Abstract
Comparative analysis of genomic sequences provides a powerful tool for identifying regions of potential biologic function; by comparing corresponding regions of genomes from suitable species, protein coding or regulatory regions can be identified by their homology. This requires the use of several specific types of computational analysis tools. Many programs exist for these types of analysis; not many exist for overall view/control of the results, which is necessary for large-scale genomic sequence analysis. Using Java, we have developed a new visualization tool that allows effective comparative genome sequence analysis. The program handles a pair of sequences from putatively homologous regions in different species. Results from various different existing external analysis programs, such as database searching, gene prediction, repeat masking, and alignment programs, are visualized and used to find corresponding functional sequence domains in the two sequences. The user interacts with the program through a graphic display of the genome regions, in which an independently scrollable and zoomable symbolic representation of the sequences is shown. As an example, the analysis of two unannotated orthologous genomic sequences from human and mouse containing parts of the UTY locus is presented.
The genome sequencing projects are producing DNA sequence data at an exponentially growing rate. Presently there are about 20 microbal genomes completely sequenced (Institute for Genomic Research 2000). Of the eukaryotic genomes, Saccharomyces cerevisiae (Goffeau et al. 1997), Caenorhabditis elegans (C. elegans Sequencing Consortium 1998), and Drosophila melanogaster (Adams et al. 2000) are completed. Several other projects are well under way, for example, Arabidopsis thaliana and Schizosaccharomyces pombe. Most important, the human genome will be available in rough-draft form in 2000, with completion by 2003 (Marshall 1999). Following the human rough draft, we can expect other large genomes to be sequenced in the next few years, including those of other vertebrates such as mouse (Collins et al. 1998; Pennisi 1999).
Once a genome sequence is available, a primary goal is to identify functional regions in the sequences, including genes and regulatory sequences. Much of this identification will require new experimental work, but some information can be obtained purely computationally, and since this is fast and cheap, we should seek to maximize it. For prokaryotic genomes, and for lower eukaryotic genomes, this has been relatively straightforward, as the gene structures are relatively simple with few, if any, introns. In higher eukaryotes, this task is much more complicated. The EST sequencing projects will most certainly provide us with the majority of genes but not necessarily with the complete gene structures. However, some genes with low expression levels will be absent from the EST collections, and ESTs do not tell us about regulatory sequences. We will then have to rely on other methods to provide us with this information. For this, there are basically two computational options, that is, ab initio prediction and finding similarity to other sequences. The accuracy of ab initio prediction methods has improved in recent years, but these methods are still not very accurate (Burge and Karlin 1998). The alternative relies on the availability of homologous sequences, either DNA or protein. A particularly attractive method is to do comparative sequence analysis of corresponding chromosomal regions from different species. Comparing genomic sequences from species with evolutionary distance of 50–100 million years, such as human–mouse (80 million years, according to Li and Graur 1991), will not only reveal protein coding regions but also sequence elements that are important for regulation of gene expression, maintaining the structural organization of the genome, and so forth.
Comparative sequence analysis is becoming increasingly popular, and in recent years, a number of comparative studies of large genomic regions have been done, mainly between human and mouse (reviewed by Hardison et al. 1997). What is evident from these studies is that gene structures are generally very well preserved, even going as far back as the split between fish and higher eukaryotes (450 million years, according to Elgar 1996). Noncoding regions, however, show a varying degree of conservation. Conservation of noncoding regions at these evolutionary distances is generally believed to imply a functional role. In a recent study, we estimated that 20% of the length of introns is conserved between mouse and human (Jareborg et al. 1999). Some parts of these conserved regions are involved in regulating splicing of the introns, but other parts (e.g., the btk gene) are probably involved in regulating gene expression (Oeltjen et al. 1997).
There are a number of analysis programs available that are applicable to comparative sequence analysis, and genome sequence analysis in general. These include alignment programs, programs for identifying repetitive elements, programs for identifying CpG islands, gene prediction programs, and database-searching programs. All these different programs use different file formats for input and output, and the results are most often not intuitively interpretable. Graphic representations of sequences and features associated with these are often more attractive for the average biologist. Our aim was to develop a graphic front end that would simplify both the execution of relevant analyses and the interpretation of the results of these analyses. Although there exist other programs that give this type of functionality, for example, the ACEDB genome database-management system (Durbin and Thierry Mieg 1991), the Genotator annotation workbench (Harris 1997), or the Java based Bioviews package (Helt et al. 1998), none existed that was tailored for comparative genome sequence analysis. Here we present such a program, which we have called Alfresco, that in its current implementation handles a pair of genomic sequences of several hundred kilobases (kb) and provides functionality for doing comparative analyses. The sequences and the features determined through different analyses are presented as graphic objects that the user can examine interactively. The user can select and investigate further specific regions that appear interesting, construct gene structures, and save and retrieve a working set of analysis and results.
RESULTS
General Overview
We decided to develop Alfresco using the Java programming language (Gosling and Arnold 1996), as this provided several advantages such as object orientation, memory management, platform independence, and the availability of several well-designed libraries, for example, that for windowing widgets and graphics programming. It also offers the possibility to run the programs through a Web browser as a so-called applet. The central idea of Alfresco was to provide a graphic front end for a variety of analysis programs. As mentioned above, there are several other programs with this functionality, but none tailored for comparative analysis. An alternative to implement a new system would have been to modify one of these existing workbenches. The need to represent two sequences has fundamental implications for the design of the program, and it seemed to us worth developing an application that built this in from the start. We also were committed to using Java and did not have an available public domain Java source code to build on when we started the project.
In it's first instance, Alfresco was implemented as a stand-alone program that handled a pair of genomic sequences and could access external programs through system calls on any computer running the UNIX operating system. However, since it might be impractical for all users to install analysis programs locally, Alfresco offers the possibility to run the analysis programs remotely on a server using the CORBA (Common Object Request Broker Architecture) technology [Object Management Group 1999, see below]. Together with Java, this provides platform independence for the program. Alfresco can thus be used on UNIX, Macintosh, and Windows systems even though the analysis programs used are UNIX based.
When Alfresco is first started, the user is presented with the main window (Fig. 1A). In the upper half, there are two buttons for selecting input sequences. Alfresco reads DNA sequence files in FASTA format. Underneath the file selection buttons are four rows for status messages: the general status of the program, current zoom levels of the sequences, and a selected-feature label. The bottom half contains the sequence map canvas. Menus provide different types of functionality, such as calling various analysis programs and opening/saving analyzed sequence pairs.
Figure 1.
Alfresco user interface. (A) The display area shows a representation of two EMBL sequence entries of the orthologous mouse and human keratin 18 genes (mouse: ID MMENDOBA, AC M22832; human: ID HSKER101, AC M24842). Conserved regions are connected by gray lines. Dark yellow boxes represent conserved regions found by BLASTN/MSPcrunch, boxes in different shades of blue represent conserved regions found by DBA (see text). Gene structures are represented by blue borders and a name tag, with “>” and “<” indicating the direction of transcription. Coding and noncoding exons are indicated in green and yellow, respectively. The shown gene structures are the result of analyses with Blastwise, blEst_genome, and Genscan, respectively. Sequence repeats found by RepeatMasker are indicated by boxes in blue (LINEs), green (SINEs), brown (low complexity), and pink (simple repeats). CpG islands predicted by Cpg are shown as yellow boxes around the start of the first introns. The attributes of selected features (colored red) can be manually edited through a dialog window (seen on the right). (B) Subregions of the sequences can be selected and subjected to further analysis. To the right of the main window are shown a window displaying alignments of conserved regions found by DBA and a Dotter window of the selected regions. Conserved blocks found by DBA can be added to the display as features of the entries. Positions of features defined in Alfresco is exported to Dotter and displayed along the sides of the dot plot. (C) Regions similar to any other region can be identified using the “Region set parameters” dialog. These regions are displayed as dark gray boxes. The Cutoff scrollbar selects the threshold of similarity. Clicking on a region (the red box in the figure) will indicate which other regions belong to the same set by displaying blue boxes above the sequence representations. A selected Region set can be permanently added to the display with a choice of colors.
Sequence Map Canvas
The central part of the Alfresco main window is the sequence map canvas. It is on this canvas that the graphic representations of the sequences are shown, and it is through this display that the user interacts with the sequences and their associated features. The sequence representations can be independently scrolled using the horizontal scrollbars above and below the map canvas. The horizontal scrollbar at the bottom of the window allows the sequences to be synchronously scrolled. Similarly, the rightmost vertical scrollbar is used for synchronous zooming of the sequence representations, and the vertical scrollbars next to each sequence are used for individual zooming.
Sequences are represented as lines and features as open or solid boxes. Features can be located on different levels, or tiers. For the upper sequence these are on or above the sequence line; for the lower sequence the higher tiers are below the sequence line. Reciprocal features in the two sequences, such as corresponding exons, are represented by connecting gray lines (reciprocals). Features can be selected by clicking with the mouse button, and several features can be selected by subsequent clicking while holding down the shift key. Ranges of the sequences can also be selected by clicking and dragging the mouse. Information about selected features or ranges is shown above the sequence map canvas. The subsequence covered by a selected feature or range can be viewed by selecting a menu option. This will bring up a separate window with that DNA sequence. This sequence can then be saved to a local file.
Alfresco implements a visualization feature termed semantic zooming (Bederson and Hollan 1994), that is, showing a graphic object only at a magnification level that is relevant for the object in question. For example, it is not relevant to show a short feature, like an initiation codon, at a magnification level where the sequence representation shows several tens of kilobases. If the view is zoomed to a higher magnification level it will become relevant to show the feature and it will be displayed. Alfresco at the moment implements semantic zooming for the scale bars, for ATG initiation codons, and for the actual DNA sequence that will be displayed at the highest magnification level.
External Programs
One of the main design ideas of Alfresco was to use already-existing analysis programs that are relevant for comparative genome sequence analysis. The external programs can be installed locally on the same computer running Alfresco or accessed as distributed software objects from a CORBA server running on a remote machine (or on the local machine, for that matter).
CORBA (Object Management Group 1999) is an open middleware standard that contains specifications for a distributed object-oriented framework. CORBA has been defined by OMG, a consortium of >700 leading software and information technology companies. Definitions for describing data and methods as objects are formulated in a formal metalanguage (Interface Definition Language [IDL]). CORBA provides interoperability between applications and data on different machines in heterogeneous distributed environments and can seamlessly interconnect multiple object systems. The mapping between IDL and several programming languages makes it very easy to implement a client-server architecture, as remote objects are used as if they were native local objects regardless of where they reside and in which programming language they were implemented. In the bioinformatics domain, databases and aplications have been developed that use CORBA to provide introperability (Rodriguez-Tome et al. 1997; Lijnzaad et al. 1998; Barillot et al. 1999; Parson et al. 1999). In our minds, CORBA provided a more flexible and more easily maintainable way to implement the remote invocations of analysis methods than other technologies, such as the use of sockets.
CORBA server wrappers have been written for several of the external analysis programs mentioned below. The IDL for the CORBA methods server is shown in Box 1. The core interface is the “Method” interface that specifies the functions needed to perform an analysis. All method wrapper objects implement the “Method” interface. However, it should be noted that the “Method” interface is very general and that for other programmers to use it, further information about specific methods, such as parameters, is needed. When the user requests an analysis method, the appropriate server will be contacted and provided with the necessary information (e.g., a DNA sequence and method parameters). When the server has performed the analysis, the result will be sent back to Alfresco in GFF (General Feature Format) and CGFF (Comparative General Feature Format) text formats (see Saving and Transferring Information section below) and the results displayed. Parameters for the different programs can be altered through dialog windows for the different methods that are also used to specify if the program should be run locally or from a CORBA server. This provides flexibility and functionality for users who might not want to install all external programs locally or who use platforms for which the analysis programs are not available. A short description of the external programs and how the results are used and displayed by Alfresco follows.
RepeatMasker
RepeatMasker finds interspersed nuclear elements (LINEs and SINEs), DNA transposons, LTR repeats, simple repeats, and low-complexity regions (Smit and Green 1995). Alfresco calls the RepeatMasker program with each sequence and displays the repeats found as different repeat objects colored depending on their type (Fig. 1A). Identified repeats will affect other subsequent analysis. The regions covered by repeats will be masked as 'N's before doing Genscan predictions, Cpg predictions, BLAST searches, BLAST/MSPcrunch alignments, and DBA alignments (see below).
Cpg
CpG islands are genomic regions with an enrichment of the CG dinucleotide. These islands are associated with coding regions, and it has been estimated that half of all human genes will be associated with CpG islands (Antequera and Bird 1993). Alfresco uses the program Cpg (G. Micklem and R. Durbin, unpubl.) for determining the presence of CpG islands. By default a CpG island is defined as a DNA stretch at least 200 bp long with a GC content > 50% and an observed to expected ratio of CpG dinucleotides > 0.6 (Gardiner-Garden and Frommer 1987).
GeneWise
Database hits from searching protein sequence databases with BLASTX (Altschul et al. 1997) are aligned to the genomic query sequence using the GeneWise program from the Wise2 package (Birney and Durbin 1997). GeneWise is a program that aligns a protein sequence to a genomic DNA sequence, allowing for introns and frameshifting errors. Alfresco uses a script, blastwise.pl, that is part of the Wise2 distribution. The blastwise script initiates a BLASTX search against a specified protein sequence database and selects the most significant nonoverlapping hits for GeneWise aligning to the genomic sequence. Predicted gene structures are then displayed as a gene object (Fig. 1A).
Est_genome
Database hits from searching RNA or EST sequence databases with BLASTN are aligned to the genomic sequence using the est_genome program (Mott 1997).Est_genome is a program that aligns an mRNA sequence to a genomic sequence allowing for introns (and sequencing errors). A Perl script, blEst_genome, was written to automate the BLASTN search against a specified nucleotide database (e.g., the EST or RNA subsections of EMBL), select significant nonoverlapping hits, and submit these to est_genome alignment. As with GeneWise, the output from est_genome for significant database hits is displayed as predicted gene structures (Fig. 1A).
Genscan
Genscan is a probabilistic gene prediction program (Burge and Karlin 1997). It predicts whole gene structures, as well as separate suboptimal coding exons. It also has a simple promoter prediction model and a polyA-site model. It was chosen as the most accurate gene prediction program at the time of the development of Alfresco. Predicted genes are displayed on the sequence map (Fig. 1A).
Dotter
Dotter (Sonnhammer and Durbin 1995) is a dot plot program with useful features such as a dynamic gray-scale ramp, an alignment viewer, and zooming. It also allows the display of sequence features as graphic objects on the sides of the plot. Two regions or sequence features can be selected in Alfresco and inspected in Dotter. Features associated with the regions in Alfresco will be exported to Dotter through the Sequence Feature Segments (SFS) format (E. Sonnhammer and J. Wootton, pers. comm.) and displayed in the dot plot window (Fig. 1B). Dotter is also used for the RegionSet method (see below).
Blast/MSPcrunch Alignments
To provide a rapid method for identifying conserved sequence regions, Alfresco uses the BLASTN program (Altschul et al. 1997). One of the sequences is converted to a blast-formatted database, which is then searched using the other sequence as a query. The blast output is then parsed and filtered by MSPcrunch (Sonnhammer and Durbin 1994) to remove insignificant matches. Matches are displayed as features in both sequences with a connecting reciprocal between them (Fig. 1A).
DBA
DBA (Jareborg et al. 1999) is a dynamic programming alignment program that finds and aligns colinear conserved blocks in two DNA sequences. This method is well suited for aligning genomic sequences, which typically contain nonalignable regions flanking the conserved portions. The aligned blocks are divided into four categories depending on the degree of identity (roughly 60%–70%, 70%–80%, 80%–90%, and 90%–100%). DBA is generally more sensitive than BLAST but assumes that the regions of similarity are colinear (for more details, see Jareborg et al. 1999). Regions or features of the sequences on the sequence map canvas can be aligned after selection of two regions or features. Repeats (see “RepeatMasker” above) that overlap the selected regions are masked and excluded. The resulting aligned blocks are shown in a separate window and can be added back on to the sequence map as highly conserved region (HCR) features in both sequences with a connecting reciprocal between them (Fig. 1B). The HCR features are color and size coded. The higher the identity of the conserved blocks the broader and more darkly blue colored is the graphic representation. The alignments displayed in the alignment window can also be saved to a local text file.
Adapting Alfresco to Other Analysis Programs
Incorporating other programs into Alfresco menus is also possible and, because of the design of Alfresco, can readily be done. It does, however, currently involve altering the source code. Output from other analysis programs can also be read by Alfresco if the output can be converted to the GFF format (see below).
Internal Implementations
To provide some further functionality, we also implemented some analysis methods internally in the Alfresco code.
Region Sets
A Region set is a group of sequence regions that are similar to each other, regardless of their position or orientation or in which sequence entry they reside. The degree of similarity detected can be altered by adjusting the cutoff through a parameter dialog box (Fig. 1C). Regions belonging to a Region set with a score above the cutoff will be displayed on the sequence canvas as dark-gray boxes. Selecting one of the boxes will indicate where the other regions in the set are located by displaying light-blue boxes above (or below) the initial regions (Fig. 1C). Such a Region set can be added to the canvas and given a custom color from the parameter dialog. Alfresco uses Dotter and DBA to calculate the Region sets. The shortest sequence is divided into 100 regions, and an approximately 100 × 100 dot plot score matrix is calculated for all three sequence combinations (two self-comparisons and one intersequence comparison) using Dotter. Only those regions with scores above the cutoff will be displayed. DBA is used to better define the regions of similarity within the set.
Global and Local Alignments
Simple global (Needleman and Wunsch 1970) and local (Smith and Waterman 1981) alignment algorithms with linear gap penalties were implemented to provide gapped alignment of short sequence regions.
Batch Mode
Some of the analysis methods available in Alfresco, such as RepeatMasker, DBA, and Region sets, will be quite time consuming when the sequences analyzed are long. In those cases, Alfresco provides a ‘batch’ command line option that will carry out a predefined set of analyses: RepeatMasker, Cpg, Blast/MSPcrunch alignment, GeneWise, est_genome, and Genscan. These analyses are typically run in the background overnight and are carried out without displaying the graphic user interface. Instead, results are saved to a file that can be opened for visual inspection later.
Editing of Features
The program provides the possibility to edit and create sequence features through a dialog window (Fig. 1A). Feature coordinates, type, tier, and strand can be set through this dialog. Genes can be constructed from a combination of selected features (exons, UTRs, promoters, or polyA sites). Reciprocals can be constructed from two features from the two different entries.
Saving and Transferring Information
Analyzed sequence pairs can be saved to a persistent file that allows the user to maintain and edit a working annotation of a sequence pair. Results can also be saved in the GFF text format (R. Durbin and D. Haussler, unpubl.; http://www.sanger.ac.uk/Software/GFF). GFF is an easily parsable text format, initially devised for describing gene-finding features but also useful for describing any simple feature in a sequence. However, it does not specify how to describe information about reciprocal features. For this purpose, an additional text format, named CGFF, was designed to hold the reciprocal information in one or more GFF files.
Alfresco can also read GFF files generated by other programs, which provides a simple way to use analysis results from other programs without the need to alter the Alfresco code.
Applet
One of the advantages of the Java programming language is the ability to distribute programs over the World Wide Web without needing to install the application locally on the executing machine. This is done by implementing a so-called applet that is embedded in a Web page that will be downloaded to the computer accessing the page in question. This feature of the Java language was one of the reasons for choosing it as a development language. However, due to security issues, applets do have some drawbacks, mainly that an applet is not generally allowed to access the local file system or execute locally installed programs. This means that an applet will need to provide other means of handling sequences to be analyzed and calling analysis programs. Currently there is an applet version of Alfresco that can be used to display preanalyzed sequences pairs. It offers the same exploratory features as the stand-alone version but does not provide any analysis methods except the internally implemented local and global alignment methods. The preanalyzed sequence pairs are obtained from a CORBA server. The applet can be reached at http://kisac.cgr.ki.se/nic/alfrescoapplet and requires a Web browser that is jdk1.1 compliant.
Analysis of Two Orthologous Mouse/Human Genome Sequences
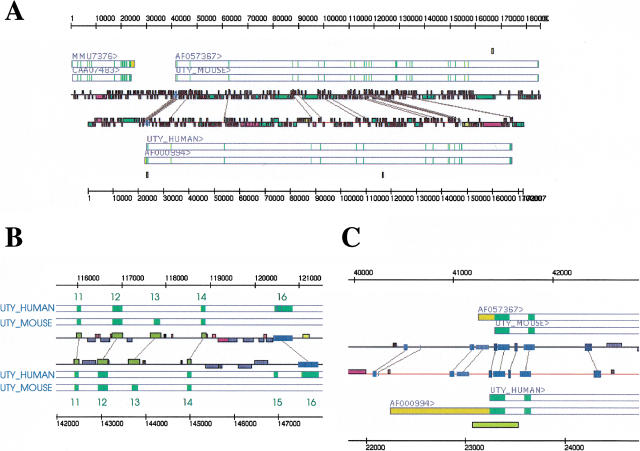
As an example of the use of Alfresco, two orthologous unannotated mouse and human genomic sequences were analyzed. Large mouse genomic sequences lacking extensive annotations were extracted from the EMBL nucleotide database (release 60). These sequences were then used to search a database of human genomic sequences > 50 kb with BLASTN. High-scoring matches were selected if they did not contain any informative annotations. Selected human entries were used to search a database with mouse genomic sequences > 3 kb with BLASTN to make sure that the highest-scoring match to the human sequence was the mouse entry used initially. This was done to select entries that were the best available candidates for an orthologous pair. From the resultant pairs, the entries AC006508 (mouse) and AC006376 (human) were chosen. The mouse entry corresponds to the reverse-complement of the human entry, so in the subsequent analysis the reverse-complement of the human entry was used. This pair of entries was then analyzed using Alfresco. AC006508 contained the genomic sequence for all 17 exons of the Dby gene and 25 exons of the mouse Uty gene (Mazeyrat et al. 1998), and AC006376 contained 16 exons of the human UTY gene (Lahn and Page 1997; Fig. 2A). Nearly the whole mouse Uty is covered, coding for 1148/1212 amino acids (Swissprot id: UTY_MOUSE). The portion of the UTY gene that is coded by the human entry is 848/1347 amino acids (Swissprot id: UTY_HUMAN).
Figure 2.
Analysis of the mouse and human UTY genomic regions. (A) Overall view of the region. The mouse sequence (AC006508) is shown at the top and the human sequence (AC006376) at the bottom. Gene structures are the result of analysis with blEst_genome and Blastwise. The sequence IDs of the RNA and protein entries from EMBL, SWISSPROT, and TREMBL that have been aligned to the genomic sequences are shown above the gene structure representations. Regions of similarity detected by BLASTN/MSPcrunch and DBA are connected by gray lines. Regions covered by repeats are represented as colored boxes on the lines representing the sequences. (B) Exons 11–16. Exon positions are derived from GeneWise alignments of the mouse and human protein sequences (Swissprot id: UTY_MOUSE and UTY_HUMAN) with the two genomic sequences as indicated. Dark-yellow blocks connected by gray lines are conserved regions found by BLASTN/MSPcrunch, blue boxes are regions found by DBA. The mouse sequence uses exon 13 but not exons 15 and 16. The human sequence uses exons 15 and 16 but not exon 13. Exon 13 is conserved in both species and parts of exon 16 is also conserved in both species, whereas exon 15 is not. The conservation of these exons indicates the possibility of alternatively spliced transcripts in both species. (C) Conservation of noncoding regions of the 5′ end of the UTY gene. Blocks in different shades of blue connected by gray lines are conserved regions found by DBA. The upstream region contains one conserved block upstream of a possible TATA box (short light-blue region). The 5′ UTR contains two conserved blocks, and the first and second introns contain one conserved region each that are separate from the conservation seen around the exons. These blocks contain several conserved transcription factor-binding sites (see text). A predicted CpG island represented by a yellow box covers parts of the first exon and intron in the human sequence.
The 17 exons of the human gene cover a considerably larger region than the corresponding mouse region, 144 kb in the human sequence versus 87 kb in the mouse sequence, corresponding to a factor of × 1.6. This difference in size is largely explained by the presence of a larger proportion of repetitive elements in the human sequence. Both entries are rich in repeats, although the human entry is more so: 64% of the human entry is made up by repetitive elements, versus 47% of the mouse entry. In the mouse sequence, the LINE1 type is the dominant type covering 24% of the entry. In the human sequence, the dominant type is also the LINE1 type covering 30% of the total sequence, and a substantial portion (12%) is made up of Alu type repeats.
Most exons are conserved in both species (Fig. 2B). Interestingly, the published mRNA sequences indicate alternative splicing. In the human entry there are 17 possible exons. The available human RNAs use all of these except exon 13. This exon is, however, identical to the mouse sequence (134/135 nucleotides; 45/45 amino acids). The mouse, however, uses this exon but does not use exons 15 and 16. The greater part of exon 16 is conserved to some degree, but alignment of the human UTY protein sequence to the mouse genomic sequence with GeneWise indicates frameshifts in the mouse sequence. This could either be a true alteration in the mouse or be caused by a sequencing error. The short exon 15 is not conserved in the mouse sequence. This indicates that, at least in human, there are two alternatively spliced transcripts. The absence of exon 15 and the frameshifts in exon 16 in mouse could indicate that the transcript reported in human is not expressed from the mouse genome.
The human gene contains a CpG island around the first exon. A corresponding CpG island is not identified by the Cpg program in the mouse sequence.
In the 5′ end only short regions are conserved as determined by DBA (Fig. 2C). One conserved block is located 135 nt upstream of the start of the longest described human mRNA (EMBL accession no. AF000994), the 5′ UTR contains two conserved regions, and the first and second introns each contain a short conserved block. The conserved blocks were further analyzed with the MatInspector program (Quandt et al. 1995) to identify transcription-factor binding sites described in the TRANSFAC database (Wingender et al. 2000) that were conserved in the sequences of the two species. The upstream block contains conserved binding sites for the C/EBPalpha, C/EBPbeta, and GATA transcription factors. Downstream of this block a possible TATA box can be observed in both species. The blocks in the 5′ UTR and the two first introns also contain conserved binding sites for factors such as AP-1, GATA, Myc/Max, and cEts-1. UTY is a ubiquitously expressed protein, and the regulation of transcription is probably not that complex. This is reflected in the relatively few conserved regions around the first exons.
Conclusions
The employment of comparative genome sequence analysis will become more important in the near future as a tool to uncover coding and regulatory regions in the genomic DNA sequences from the model organisms now being sequenced. It will also tell us a lot about the evolution of the different species in question. There are many ways of analysing two orthologous, or paralogous, regions. Alfresco provides one approach, where the actual running of different analysis programs is hidden from the user and the results are presented through an interactive graphic front end that allows selection and further investigation. We believe that this is an important aspect of bioinformatics. The results of bioinformatics methods must be presented in a way that makes sense to the biologists who are to interpret the data. This is especially true when the information content associated with a particular sequence increases. Alfresco is useful as a tool for the biologist or bioinformatician who is interested in analyzing a specific genomic region to a greater extent. This is in contrast to the global, automated comparative analysis that will undoubtedly be performed as large amounts of comparative data appear during the next few years. One of the primary goals with the mouse genome sequencing project should be to better annotate the human genome sequence.
An alternative to Alfresco is the recently described PipMaker server (Schwartz et al. 2000). PipMaker provides displays of alignments between two DNA sequences in the form of static percentage identity plots that give a good overview of the degree of similarity between the sequences. The locations of features such as repeats and exons can also be displayed, but these have to be provided by the user.
In the short term there is still functionality missing in Alfresco. For example, it would be nice to search conserved regions, for example, DBA blocks, for conserved transcription factor binding sites from a library such as TRANSFAC (Wingender et al. 2000), to help predict functional promoter elements. It will also be of importance to provide ways of accessing other nonlocal analysis and database resources on the network. The CORBA architecture provides a potential means to do this. Finally, it would be nice to analyze more than two homologous sequences at a time. Alfresco is now being tested by biologists at several sites, and it has been found to be a useful tool for comparative genome sequence analysis.
METHODS
Java
Alfresco was developed using the Java Development Kit version 1.1 (Sun Microsystems 1997). It has been tested on Digital UNIX, Irix, Solaris, Linux, and to some degree on computers running Mac OS and Windows 95. Source code and precompiled Java classes can be downloaded from http://www.sanger.ac.uk/Software/Alfresco.
CORBA
The ORBacus for Java (Object Oriented Concepts, http://www.orbacus.com/ob) object request broker was used to provide remote connectivity through the CORBA architecture.
External Analysis Programs
The Alfresco program employs a number of external programs. RepeatMasker (Smit and Green 1995) and Genscan (Burge and Karlin 1997) were obtained from the authors; Dotter (Sonnhammer and Durbin 1995) was obtained from http://www.sanger.ac.uk/Software/Dotter; Cpg was developed at the Sanger Centre (Micklem and R. Durbin, unpubl.); DBA (Jareborg et al. 1999) is available from http://www.sanger.ac.uk/Software/Wise2; GeneWise and BlastWise (Birney and Durbin 1997) were obtained from http://www.sanger.ac.uk/Software/Wise2/; and the BLAST programs (Altschul et al. 1997) were obtained from ftp://ftp.ncbi.nlm.nih.gov/blast.
Sequence Data
Mouse and human genomic sequences were obtained from the EMBL nucleotide sequence database, release 60 (Stoesser et al. 1999).
Table.
Box 1.
| module alfresco{ |
| module corba_wrappers{ |
| exception NoSuchMethodException { |
| string reason; |
| }; |
| exception NoOutputException { |
| string reason; |
| }; |
| typedef sequence〈string〉 string_seq: // array of strings |
| struct InputStruct { |
| string_seq names; |
| string_seq seqs; |
| string_seq parameters; |
| }; |
| struct GffDataStruct{ |
| string gff;// primary feature information |
| string cgff;// reciprocal feature pair data |
| string_seq suplements;// e.g. alignment info |
| }; |
| interface Method { |
| void input (in InputStruct input); |
| void run (); |
| GffDataStruct getGffData () raises (NoOutputException); |
| }; |
| interface Server { |
| Method getMethod (in string methodName) raises |
| (NoSuchMethodException); |
| string_seq getAvailableMethods (); |
| }; |
| }; |
| }; |
Acknowledgments
N.J. was supported by a Marie Curie Fellowship grant (contract FMBICT961755) from the European Commission and by a grant from the Wenner-Gren Foundation. R.D. was supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL niclas.jareborg@cgr.ki.se; FAX 46 8 337983.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. .1995–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillot E, Leser U, Lijnzaad P, Cussat-Blanc C, Jungfer K, Guyon F, Vaysseix G, Helgesen C, Rodriguez-Tome P. A proposal for a standard CORBA interface for genome maps. Bioinformatics. 1999;15:157–169. doi: 10.1093/bioinformatics/15.2.157. [DOI] [PubMed] [Google Scholar]
- Bederson, B.B. and Hollan, J.D. 1994. Pad++: A zooming graphical interface for exploring alternate interface physics. Proceedings of the ACM Symposium on User Interface Software and Technology, November 2–4, 1994, Marina del Ray, CA USA. p. 17–26.
- Birney, E. and Durbin, R. 1997. Wise2. http://www.sanger.ac.uk/Software/Wise2.
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- ————— Finding the genes in genomic DNA. Curr Opin Struct Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U.S. Human Genome Project: 1998–2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- Durbin, R. and Thierry Mieg, J. 1991. A C. elegans database. http://www.acedb.org.
- Elgar G. Quality not quantity: The pufferfish genome. Hum Mol Genet. 1996;5:1437–1442. doi: 10.1093/hmg/5.supplement_1.1437. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Aert R, Agostini-Carbone ML, Ahmed A, Aigle M, Alberghina L, Albermann K, Albers M, Aldea M, Alexandraki D, et al. The yeast genome directory. Nature. 1997;387(Suppl.):5–105. [Google Scholar]
- Gosling J, Arnold K. The Java programming language. Reading, MA: Addison-Wesley; 1996. [Google Scholar]
- Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Harris NL. Genotator: A workbench for sequence annotation. Genome Res. 1997;7:754–762. doi: 10.1101/gr.7.7.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helt GA, Lewis S, Loraine AE, Rubin GM. BioViews: Java-based tools for genomic data visualization. Genome Res. 1998;8:291–305. doi: 10.1101/gr.8.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute for Genomic Research. 2000. TIGR Microbial Database: A listing of microbial genomes and chromosomes completed and in progress. http://www.tigr.org/tdb/mdb/mdb.html.
- Jareborg N, Birney E, Durbin R. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 1999;9:815–824. doi: 10.1101/gr.9.9.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Li W-H, Graur D. Fundamentals of molecular evolution. Sunderland, MA: Sinauer; 1991. [Google Scholar]
- Lijnzaad P, Helgesen C, Rodriguez-Tome P. The Radiation Hybrid Database. Nucleic Acids Res. 1998;26:102–105. doi: 10.1093/nar/26.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E. Human Genome Project: Sequencers endorse plan for a draft in 1 year. Science. 1999;284:1439–1441. doi: 10.1126/science.284.5419.1439. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Sargent CA, Grimmond S, Longepied G, Ehrmann IE, Ellis PS, Greenfield A, Affara NA, Mitchell MJ. The mouse Y chromosome interval necessary for spermatogonial proliferation is gene dense with syntenic homology to the human AZFa region. Hum Mol Genet. 1998;7:1713–4724. doi: 10.1093/hmg/7.11.1713. [DOI] [PubMed] [Google Scholar]
- Mott R. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci. 1997;13:477–478. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Object Management Group. 1999. The Common Object Request Broker: Architecture and specification. http://www.omg.org/cgi-bin/doc?formal/99-10-07.
- Oeltjen JC, Malley TM, Muzny DM, Miller W, Gibbs RA, Belmont JW. Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Parsons JD, Buehler E, Hillier L. DNA sequence chromatogram browsing using JAVA and CORBA. Genome Res. 1999;9:277–281. [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Mouse genome added to sequencing effort. Science. 1999;286:210. doi: 10.1126/science.286.5438.210. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Tome P, Helgesen C, Lijnzaad P, Jungfer K. A CORBA server for the Radiation Hybrid DataBase. ISMB. 1997;5:250–253. [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A.F. and Green, P. 1995. RepeatMasker. http://ftp.genome.washington.edu/RM/RepeatMasker.html.
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A workbench for large-scale sequence homology analysis. Comput Appl Biosci. 1994;10:301–307. doi: 10.1093/bioinformatics/10.3.301. [DOI] [PubMed] [Google Scholar]
- ————— A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Stoesser G, Tuli MA, Lopez R, Sterk P. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 1999;27:18–24. doi: 10.1093/nar/27.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Microsystems. 1997. Java development Kit 1.1: The universal internet/intranet software development and deployment platform. http://www.java.sun.com/marketing/collateral/jdk_sc.html.
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pr M, Reuter I, Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]