (R.M. van Spaendonk, C.J. Janse, and A.P. Waters, unpubl.), which is located in an internal region of chromosome 5 (van Lin et al. 1997) and is silent during the schizogonic cycle of the parasite (Waters et al. 1997b). The two versions of the dhfr/ts gene again appeared to be transcribed at the same level in the presence or in the absence of pyrimethamine selection (Fig. 3) in asynchronous asexual populations.
Chromosome 5 lacks the subtelomeric 2.3-kb repeats that are present at the distal portion of most P. berghei chromosomes (Ponzi et al. 1990) and bear internal telomeric motifs (Pace et al. 1987). To investigate whether these telomere-associated, noncoding sequences might influence the expression of the dhfr/tsR gene we analyzed RNA from transfected parasites having the mutant copy integrated within a 2.3-kb unit in chromosome 7 or 11 (van Dijk 1996). These regions contain up to 200 tandemly arranged copies of the 2.3-kb repeat (Ponzi et al. 1990). As shown in Figure 3, once more, the chromosome context did not appear to influence the transcription of the transfected gene.
DISCUSSION
Genome plasticity in Plasmodium, extensively studied during the past years (Lanzer et al. 1995), is mainly confined to the highly recombinogenic subtelomeric portions of the parasite's chromosomes. Spontaneous large terminal deletions, which involve in some cases the loss of subtelomeric genes, occur frequently in P. berghei and in P. falciparum laboratory strains. In a number of cases, association between the loss of specific functions and the occurrence of spontaneous terminal deletions at specific chromosomes, has been observed. Terminal deletions of chromosome 9 in P. falciparum (Day et al. 1993) and of chromosome 5 in P. berghei (van Lin et al. 1997) appear to be linked to the loss of ability to produce gametocytes, the only forms infective for mosquito vector. However, with the exception of some well-characterized cases (Pologe and Ravetch 1986; Cappai et al. 1989; Scherf et al. 1992), the large size of spontaneous deletions, along with the possible presence of additional independent mutations in the genome of these parasites, might complicate the identification of gene(s) responsible for the phenotype under study.
In this work we describe a transfection system designed to induce subtelomeric deletions at specific sites in the Plasmodium genome. The linear construct (pTDEL-1) used in these experiments has been designed to induce a 60-kb deletion at one extremity of P. berghei chromosome 5. The construct contains the P. berghei dihydrofolate reductase–thymidylate synthetase mutant gene, conferring pyrimethamine resistance (dhfr/tsR), a properly oriented array of telomeric repeats at one end, and the target parasite sequence for recombination at the other end. Results show that the intended deletion of the telomere-proximal portion of chromosome 5 did in fact readily occur, as a consequence of the expected recombination event, in three independent transfection experiments.
This approach opens the possibility to obtain, in a single selection step, parasites bearing terminal deletions with well-defined breakpoints and to compare the resulting parasite phenotype directly with that of the isogenic parental clone. This provides an important tool for the functional genomics studies that are necessary to interpret the rapidly accumulating data from genomic sequencing projects. The approach may have interesting applications; for instance, by using suitable combinations of selectable markers that are being developed (Fidock and Wellems 1997; Mamoun et al. 1999; de Konig-Ward et al. 2000), the sequential and systematic deletion might be attempted of the subtelomeric multigene families that are thought to be associated with blood-stage antigenic variation in P. falciparum. This could provide a simplified model to study mechanisms of switching of gene expression within these families. There are some hints that suggest that plasmodial genes are clustered according to the stage of their expression in the life cycle (L. van Lin, T. Pace, C.J. Janse, C. Birago, J. Ramesar, L. Picci, M. Ponzi, and A.P. Waters, in prep.); the type of vector described here might be applied to investigate such clustering, should it occur on a scale that occupies significant chromosomal regions. One current limitation of this approach is the fact that no large-scale deletion can be attempted that involves the centromere of the target chromosome. The recent description of candidate centromeres may allow the design of vectors that could eliminate larger regions of a chosen chromosome. A second constraint is that the deleted region must not contain a gene essential for blood-stage parasite survival. Centromere-containing vectors would allow the direct demonstration of the presence of essential genes on chromosome fragments that are candidates for deletion.
Cases have been described in P. falciparum where a terminal deletion is accompanied by the formation of a new functional telomere (Pologe and Ravetch 1988; Mattei and Scherf 1994). In P. berghei blood stages the array of telomeric repeats is maintained at an average length of ∼ 1–1.2 kb (Dore et al. 1994), most probably by a mechanism involving telomere turnover determined by shortening events and telomerase-mediated distributive addition of the two types of telomeric repeats (Ponzi et al. 1992). Consistent with this was the recent demonstration of telomerase activity in P. falciparum (Bottius et al. 1998). The telomeric tract present in the transfection construct (pTDEL-1) consists of a 550-bp array of telomeric repeats marked by a rare-cutting NotI site at the inner extremity. This allowed us to show de novo addition of telomeric repeats at the newly formed extremity, up to 1.2 kb length as an average. This demonstration will provide the basis for an in vivo investigation of telomerase activity and substrate specificity.
By analogy with the case of yeast, one might expect that telosome proximity and propagation of a modified chromatin structure would influence transcription of subtelomeric Plasmodium genes. Some similarity exists between three P. falciparum deduced proteins (A.P. Waters, unpubl.) and Sir 2 and Sir3, two telomere-binding proteins of S. cerevisiae that are known to mediate the establishment of the heterochromatin typical of telosomes (Grunstein 1997; Haber 1999) and to be involved in the telomere position effect that modulates and represses transcription of subtelomeric genes in direct proportion to the proximity to the telomere (Gotta et al. 1997).
Taking advantage of the deletion construct described in the present work, we studied the transcription pattern of the P. berghei dhfr/tsR mutant gene used as a selectable reporter gene in differently transfected parasite lines. The transfected copy, integrated in different genomic positions, is in all cases under the control of a regulatory region identical to that of the resident gene (located on chromosome 7). No transcriptional silencing is observed when the regulatory region of the selectable reporter gene is adjacent to the telomeric repeats of chromosome 5. Both versions of the dhfr/ts gene are, in fact, transcribed at comparable levels in parasites grown either in the presence or in the absence of selective pressure. Similar results have been obtained when the mutant copy of the gene is integrated (A.P. Waters, unpubl.) in an internal position of chromosome 5, within the C-unit rRNA locus repressed during asexual growth or within the transcriptionally inactive 2.3-kb subtelomeric tandem repeats (van Dijk et al. 1996) present at many chromosomal ends.
Expression of the selectable marker is driven by the 2.5-kb regulatory region normally present upstream of the P. berghei dhfr/ts gene on chromosome 7. Regions < 2.5 kb have proven unable to drive adequate expression of the selectable marker. It might be suggested that, along the 2.5 kb, a region of chromatin organization is established that is independent of chromosome position and immune to possible position effects. Furthermore, the data presented here suggest that simple proximity to the telomere is insufficient to mediate inactivation through Sir protein-mediated mechanisms.
METHODS
Construction of pTDEL-1 Transfection Vector
orfA, a member of a three-gene family (orfA, orfB, and orfC) located at the terminal portions of P. berghei chromosome 5 and abundantly transcribed in asexual stages, was chosen as a target sequence for transfection-mediated terminal deletion of chromosome 5. The complete coding region of orfA and its flanking sequences are contained in the 4.2-kb insert of clone Z11 isolated from a P. berghei genomic library in λ-ZAP (Birago et al. 1996). pMD204, used as a transfection vector (Waters et al. 1997a), contains the P. berghei dhfr/tsR gene including the 2.5 kb of the upstream regulatory region and 0.5 kb of the downstream region. A point mutation within the coding region confers pyrimethamine resistance to the parasites (van Dijk et al. 1995). A 2.35-kb BalI–NotI fragment, containing the terminal portion of the coding region of orfA and its 3′ UTR, was cloned into the SmaI–NotI sites of the pMD204 vector, downstream of the dhfr/tsR gene (pMD204–orfA). A previously cloned 550 bp of Plasmodium-specific telomeric repeats (Ponzi et al. 1990) was recloned in the TA vector (Invitrogen), recovered through Kpn–XhoI double digestion, and finally cloned in the same sites of the PMD204–orfA construct, upstream of the dhfr/tsR gene, thus obtaining the final transfection construct pTDEL-1. The dhfr/tsR flanked by the telomeric repeats at one side and by the target sequence (orfA) at the other side, was recovered as an 8-kb SacI fragment.
Transfection and Parasite Selection
SacI-digested pTDEL-1 (50 μg) was used to transfect 2.108 mature schizonts from P. berghei clone 8417HP as described (Waters et al. 1997a). Electroporated parasites were injected into the tail vein of two mice. Pyrimethamine selection (10 mg/kg) was applied 48 hr post infection and maintained during 4 days. Resistant parasites (HPΔ5), harvested 6 days later, were mechanically passaged into 4 mice and maintained under drug pressure until bleeding for further analysis. Individual clones (HPΔ5 1–6) were obtained by end dilution.
Karyotype Analysis of Transfected Parasites
Blocks containing intact total DNA, obtained by incubating agarose-embedded parasites with proteinase K (2 mg/ml) for 48 hr were used for chromosome separation. Karyotypes of parental (8417HP) parasites and of transfected (HPΔ5) line and clones (HPΔ5 1–6) were obtained using a CHEF (LKB 2015 Pulsaphor apparatus), 1% agarose gel, 0.5× TBE buffer, at 10°C. Seakem–GTG agarose (FMC Bio-Products) was used when chromosomes had to be recovered from the gel and digested with restriction enzymes. Electrophoretic conditions are specified in the figure legends.
Primers
Primers used in this work are as follows: orfa1, 5′-CTCAGCATCAGATAATGACA-3′]; orfa2, 5′-TCTGCTTCAATGTAACATAG-3′; orfb1, 5′-ACCAATAAGACGACAATG-3′;, orfb2, 5′-TAGTTTATTATAATGTTACA-3′; dhfr1, 5′-ATTAGGTACTTCCTCATTTGGA-3′;, dhfr2, 5′-TGATGAAAAGGTTAGATGCT-3′; dhfrSQ, 5′-AGATTGAAATGGAAAAGAG-3′; L41-SEN, 5′-GAATACTTTCCCAACTTTTTTTTC-3′; L40-RES, 5′-GAATACTTTCCCAATTTTTTTTTC-3′.
Probes
The DNA fragment containing telomeric repeats used as a probe in this work was recovered by EcoRI–HindIII restriction of the original pTb4.1 plasmid (Ponzi et al. 1990). The 5-kb insert corresponding to P. berghei dhfr/tsR gene was obtained from the pMD204 vector (Waters et al. 1997a) by double digestion with HincII–SmaI. Probe A, a 432-bp PCR fragment amplified with primers orfa1 and orfa2, contains the 3′ end of the orfA gene present in the pTDEL-1 construct. Probe B, a 565-bp PCR fragment amplified with primers orfb1 and orfb2, contains the 3′ end of the orfB gene. PCR amplifications were performed for 1 min at 90°C, 1 min at 55°C, and 2 min at 68°C for 35 cycles. End-labeled oligonucleotides L41-SEN and L40-RES differing for a single base-pair substitution (C→T) were used as probes either selective for sensitive or resistant version of dhfr/ts transcript (van Dijk et al. 1995). Oligonucleotide SQ recognizing both versions of dhfr/ts was used to normalize blotted DNA.
Detection of P. berghei dhfr/ts transcript
Total RNA from P. berghei parasites was extracted using RNeasy kit (Promega). Samples used for RT-PCR experiments were treated previously with DNaseI (RNase free, Promega). Absence of residual DNA fragments still able to function as templates was checked by direct PCR using the same primers as for RT-PCR. RT-PCR amplification of dhfr/ts transcript was performed on transfected parasites and on parental clone starting from 0.5 μg of total RNA. cDNA synthesis for 45 min at 45°C was followed by 40 cycles of PCR amplification (45 sec at 94°C, 1 min at 55°C, 90 sec at 68°C). Primers dhfr1 and dhfr2 amplify a 682-bp fragment encompassing the single base substitution (G→A) that confers the pyrimethamine resistance. The relative amount of each transcript was measured by the use of a PhosphorImager after hybridization, at high stringency (van Dijk et al. 1995), with oligonucleotides L40-SEN and L41-RES, which are able to distinguish between the two transcripts.
Acknowledgments
We thank Dr. Clara Frontali (Laboratorio di Biologia Cellulare, Istituto Superiore di Sanita) for critical reading of the manuscript. We acknowledge Jai Ramesar (Department of Parasitology, Leiden University Medical Centre) for his invaluable technical assistance, Resie van Spaendonk for the gift of the mutant parasites bearing the dhfr/tsR gene in the C tRNA locus, and Milly van Dijk for advice on the use of the dhfr/ts oligonucleotides. This work received support from the Commission of the European Communities in the framework of the INCO DC Program IC18-CT96–0052, IC18-CT96–0092 and IC18-CT96–0071.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL frontali@iss.infn.it; FAX 39-06-49387143.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.1400R00.
REFERENCES
- Birago C, Pace T, Barca S, Picci L, Ponzi M. A chromatin-associated protein is encoded in a genomic region highly conserved in the Plasmodium genus. Mol Biochem Parasitol. 1996;80:193–202. doi: 10.1016/0166-6851(96)02680-1. [DOI] [PubMed] [Google Scholar]
- Bottius E, Bakhsis N, Scherf A. Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol Cell Biol. 1998;18:919–925. doi: 10.1128/mcb.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher CM, Craig A, Davies RM, Devlin K, Feltwell T, et al. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature. 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- Cappai R, van Schravendijk MR, Anders RF, Peterson MG, Thomas LM, Cowman AF, Kemp DJ. Expression of the resa gene in Plasmodium falciparum isolate FCR3 is prevented by a subtelomeric deletion. Mol Cell Biol. 1989;9:3584–3587. doi: 10.1128/mcb.9.8.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Fernandez V, Sundström A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. Developmental selection of var gene expression in Plasmodium falciparum. Nature. 1998;394:392–395. doi: 10.1038/28660. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Cloonan N, Fischer K, Thompson J, Waine G, Lanzer M, Saul A. Stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol Biochem Parasitol. 1998;97:161–176. doi: 10.1016/s0166-6851(98)00144-3. [DOI] [PubMed] [Google Scholar]
- Corcoran LM, Thompson JK, Walliker D, Kemp D. Homologous recombination within subtelomeric repeat sequences generates chromosome size polymorphisms in P. falciparum. Cell. 1988;53:807–813. doi: 10.1016/0092-8674(88)90097-9. [DOI] [PubMed] [Google Scholar]
- Day KP, Karamalis F, Thompson J, Barnes DA, Peterson C, Brown H, Brown GV, Kemp DJ. Genes necessary for expression of a virulence determinant and for transmission of Plasmodium falciparum are located on a 0.3-megabase region of chromosome 9. Proc Natl Acad Sci. 1993;90:8292–8296. doi: 10.1073/pnas.90.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward TF, Fidock DA, Thathy V, Menard R, van Spaendonk RM, Waters AP, Janse CJ. The selectable marker human dihydrofolate reductase enables sequential genetic manipulation of the Plasmodium berghei genome. Mol Biochem Parasitol. 2000;106:192–212. doi: 10.1016/s0166-6851(99)00189-9. [DOI] [PubMed] [Google Scholar]
- Dore E, Pace T, Picci L, Pizzi E, Ponzi M, Frontali C. Dynamics of telomere turnover in Plasmodium berghei. Mol Biol Rep. 1994;20:27–33. doi: 10.1007/BF00999852. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Aravind L, Koonin EV, Shallom S, Mason T, Yu K, Fujii C, et al. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- Gotta M, Gasser SM. Nuclear organisation and transcriptional silencing in yeast. Experientia. 1996;52:1136–1147. doi: 10.1007/BF01952113. [DOI] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localisation of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- Haber JE. Sir-Ku-itous routes to make ends meet. Cell. 1999;97:829–832. doi: 10.1016/s0092-8674(00)80795-3. [DOI] [PubMed] [Google Scholar]
- Hernandez-Rivas R, Hinterberg K, Scherf A. Compartmentalisation of genes coding for immunodominant antigens to fragile chromosome ends leads to dispersed subtelomeric gene families and rapid gene evolution in Plasmodium falciparum. Mol Biochem Parasitol. 1996;78:137–148. doi: 10.1016/s0166-6851(96)02618-7. [DOI] [PubMed] [Google Scholar]
- Janse CJ, Ramesar J, van den Berg FM, Mons B. Plasmodium berghei: In vivo generation and selection of karyotype mutants and non-gametocyte producer mutants. Exp Parasitol. 1992;74:1–10. doi: 10.1016/0014-4894(92)90133-u. [DOI] [PubMed] [Google Scholar]
- Kyes SA, Rowe JA, Kriek N, Newbold CL. Rifins: A second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc Natl Acad Sci. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzer M, Fischer K, Le Blancq SM. Parasitism and chromosome dynamics in protozoan parasites: Is there a connection? Mol Biochem Parasitol. 1995;70:1–8. doi: 10.1016/0166-6851(95)00021-r. [DOI] [PubMed] [Google Scholar]
- Mamoun CB, Gluzman IY, Goyard Beverley S, SM, Goldberg DE. A set of independent selectable markers for transfection of the human malaria parasite Plasmodium falciparum. Proc Natl Acad Sci. 1999;96:8716–8720. doi: 10.1073/pnas.96.15.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei D, Scherf A. Subtelomeric chromosome instability in Plasmodium falciparum: Short telomere-like sequence motifs found frequently at healed chromosome breakpoints. Mutat Res. 1994;324:115–120. doi: 10.1016/0165-7992(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Muniyappa K, Kironmai KM. Telomere structure, replication and length maintenance. Crit Rev Biochem Mol Biol. 1998;33:297–336. doi: 10.1080/10409239891204242. [DOI] [PubMed] [Google Scholar]
- Oquendo P, Goman M, MacKay M, Langsley G, Walliker D, Scaife J. Characterisation of a repetitive DNA sequence from the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1986;18:89–101. doi: 10.1016/0166-6851(86)90053-8. [DOI] [PubMed] [Google Scholar]
- Pace T, Ponzi M, Dore E, Frontali C. Telomeric motifs are present in a highly repetitive element in the Plasmodium berghei genome. Mol Biochem Parasitol. 1987;24:193–202. doi: 10.1016/0166-6851(87)90106-x. [DOI] [PubMed] [Google Scholar]
- Pologe LG, Ravetch JV. A chromosomal rearrangement in a P. falciparum histidine-rich protein gene is associated with the knobless phenotype. Nature. 1986;322:474–477. doi: 10.1038/322474a0. [DOI] [PubMed] [Google Scholar]
- ————— Large deletions results from breakage and healing of P. falciparum chromosomes. Cell. 1988;55:869–874. doi: 10.1016/0092-8674(88)90142-0. [DOI] [PubMed] [Google Scholar]
- Ponzi M, Pace T, Dore E, Frontali C. Identification of a telomeric DNA sequence in Plasmodium berghei. EMBO J. 1985;4:2991–2995. doi: 10.1002/j.1460-2075.1985.tb04034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzi M, Janse CJ, Dore E, Scotti R, Pace T, Reterink TJF, van der Berg FM, Mons B. Generation of chromosome size polymorphism during in vivo mitotic multiplication of Plasmodium berghei involves both loss and addition of subtelomeric repeat sequences. Mol Biochem Parasitol. 1990;41:73–82. doi: 10.1016/0166-6851(90)90098-7. [DOI] [PubMed] [Google Scholar]
- Ponzi M, Pace T, Dore E, Picci E, Pizzi E, Frontali C. Extensive turnover of telomeric DNA at a Plasmodium berghei chromosomal extremity marked by a rare recombinational event. Nucleic Acids Res. 1992;20:4491–4497. doi: 10.1093/nar/20.17.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Carter R, Petersen C, Alano P, Nelson R, Aikawa M, Mattei D, Pereira da Silva L, Leech J. Gene inactivation of Pf11.1 of Plasmodium falciparum by chromosome breakage and healing: Identification of a gametocyte- specific protein with a potential role in gametogenesis. EMBO J. 1992;11:2293–2301. doi: 10.1002/j.1460-2075.1992.tb05288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnis P, Wellems T. Long-range restriction maps of Plasmodium falciparum chromosomes: Crossingover and size variation among geographically distant isolates. Genomics. 1988;3:287–295. doi: 10.1016/0888-7543(88)90117-6. [DOI] [PubMed] [Google Scholar]
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MR, Waters AP, Janse CJ. Stable transfection of malaria parasite blood stages. Science. 1995;298:1358–1362. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- van Dijk MR, Janse CJ, Waters AP. Expression of a Plasmodium gene introduced into subtelomeric regions of Plasmodium berghei chromosomes. Science. 1996;271:662–665. doi: 10.1126/science.271.5249.662. [DOI] [PubMed] [Google Scholar]
- van Lin LKM, Pace T, Janse CJ, Scotti R, Ponzi M. A long range restriction map of chromosome 5 of Plasmodium berghei demonstrates a chromosome specific symmetrical subtelomeric organisation. Mol Biochem Parasitol. 1997;86:111–115. [PubMed] [Google Scholar]
- Vernick KD, McCutchan TF. Sequence and structure of a Plasmodium falciparum telomere. Mol Biochem Parasitol. 1988;28:85–94. doi: 10.1016/0166-6851(88)90055-2. [DOI] [PubMed] [Google Scholar]
- Waters AP, Thomas A, van Dijk MR, Janse CJ. Transfection of malaria parasites. Methods: Comp to Meth Enzym. 1997a;13:134–147. doi: 10.1006/meth.1997.0506. [DOI] [PubMed] [Google Scholar]
- Waters AP, van Spaendonk RM, Ramesar J, Vervenne RA, Dirks RW, Thompson J, Janse CJ. Species-specific regulation and switching of transcription between stage-specific ribosomal RNA genes in Plasmodium berghei. J Biol Chem. 1997b;272:3583–3589. doi: 10.1074/jbc.272.6.3583. [DOI] [PubMed] [Google Scholar]