Abstract
We measured the ability of Staphylococcus epidermidis to form biofilms in the presence of subminimal inhibitory (sub-MIC) concentrations of vancomycin, tigecycline, linezolid and novobiocin. Six strains that produce different amounts of biofilm were tested. The three strains that produced the highest amounts of biofilm exhibited steady-state or decreased biofilm formation in the presence of sub-MIC antibiotics, whereas the three strains that produced lower amounts of biofilm exhibited up to 10-fold-increased biofilm formation in the presence of sub-MIC antibiotics. In two of the inducible strains (9142 and 456a), antibiotic-induced biofilm formation was inhibited by dispersin B, an enzyme that degrades poly-N-acetylglucosamine (PNAG) biofilm polysaccharide. In the third inducible strain (RP62A), dispersin B inhibited biofilm formation in response to sub-MIC vancomycin, but not to sub-MIC tigecycline. In contrast, DNase I efficiently inhibited biofilm formation by strain RP62A in response to sub-MIC tigecycline and vancomycin. DNase I had no effect on antibiotic-induced biofilm formation in strains 9142 and 456a. Our findings indicate that antibiotic-induced biofilm formation in S. epidermidis is both strain- and antibiotic-dependent and that S. epidermidis RP62A utilizes an extracellular DNA-dependent mechanism to form biofilms in response to sub-MIC antibiotics.
Keywords: Biofilm matrix, Crystal violet binding assay, Pulmozyme, rhDNase, Staphylococcus epidermidis NJ9709, Staphylococcus epidermidis 1457
1. Introduction
Staphylococcus epidermidis is a leading cause of prosthetic device infections and catheter-related bacteremia (Huebner and Goldman, 1999). Evidence suggests that the ability of S. epidermidis to form biofilms on abiotic surfaces contributes to its virulence (Christensen et al., 1982). S. epidermidis biofilms are comprised of clusters of cells encased in a self-synthesized extracellular polymeric matrix. The biofilm matrix holds the cells together, attaches them to the surface and protects them from killing by host defenses and antimicrobial agents. A major component of the S. epidermidis biofilm matrix is poly-N-acetylglucosamine (PNAG), a cell surface polysaccharide that mediates various biofilm-related functions including intercellular adhesion (Mack et al., 1994) and resistance to killing by antimicrobial peptides and phagocytes (Vuong et al., 2004a, 2004b). The genetic locus that encodes the production of PNAG (icaADBC) is present in strains isolated from infections more frequently than in commensal skin isolates, suggesting that PNAG plays a role in human infections (Kozitskaya et al., 2005; Rogers et al., 2008). Other structural components of the S. epidermidis biofilm matrix include extracellular DNA (ecDNA) (Izano et al., 2008; Qin et al., 2007), proteins (Hussain et al., 1997) and teichoic acids (Sadovskaya et al., 2004). Because of the protective nature of biofilms, antibiotics usually fail to eradicate S. epidermidis device-related infections (O’Gara and Humphreys, 2001).
Numerous in vitro studies have shown that various antibiotics, when present in the culture medium at subminimal inhibitory (sub-MIC) concentrations, can induce bacterial biofilm formation. Antibiotics that have been shown to induce S. epidermidis biofilm formation include cefamandole (Dunne, 1990), vancomycin (Cargill and Upton, 2009; Dunne, 1990), linezolid (Frank et al., 2007), rifampicin (Schadow et al., 1988), tetracycline (Rachid et al., 2000), the macrolides azithromycin, clarithromycin and erythromycin (Wang et al., 2010) and the fluoroquinolones ciprofloxacin, ofloxacin and sparfloxacin (Pérez-Giraldo et al., 1994). Sub-MIC concentrations of tetracycline, erythromycin and quinpristin-dalfopristin have been shown to increase expression of the ica genes (Rachid et al., 2000), suggesting that antibiotic-induced biofilm formation may depend on increased PNAG production. However, Wang et al. (2010) showed that enhancement of biofilm formation by sub-MIC concentrations of macrolides was independent of the ica status of the strain. In their study, 25% of ica-positive strains (3 out of 12) and 50% of ica-negative strains (5 out of 10) exhibited increased biofilm formation in the presence of macrolides at 1/4 MIC (Wang et al., 2010). Thus, the precise role of PNAG in antibiotic-induced biofilm formation is unclear, and PNAG-independent mechanisms of induction must exist. The process of antibiotic-induced biofilm formation may have clinical relevance because bacteria are exposed to sub-MIC concentrations of antibiotics at the beginning and end of a dosing regimen, between doses, or continuously during low-dose therapy (Craig, 1998). In addition, cells buried deep within a biofilm colony may be exposed to sub-MIC concentrations of some antibiotics because of diffusion gradients (Singh et al., 2010).
In this study, we investigated the effects of sub-MIC levels of four structurally distinct antibiotics (vancomycin, tigecycline, novobiocin and linezolid) on biofilm formation by six ica-positive strains of S. epidermidis. These strains included the commonly used reference strain RP62A, as well as five other strains that produce different amounts of biofilm in vitro. Here we show that sub-MIC concentrations of all four antibiotics can induce S. epidermidis biofilm formation, but the amount of induction depends on the antibiotic and the amount of biofilm produced by the test strain. We also show that biofilm formation by strain RP62A in response to sub-MIC vancomycin and tigecycline depends on the production of ecDNA.
2. Materials and methods
2.1. Reagents
Vancomycin was purchased from MP Biochemicals, novobiocin was purchased from Sigma, and linezolid and tigecycline were obtained from Pfizer and Wyeth, respectively. Dispersin B (DspB), a PNAG-degrading enzyme (Kaplan et al., 2004), was obtained from Kane Biotech. Recombinant human DNase I (rhDNase) was obtained from Genentech.
2.2. Bacterial strains
The S. epidermidis strains used in this study were are listed in Table 1. All strains were isolated from infected catheters or prosthetic joints and all strains carried the icaADBC locus responsible for PNAG production (Gerke et al., 1998). Bacteria were passaged weekly on tryptic soy agar and stored at 4°C.
Table 1.
Maximum-fold induction of S. epidermidis biofilm formation in response to sub-MIC concentrations of four antibiotics.a
| Antibiotic | |||||
|---|---|---|---|---|---|
| Strainb | Biofilm phenotypec |
||||
| Vancomycin | Tigecycline | Linezolid | Novobiocin | ||
| 5 | 302 | 1.0 ± 0.0 | 1.1 ± 0.0 | 1.0 ± 0.0 | 1.2 ± 0.1 |
| 1457 | 170 | 1.0 ± 0.0 | 1.5 ± 0.1** | 1.6 ± 0.4 | 1.6 ± 0.3* |
| NJ9709 | 123 | 1.3 ± 0.0* | 1.4 ± 0.0** | 1.3 ± 0.0* | 1.1 ± 0.0 |
| RP62A | 100 | 1.9 ± 0.5* | 2.3 ± 0.2** | 1.4 ± 0.1** | 4.0 ± 0.1** |
| 9142 | 60 | 1.9 ± 0.8 | 6.8 ± 0.6** | 6.8 ± 0.8** | 6.5 ± 1.5** |
| 456a | 34 | 2.1 ± 0.0** | 9.1 ± 1.9** | 5.8 ± 0.7** | 7.8 ± 0.8** |
Values show mean maximum biofilm induction (± sd) from at least three independent crystal violet binding assays. Biofilm induction = A[with antibiotic]/A[without antibiotic].
P < 0.05;
P < 0.01.
References: Strains 5 and 456a (Chokr et al., 2006); 1457 (Mack et al., 1992); NJ9709 (Kaplan et al., 2004); RP62A (Christensen et al., 1985); 9142 (Mack et al., 1994).
Average amount of crystal violet binding in the absence of antibiotics compared to reference strain RP62A. Values = A[test strain]/A[strain RP62A] × 100. All values are ± 10%.
2.3. Biofilm formation assay
Biofilms were cultured in 96-well polystyrene microtiter plates as previously described (Izano et al., 2008). Briefly, cells were diluted to 104 to 105 CFU/ml in tryptic soy broth supplemented with antibiotics and enzymes at the indicated concentrations. Aliquots of cells (200 µl each) were transferred to the wells of a microtiter plate and the plate was incubated at 37°C for 16 ± 2 h. For the crystal violet binding assay, biofilms were washed extensively with water and then stained for 1 min with 200 µl of Gram’s crystal violet. Stained biofilms were rinsed with water and dried. The amount of biofilm biomass was quantitated by destaining the biofilms for 10 min with 200 µl of 33% acetic acid (by vol) and then measuring the absorbance of the crystal violet solution at 595 nm. In some cases, the absorbance value of the crystal violet solution was outside the range of the instrument. In these cases, absorbance values were read at 570 or 655 nm. Calibration measurements indicated a linear correlation between absorbance values at 595 nm and 655 nm (r = 0.988) and at 595 nm and 570 nm (r = 0.996). For enumeration of biofilm CFUs, biofilms were rinsed twice with sterile saline to remove loosely adherent cells and then treated with 200 µl of a mixture of DspB and rhDNase (10 µg/ml each) in enzyme buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2). After 10 min at 37°C, biofilm cells were mechanically detached from the wells with a pipette tip, transferred to a microcentrifuge tube, disaggregated by vortex agitation for 30 s and then diluted and plated on agar.
2.4. Biofilm detachment assay
Biofilms were cultured in 96-well microtiter plates as described above. They were rinsed extensively with water and then treated with 200 µl of 10 µg/ml DspB or rhDNase in enzyme buffer. Control wells were treated with enzyme buffer alone. After 30 min at 37°C, biofilms were rinsed with water and stained with crystal violet as described above.
2.5. Total hexosamine assay
Biofilms were cultured in 150-mm-diam tissue culture-treated Petri dishes in a volume of 50 ml. Media were supplemented with antibiotics at the indicated concentrations. After growth at 37°C for 18–20 h, media and non-adherent cells were decanted and the biofilm was scraped from the surface with a cell scraper into 5 ml of 0.9 % NaCl and then transferred to a tube. The cell suspension was sonicated on ice (2 × 30 s) using an IKA Labortechnik sonicator set to 50% power and 50% duty cycle. Cells were collected by centrifugation at 5,000 × g for 15 min. The supernatant was clarified by one additional centrifugation at 10,000 × g for 10 min. The amount of total hexosamine was measured using the Morgan-Elson assay (Enghofer and Kress, 1979) with GlcNAc (Acros Organic) as a standard.
2.6. Reproducibility of results and statistics
Microtiter plate biofilm assays were performed in duplicate wells, which exhibited <10% average variation in absorbance values. Biofilm induction experiments were repeated at least three times and biofilm detachment assays were repeated 2–3 times. In all cases, the observed patterns of biofilm induction and detachment were reproducible. The significance of differences between means was analyzed using Student’s t-test. Correlation was analyzed using the Pearson correlation coefficient.
3. Results
3.1. Characterization of S. epidermidis strains
The six S. epidermidis strains employed in this study are listed in Table 1. In the absence of antibiotics, these strains exhibited a range of biofilm phenotypes in a 96-well microtiter plate crystal violet binding assay (Table 1).
The MIC values of four antibiotics against all six strains were measured using the broth microdilution method. For all six strains, MIC values were in the range of 0.8–3.1 µg/ml for vancomycin, 0.05–0.2 µg/ml for tigecycline, 0.05–0.2 µg/ml for novobiocin and 0.8–3.1 µg/ml for linezolid.
3.2 Sub-MIC antibiotics induce biofilm formation
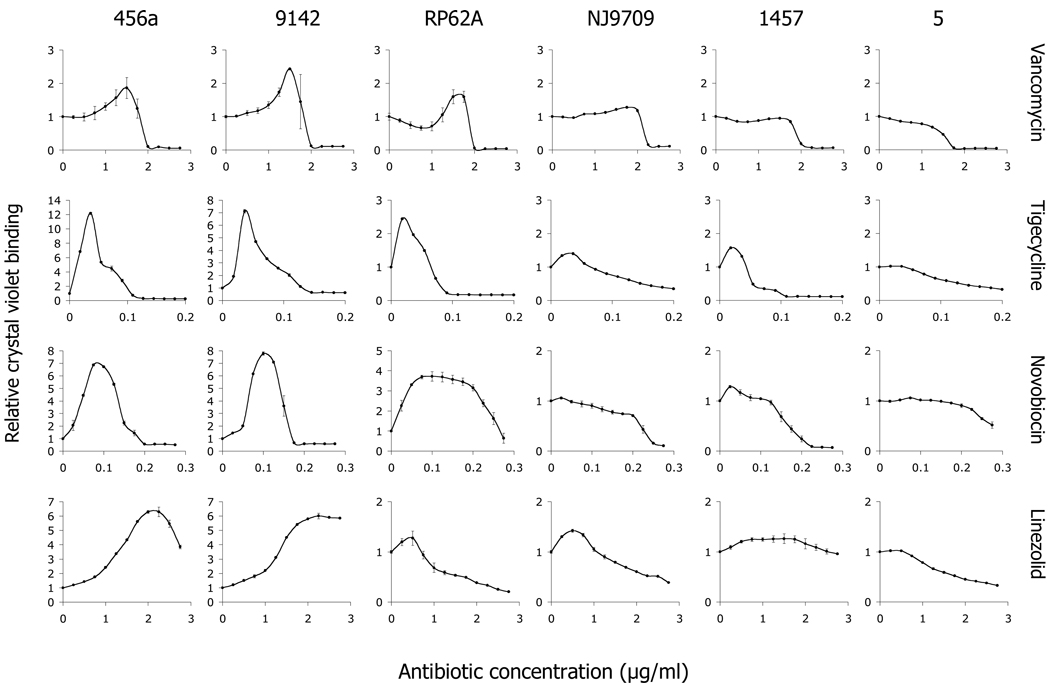
To determine whether sub-MIC concentrations of antibiotics induce biofilm formation, S. epidermidis strains were cultured in broth supplemented with vancomycin, tigecycline, novobiocin or linezolid at concentrations ranging from 0–1 MIC, and the amount of biofilm was quantitated using the crystal violet binding assay (Fig. 1). All four antibiotics induced biofilm formation in some strains, but the amount of induction was both antibiotic- and strain-dependent. The average maximum biofilm induction for each antibiotic against each of the six strains is shown in Table 1. There was a significant negative correlation between the average maximum biofilm induction and the biofilm phenotype of the strain. The r values were −0.914 for vancomycin (P < 0.01), −0.943 for tigecycline (P < 0.01), −0.771 for linezolid (P < 0.01) and −0.829 for novobiocin (P < 0.01).
Fig. 1.
Biofilm formation in 96-well microtiter plates by six strains of S. epidermidis in the presence of subminimal inhibitory concentrations of four antibiotics. The names of the strains are indicated along the top, and the antibiotics are indicated on the right. The x-axis of each graph shows the antibiotic concentration (in µg/ml), and the y-axis indicates the relative crystal violet binding compared to the drug-free control (A[plus antibiotic]/A[minus antibiotic]). Values show mean and range for duplicate wells.
Different antibiotics caused maximal biofilm induction at different concentrations relative to their MICs. Combining all data from strains 456a, 9142 and RP62A, for example, vancomycin caused maximum induction at 74% ± 8% of the MIC (n = 11), whereas tigecycline caused maximum induction at 25% ± 4% of the MIC (n = 11). This difference was statistically significant (P < 0.01).
To determine whether the observed antibiotic-induced increases in crystal violet binding corresponded to increased biofilm CFUs, we measured the number of CFUs in biofilms formed by strain 9142 cultured in 0 or 0.02 µg/ml (1/4 MIC) of tigecycline (Table 2). There were more CFUs in biofilms cultured in tigecycline than in control biofilms. In addition, there was more total hexosamine in the matrix of 9142 biofilms cultured in sub-MIC tigecycline than in the matrix of control biofilms, consistent with increased PNAG production. In contrast, the broth OD and CFU/well values were decreased in cultures grown in tigecycline. The total CFUs per well (biofilm plus broth) were not significantly different in tigecycline and control cultures, indicating that tigecycline at 1/4 MIC did not inhibit bacterial growth. Similar increases in biofilm CFUs, and similar decreases in broth OD and CFU values were observed in biofilms formed by strain 9142 cultured in 1/4 MIC vancomycin and in biofilms formed by strain RP62A cultured in 1/4 MIC tigecycline or 3/4 MIC vancomycin (data not shown). These data confirm that the observed antibiotic-induced increases in crystal violet binding were due to increased biofilm CFUs and biofilm biomass, and not merely to increased dye penetration.
Table 2.
Characteristics of S. epidermidis strain 9142 cultures grown in a sub-MIC concentration of tigecycline.
| Tigecycline concentration (µg/ml)a | ||
|---|---|---|
| 0 | 0.02 | |
| Biofilm | ||
| Crystal violet binding (A655) | 0.12 ± 0.00 | 0.72 ± 0.02 |
| CFU/well (× 106) | 22.5 ± 4.2 | 37.5 ± 5.1 |
| Biofilm matrix (slime layer)b | ||
| Total hexosamine (µg/dish) | 101 ± 3 | 260 ± 20 |
| Broth | ||
| Optical density (A490) | 0.27 ± 0.01 | 0.14 ± 0.01 |
| CFU/well (× 106) | 26.5 ± 2.5 | 9.7 ± 1.3 |
Values show mean and range for duplicate determinations.
Values for biofilm matrix (slime layer) are from biofilms cultured in 150-mm-diam Petri dishes. Other values are from biofilms cultured in 96-well microtiter plates.
3.3. Antibiotic-induced biofilm formation in strain RP62A depends on ecDNA production
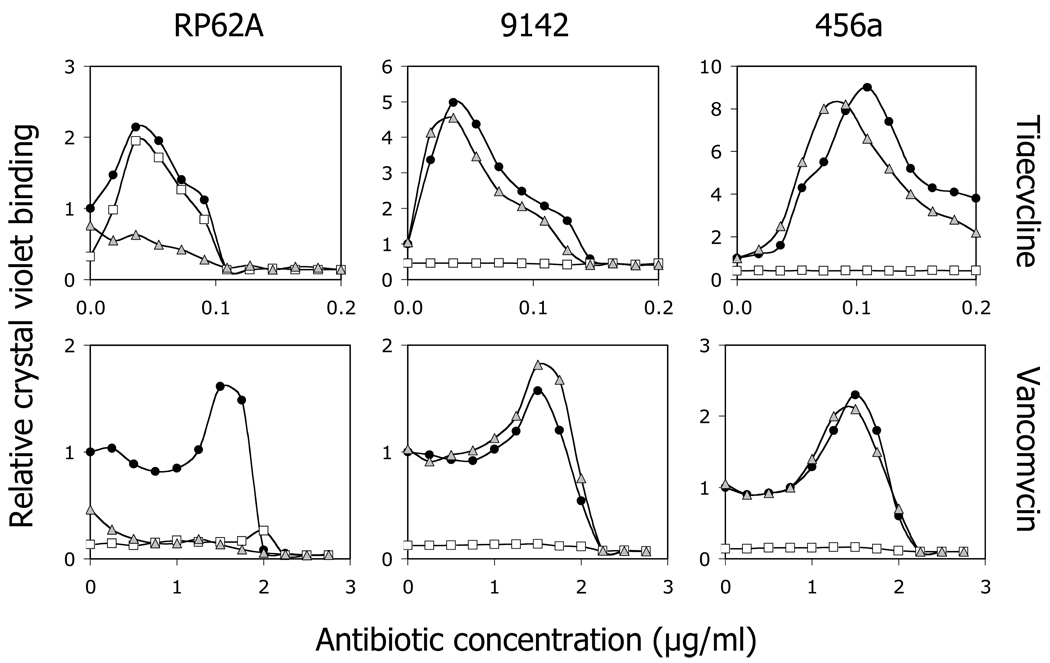
To determine whether PNAG and ecDNA play a role in antibiotic-induced biofilm formation, cultures of strains RP62A, 9142 and 456a containing sub-MIC concentrations of tigecycline or vancomycin were supplemented with 10 µg/ml of DspB or rhDNase. After incubation, biofilm formation was measured using the crystal violet binding assay (Fig. 2). In the absence of antibiotics, DspB inhibited biofilm formation by all three strains, whereas rhDNase partially inhibited biofilm formation by strain RP62A, but had no effect on biofilm formation by strains 9142 and 456a. In the presence of sub-MIC antibiotics, DspB inhibited biofilm formation by strains 9142 and 456a cultured in both antibiotics, and by strain RP62A cultured in vancomycin, but not by strain RP62A cultured in tigecycline. In contrast, rhDNase efficiently inhibited biofilm formation by strain RP62A in response to both vancomycin and tigecycline. These results suggest that antibiotic-induced biofilm formation by strain RP62A depends on the production of ecDNA.
Fig. 2.
Effects of DspB and rhDNase on biofilm formation by S. epidermidis strains RP62A (left panels), 9142 (middle panels), and 456a (right panels) in the presence of 1/4 MIC tigecycline (top panels) or 3/4 MIC vancomycin (bottom panels). Cultures were grown in the absence of enzymes (filled circles), or in the presence of 10 µg/ml DspB (open squares) or 10 µg/ml rhDNase (gray triangles). The y-axis indicates the relative crystal violet binding compared to the drug-free control, calculated as described in the legend to Fig. 1. Graphs show mean values for duplicate wells. Error bars were omitted for clarity.
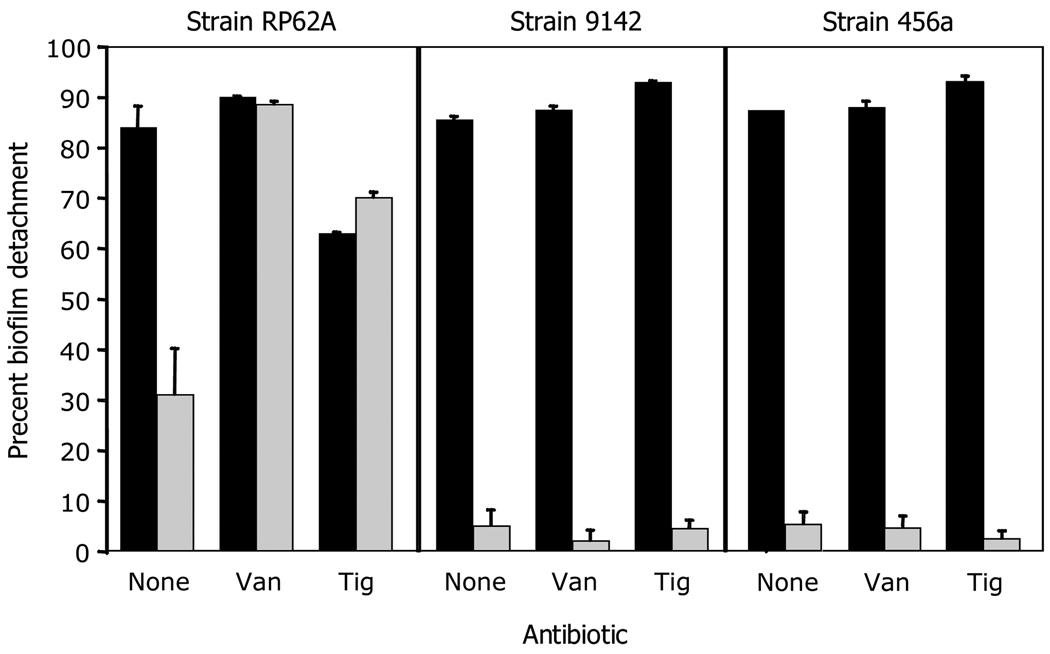
To confirm that RP62A biofilms contain ecDNA, we measured the ability of DspB and rhDNase to detach biofilms formed by strains RP62A, 9142 and 456a cultured in sub-MIC tigecycline or vancomycin (Fig. 3). Biofilms formed by strain RP62A in the absence of antibiotics were detached by both DspB and rhDNase, whereas biofilms formed by strains 9142 and 456a in the absence of antibiotics were detached by DspB, but not by rhDNase. Biofilms formed by strain RP62A in the presence of sub-MIC vancomycin or tigecycline were detached by rhDNase more efficiently than biofilms formed in the absence of antibiotics (P < 0.05). In contrast, rhDNase did not detach biofilms formed by strains 9142 or 456a in the presence of sub-MIC vancomycin or tigecycline. These findings confirm that ecDNA is an important adhesive component of biofilms formed by strain RP62A both in the absence and presence of sub-MIC antibiotics.
Fig. 3.
Detachment of antibiotic-induced S. epidermidis RP62A, 9142 and 456a biofilms by DspB and rhDNase. Biofilms were cultured in 1/4 MIC (0.02 µg/ml) tigecycline (Tig), 3/4 MIC (1.7 µg/ml) vancomycin (Van) or no antibiotic as a control (None). Biofilms were then rinsed and treated with 10 µg/ml DspB (filled bars) or 10 µg/ml rhDNase (gray bars). Control biofilms were treated with enzyme buffer alone. Values show mean percent biofilm detachment for duplicate wells and error bars indicate range. Percent biofilm detachment values were calculated the formula 1 − (A[plus enzyme]/A[minus enzyme]) × 100.
4. Discussion
Our findings are consistent with the results of previous studies demonstraing that sub-MIC concentrations of a variety of antibiotics can induce S. epidermidis biofilm formation (Cargill and Upton, 2009; Dunne, 1990; Frank et al., 2007; Pérez-Giraldo et al., 1994; Rachid et al., 2000; Schadow et al., 1988; Wang et al., 2010). Our results show that the amount of antibiotic-induced biofilm formation is antibiotic-specific and is inversely proportional to the amount of biofilm produced by the test strain. Our results also show that strain RP62A can utilize ecDNA-dependent mechanisms to form biofilms in response to sub-MIC antibiotics.
Several previous studies investigated the effects of sub-MIC concentrations of vancomycin on S. epidermidis biofilm formation (Cargill and Upton, 2009; Carsenti-Etesse et al., 1993; Cerca et al., 2005; Dunne, 1990; Polonio et al., 2001; Rupp et al., 1998; Schadow et al., 1988; Wang et al., 2010). These studies utilized vancomycin at concentrations that were • 1/2 MIC. Our data suggest that vancomycin exhibits maximal biofilm induction at a concentration of 3/4 MIC, which may explain why most previous studies found little or no effect of sub-MIC vancomycin on S. epidermidis biofilm formation.
Our results indicate that linezolid at 2/3 MIC induces S. epidermidis biofilm formation by 6- to 7-fold in strains 9142 and 456a (Fig. 1 and Table 2). Linezolid caused weak biofilm induction (ca. 1.4-fold) in strain RP62A at 1/4 MIC. Frank et al. (2007) found that linezolid induced S. epidermidis RP62A biofilm formation by 2.7-fold at 1/4 MIC. Linezolid has also been shown to induce biofilm formation in S. aureus (Frank et al., 2007) and E. coli (Boehm et al., 2009).
Our findings are the first to demonstrate that sub-MIC concentrations of tigecycline and novobiocin induce bacterial biofilm formation. In S. epidermidis strains 456a and 9142, tigecycline induced biofilm formation by 7- to 9-fold at 1/4 MIC, whereas novobiocin induced biofilm formation by 6- to 8-fold at 1/2 MIC (Fig. 1 and Table 2).
Our results indicate that the amount of biofilm formation induced by sub-MIC antibiotics is inversely proportional to the amount of biofilm produced by the S. epidermidis test strain in the absence of antibiotics. These results are consistent with those of Pérez-Giraldo et al. (1994), who showed that fluoroquinolone-induced biofilm formation by 12 S. epidermidis clinical isolates of unknown ica status was inversely proportional to the amount of biofilm formation in the absence of antibiotic. In the present study, none of the four antibiotics induced biofilm formation in strain 5, which produces large amounts of PNAG (Sadovskaya et al., 2006). In contrast, biofilm formation by strain 456a, which produces no detectable PNAG as determined by gel permeation chromatography (Sadovskaya et al., 2006), was highly inducible by sub-MIC antibiotics (Table 1). These results suggest that expression of the ica genes may be suppressed in strains that produce low amounts of biofilm and that ica expression can be induced by sub-MIC antibiotics. Consistent with this hypothesis, Boehm et al. (2009) showed that sub-MIC concentrations of chloramphenicol, tetracycline and streptomycin induce expression of the PNAG biosynthetic genes pgaA and pgaD in E. coli.
Previous studies showed that DNase I inhibits S. epidermidis biofilm formation, which suggests that ecDNA can function as an adhesive component of the S. epidermidis biofilm matrix (Izano et al., 2008, Qin et al., 2007). Evidence suggests that ecDNA is generated in S. epidermidis populations through AtlE-mediated lysis of a subpopulation of the bacteria, and the extracellular DNA promotes biofilm formation of the remaining population (Qin 2007). In the present study, we found that rhDNase efficiently inhibited biofilm formation by S. epidermidis strain RP62A, both in the absence of antibiotics, and in the presence of sub-MIC tigecycline or vancomycin (Fig. 2). In addition, rhDNase efficiently detached RP62A biofilms grown in the presence of sub-MIC concentrations of either antibiotic (Fig. 3). These findings suggest that antibiotic-induced biofilm formation by strain RP62A, but not by strains 9142 and 456a, depends on the production of ecDNA. These data demonstrate that S. epidermidis can utilize both PNAG-dependent and ecDNA-dependent mechanisms to form biofilms in response to sub-MIC antibiotics. Wang et al. (2010) showed that ica-negative strains of S. epidermidis exhibit biofilm induction upon exposure to sub-MIC macrolides. It is possible that antibiotic-induced biofilm formation in ica-negative strains depends on the production of ecDNA.
A growing number of studies have shown that sub-MIC concentrations of antibiotics induce bacterial biofilm formation. It remains to be seen whether this phenomenon has clinical relevance, and whether it contributes to the inconsistent success of antimicrobial therapy for device infections. If so, a better understanding of this process may help guide antibiotic therapy.
Acknowledgements
We thank Pierre Hardouin and Thierry Grard (Université du Littoral-Côte d’Opale) for helpful support. This study was funded by grants to J.B.K. from the U.S. Department of State Fulbright Commission, the Nord-Pas de Calais Regional Council (France), the National Institute of Allergies and Infections Diseases (award no. AI82392) and Genentech, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jeffrey B. Kaplan, Email: kaplanjb@umdnj.edu.
Saïd Jabbouri, Email: sjabbour@univ-lr.fr.
Irina Sadovskaya, Email: irina.sadovskaya@univ-littoral.fr.
References
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 2009;72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- Cargill JS, Upton M. Low concentrations of vancomycin stimulate biofilm formation in some clinical isolates of Staphylococcus epidermidis. J. Clin. Pathol. 2009;62:1112–1116. doi: 10.1136/jcp.2009.069021. [DOI] [PubMed] [Google Scholar]
- Carsenti-Etesse H, Durant J, Mondain EV, Pradier C, Bernard E, Dellamonica P. Effects of subinhibitory concentrations of vancomycin and teicoplanin on adherence of staphylococci to tissue culture plates. Antimicrob. Agents Chemother. 1993;37:921–923. doi: 10.1128/aac.37.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerca N, Martins S, Pier GB, Oliveira R, Azeredoa J. The relationship between inhibition of bacterial adhesion to a solid surface by sub-MICs of antibiotics and subsequent development of a biofilm. Res. Microbiol. 2005;156:650–655. doi: 10.1016/j.resmic.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokr A, Watier D, Eleaume H, Pangon B, Ghnassia J-C, Mack D, Jabbouri S. Correlation between biofilm formation and production of polysaccharide intercellular adhesin in clinical isolates of coagulase-negative staphylococci. Int. J. Med. Microbiol. 2006;296:381–388. doi: 10.1016/j.ijmm.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 1982;37:318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig WA. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 1998;26:1–10. doi: 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- Dunne WM., Jr Effects of subinhibitory concentrations of vancomycin or cefamandole on biofilm production by coagulase-negative staphylococci. Antimicrob. Agents Chemother. 1990;34:390–393. doi: 10.1128/aac.34.3.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enghofer E, Kress H. An evaluation of the Morgan-Elson assay for 2-amino-2-deoxy sugars. Carbohydr. Res. 1979;76:233–238. doi: 10.1016/0008-6215(79)80022-1. [DOI] [PubMed] [Google Scholar]
- Frank KL, Reichert EJ, Piper KE, Patel R. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus lugdunensis clinical isolates. Antimicrob. Agents Chemother. 2007;51:888–895. doi: 10.1128/AAC.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke C, Kraft A, Sussmuth R, Schweitzer O, Götz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- Huebner J, Goldman DA. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 1999;50:223–236. doi: 10.1146/annurev.med.50.1.223. [DOI] [PubMed] [Google Scholar]
- Hussain C, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2004;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 2005;43:4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Siemssen N, Laufs R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 1992;60:2048–2057. doi: 10.1128/iai.60.5.2048-2057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gara JP, Humphreys H. Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 2001;50:582–587. doi: 10.1099/0022-1317-50-7-582. [DOI] [PubMed] [Google Scholar]
- Pérez-Giraldo C, Rodríguez-Benito A, Morán FJ, Hurtado C, Blanco MT, Gómez-García AC. In-vitro slime production by Staphylococcus epidermidis in presence of subinhibitory concentrations of ciprofloxacin, ofloxacin and sparfloxacin. J. Antimicrob. Chemother. 1994;33:845–848. doi: 10.1093/jac/33.4.845. [DOI] [PubMed] [Google Scholar]
- Polonio RE, Mermel LA, Paquette GE, Sperry JF. Eradication of biofilm forming Staphylococcus epidermidis (RP62A) by a combination of sodium salicylate and vancomycin. Antimicrob. Agents Chemother. 2001;45:3262–3266. doi: 10.1128/AAC.45.11.3262-3266.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. Effects of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilmforming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000;44:3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KL, Rupp ME, Fey PD. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl. Environ. Microbiol. 2008;74:6155–6157. doi: 10.1128/AEM.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp ME, Hamer KE. Effect of subinhibitory concentrations of vancomycin, cefazolin, ofloxacin, Lofloxacin and D-ofloxacin on adherence to intravascular catheters and biofilm formation by Staphylococcus epidermidis. J. Antimicrobial. Chemother. 1998;41:155–161. doi: 10.1093/jac/41.2.155. [DOI] [PubMed] [Google Scholar]
- Sadovskaya I, Chaignon P, Kogan G, Chokr A, Vinogradov E, Jabbouri S. Carbohydrate-containing components of biofilms produced in vitro by some staphylococcal strains related to orthopaedic prosthesis infections. FEMS Immunol. Med. Microbiol. 2006;47:75–82. doi: 10.1111/j.1574-695X.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Li J, Jabbouri S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus epidermidis RP62A, a reference biofilm-positive strain. Carbohydr. Res. 2004;339:1467–1473. doi: 10.1016/j.carres.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Schadow KH, Simpson WA, Christensen GD. Characteristics of adherence to plastic tissue culture plates of coagulase-negative staphylococci exposed to subinhibitory concentrations of antimicrobial agents. J. Infect. Dis. 1988;157:71–77. doi: 10.1093/infdis/157.1.71. [DOI] [PubMed] [Google Scholar]
- Singh R, Ray P, Das A, Sharma M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 2010;65:1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion and virulence. J. Biol. Chem. 2004a;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004b;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sun F-J, Liu Y, Xiong L-R, Xie L-L, Xia P-Y. Enhancement of biofilm formation by subinhibitory macrolides in icaADBC-positive and -negative clinical isolates of Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2010;54:2707–2711. doi: 10.1128/AAC.01565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]