Abstract
Targeted sequencing of the mouse t-complex has started with a 176-kb, gene-rich BAC localized with six PCR-based markers in inversion 2/3 of the highly duplicated region. The sequence contains 11 genes recovered primarily as cDNAs from early embryonic collections, including Igfals (previously placed on chromosome 17), Nubp2 (a fully characterized gene), Jsap1 (a JNK-binding protein), Rsp29 (the mouse homologue of the rat gene), Ndk3 (a nucleoside diphosphate kinase), and six additional putative genes of unknown function. With 50% GC content, 75% of the DNA transcribed, and one gene/16.0 kb (on average), the region may qualify as one of the most gene-dense segments in the mouse genome and provides candidates for dosage-sensitive phenotypes and mouse embryonic lethals mapped to the vicinity.
[The sequence data described in this paper have been submitted to the GenBank data library under accession no. AF220294.]
About 1% of the mouse genome is included in the t-complex region of chromosome 17, which is known to encode genes involved in embryonic development, distortion of male transmission ratio, male sterility, and genomic imprinting (Bennett 1975; Silver 1993). Mapping of many mutant loci to the region has fascinated geneticists and developmental biologists for much of the century (Silver 1993), but very few associations between genes and mutant phenotypes have been made. Examples of associations that have been made are T (Herrmann et al. 1990), qk (Ebersole et al. 1996), Tme (Barlow et al. 1991), and Tcr (Herrmann et al. 1999). The situation is particularly unsatisfactory for the large number of recessive lethal mutations that have been mapped to the t-complex: They have been grouped into at least eight complementation groups (Bennett 1975), but none of the responsible genes have been found.
One possible reason for the paucity of successful positional cloning and gene finding in the region is that genes might be expressed selectively in early mouse embryogenesis, a stage from which very few transcripts have yet to be recovered. The study of a cDNA cohort derived from E7.5 mouse extraembryonic tissues (Ko et al. 1998) and preimplantation embryos (Ko et al. 2000) supports such a possibility. When the cDNAs were sequenced and mapped systematically, a growing number of ESTs were localized to the t-complex.
One way to define the full complement of t-complex genes and to accelerate positional cloning and expression studies would be to map and sequence the entire region. However, a further source of difficulty for the analysis of gene content in the t-complex arises from its unusual structure. Several duplications and inversions, which affect the copy number of local genes, are known to exist in the t-complex (Schimenti et al. 1987; Schweifer and Barlow 1992). Therefore, sequencing approaches, such as shotgun sequencing on a chromosome-wide basis, can be stymied by these large duplications and inversions. Furthermore, this type of arrangement, such as the tandem repeats in centromeres, also tends to disarm standard mapping techniques based on unique sequences as probes for BAC clones across a genomic sequence region.
Nevertheless, some regions of the t-complex have been mapped in yeast artificial chromosomes (Forejt et al. 1999). Focused analysis that uses large-insert clones with auxiliary pulsed-field gel data and genetic information should permit unequivocal mapping, or at least the determination, of gene copy number. We have started such an effort aimed at the sequence analysis of an STS/BAC-based physical map of the t-complex. We report here the analysis of the first BAC sequenced with the genomic structure of five known and six new genes. These genes add to the list of candidates for studies of t-phenotypes and dosage control of expression in the region.
RESULTS
The combination of computer-aided predictions and analysis of EST, cDNA, and amino acid sequence homology permits the identification of genes and the inference of their intron-exon structures, as well as the determination of some alternatively spliced isoforms.
Global Analysis
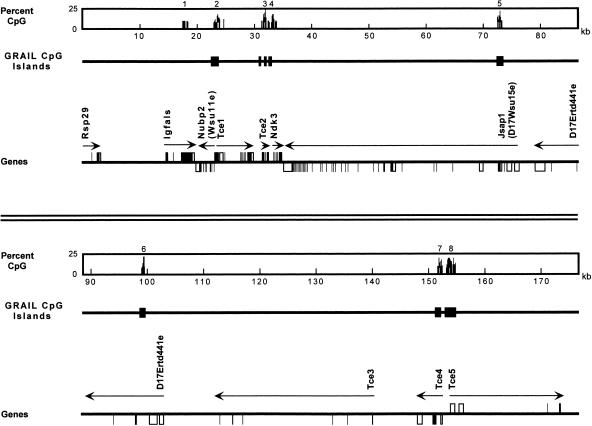
The BAC contains 175,759 nucleotides (GenBank accession number AF220294; see also http://lgsun.grc.nia.nih.gov), with overall 47.74% GC (about 50% in the first half, 46.7% in the last half). This GC content places the BAC DNA sequence among the densest isochores of non-satellite mouse DNA, suggesting a high content of genes in the region (Saccone et al. 1997). This notion is supported by the presence of eight putative CpG islands as well as by the analysis of EST/gene content (Fig. 1). Predicted genes that do not have significant matches to known genes are named t-complex expressed genes (Tce) with sequential numbers.
Figure 1.
Top and bottom panels illustrate features of BAC 126c8 for the first and second halves of the sequence, respectively. CpG percentage was calculated with a 50-bp window (clusters of signals 8% CpG or higher are shown to illustrate CpG Islands). GRAIL-predicted CpG islands are also shown. Known and putative gene structures are demonstrated with arrows indicating expected direction of transcription. Untranslated sequences are shown as open boxes.
Figure 1 illustrates the CpG islands (1–8) observable by inspection as peaks in a plot of CpG dinucleotide content across the sequence. Of the eight putative CpG islands, four fall within a 20-kb segment, another indication of gene richness. All of the islands are associated with coding units (e.g., CpG island 1 with Igfals). Furthermore, CpG island 2 coincides with the 5′ ends of two genes encoded on opposite strands of DNA: Nubp2 and Tce1 (see below). Thus, CpG island 2 may be shared between them. CpG island 3 is located less than 3 kb beyond Tce1 and is transcribed in the same orientation. The other islands are spaced according to the position of corresponding genes.
Repeat sequences were masked to focus on individual tracts of unique sequence and potential genes. RepeatMasker (Smit and Green 1999) specified 276 repetitive elements that collectively constitute 22.6% of the sequence, including 2.5% LINES, 12.66% SINES, 4.67% LTRs, and 1.13% simple sequence repeats.
Genes, cDNAs, and Isoforms
GRAIL (Uberbacher et al. 1996) analysis predicted a total 106 exons, all the CpG islands (except for that associated with Igfals), 25 potential polyadenylation signals, and 15 putative promoter motifs. Definitive identification of genes was based on BLAST searches (Altschul et al. 1997), which revealed five genes homologous to the published genes: Rsp29, Igfals, Nubp2, Ndk3, and Jsap1 (Ji et al. 1997; Boisclair et al. 1996; Nakashima et al. 1999; Venturelli et al. 1995; Ito et al. 1999). The search also revealed six putative transcripts identified by homology to ESTs. The total of 11 genes, or one gene/16.0 kb, confirms the high gene content of the region. The novel genes are labeled in Figure 1 as D17Ertd441e and Tce1–Tce5 (Tce stands for t-complex expressed), where the gene content is represented from 5′ to 3′ in the reported sequence (AF220294).
Mouse Homolog of Rat Round Spermatid Gene 29 (Rsp29)
The RSP29 protein is secreted by rat round spermatids and stimulates secretion from Sertoli cells. Its high abundance in testis is consistent with a role in spermatogenesis. Southern analysis indicates a high degree of conservation in eukaryotes (Ji et al. 1997). An EST from a mouse embryo cDNA library (W98537) identified two exons homologous to the 3′ end of rat Rsp29. GRAIL also predicted these same two exons (Uberbacher et al. 1996). Homology to the rat 3′ end translation product (U97667; 59/60 amino acids, or 98.3%) was sufficient to define the exon/intron boundaries.
Acid-labile Subunit of IGF/IGFB Protein Complex (Igfals)
This known gene, which had been reported to map on chromosome 17 (Boisclair et al. 1996), now is localized more precisely. It encodes a protein that stabilizes IGF/IGF-binding protein complexes and extends their half-life in circulation (Boisclair et al. 1996). Associated with CpG island 1, the mRNA includes three exons, two of which (the first and third exons in Fig. 1) are predicted by GRAIL and conform to the sequence of an EST (AI550843).
In addition to providing the exon/intron boundaries of the gene as described above, two additional ESTs predict alternative isoforms. One EST (AI550843) suggests an exon further upstream, which replaces the reported first exon, splicing in frame with exon 2 of the reported Igfals sequence (Boisclair et al. 1996). This EST shows very strong homology from nt 14987 to 15253 and from nt 17277 to 17644. The open reading frame in the genomic sequence, however, extends 5′ to nt 14959. Therefore, the gene likely begins further upstream than the available cDNA homology data suggest. Another EST (AI226065) implies a splicing variant in which part of the coding sequence at the 5′ end of the published cDNA sequence is missing.
Nucleotide Binding Protein 2 (Nubp2)
This gene originally was identified as an anonymous cDNA clone (D17Wsu11e) mapped to the region (Ko et al. 1998) and was used to screen for the BAC clone. Its 5′ end is associated with CpG island 2 (Fig. 1). The CpG island contains the entire coding sequence of the first exon (including the initiation codon and the first part of the open reading frame). It is transcribed from the reverse strand, with the 3′ end of the gene very near to the 3′ end of the Igfals gene (between coordinates 19982 and 23657), and has been further characterized as defining a new subfamily of nucleotide binding proteins (Nakashima et al. 1999). The genomic sequence and cDNA show a translation start site followed by seven exons (exons 2 and 3 strongly predicted by GRAIL) and a stop codon, with a tail of 400 bp of 3′ untranslated sequence containing the polyadenylation signal (Nakashima et al. 1999).
t-complex Expressed Gene 1 (Tce1)
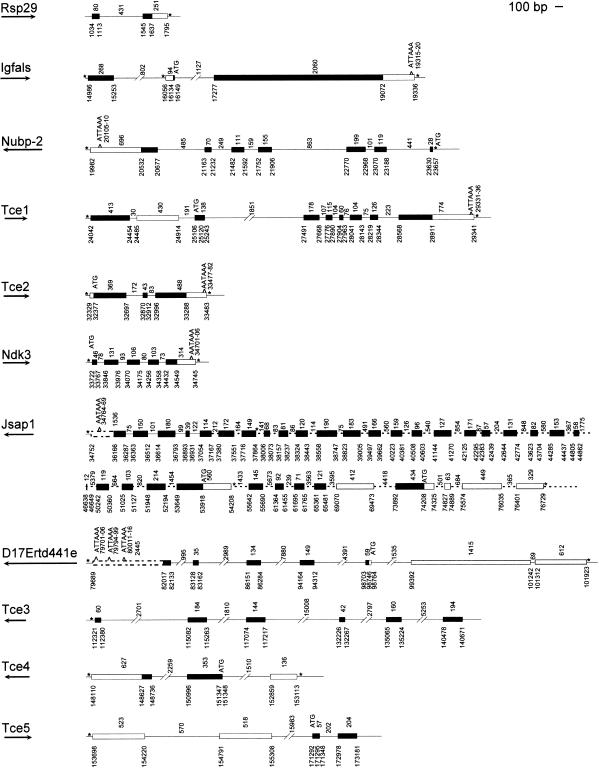
This novel gene was identified by cDNA clone L0234D06 (AW553987) derived from a mouse newborn ovary library (unpublished data) and by three EST matches in dbEST (W78483, W89288, and AI94458). The 5′ and 3′ sequence of L0234D06 identified one of the splice variants (spanning 5 kb of genomic sequence) containing 5′ and 3′ untranslated regions. Based on homology to ESTs, at least nine exons (2200 bp), their splice junctions, and translational start/stop signals are inferred among several isoforms (Fig. 2). This gene likely contains more internal coding exons not yet identified because 5′ and 3′ end cDNA sequencing of L0234D06 elucidated only 1101 bp of the 1.9-kb transcript (data not shown).
Figure 2.
Gene structures for all known and putative genes. Coordinates are placed according to the forward sense of BAC sequence (AF220294). All exons identified are shown as boxes. Alternative exon splicing likely occurs with Igfals, Tce1, D17Wsu15e (Jsap1), and D17Ertd441e. Tce2 has complete connecting EST coverage and is likely a complete gene structure. The remaining unknown genes are likely to be incomplete gene structures. Splice variants are not distinguished in the above diagram. Exons and distances between exons are drawn to scale, unless indicated otherwise by dashed or broken lines. An arrow under the gene name indicates the expected direction of transcription. Additional features are indicated by an asterisk (*), no splice site identified; filled bar, open reading frame; open bar, no open reading frame; dashed open bar, exon not drawn to scale; dashed line, distance not to scale.
The reading frame of at least one splice variant remains open at the 5′ end (W78483 and W89288). Thus, it is likely that additional 5′ sequences exist, but would probably be restricted by the start site of Nubp2, less than 400 bp upstream on the reverse strand. CpG island 2 (also transcribed in reverse sense into the first exon of Nubp2 as shown above) contains the entire coding sequence of the first exon of Tce1 transcribed in the forward sense. Thus, both strands of the DNA in this region are transcribed into mRNAs from a shared CpG island (see Discussion).
Northern blot analysis (data not shown) shows this gene to be expressed ubiquitously in fetal and adult tissues, with a predominant transcript of 1.9 kb that is consistent with the extent of the coding shown (Fig. 1 and Fig. 2). Weaker signals also are observed at 5.5 and 2.6 kb, but it is unclear if these are longer transcripts, or if they arise from cross-hybridizing RNA species.
t-complex Expressed Gene 2 (Tce2)
The second putative novel gene can be reconstructed from three GenBank EST entries (AA794791, AA013636, AA415688). CpG island 3, which is predicted by GRAIL to extend 669 bp, lies at the 5′ end of this gene and partially overlaps the coding sequence established by these ESTs. The resultant virtual cDNA includes 15 bp of 5′ untranslated sequence, three exons, also strongly identified by GRAIL, and 189 bp of 3′ untranslated sequence. Interestingly, the human equivalent to Tce2 is present (though not noted) in a reported sequence tract of human chromosome 16 containing human Ndk3 (U80813). The mouse homolog of Ndk3 is similarly adjacent in mouse as described below.
Mouse Homolog of Human Nucleoside Diphosphate Kinase 3 (Ndk3)
The mouse cDNA first was identified by homology to an EST, and the exon/intron structure was reconstructed with the aid of the human NDK3 protein sequence (Q13232). The two genes are 87% homologous (146/168 identical amino acids). The mouse gene contains five exons. In contrast, human NDK3 is reported to have six exons (Martinez et al. 1997), but the genomic sequence entry (U80813) lacks the correct donor-acceptor splice sites between exon 4 and exon 5, suggesting that there may be five exons in the human gene (U29656). CpG island 4 (indicated as two separate but closely adjacent CpG islands by GRAIL) is a candidate promoter region for this gene. This CpG island starts about 500 bp upstream of the translation start (which may indicate the extent of the 5′ untranslated sequence for this gene), and extends 477 bp into the open reading frame through exons 1, 2, and part of exon 3.
Jun N-terminal Protein Kinase (Jsap1)
This large gene is transcribed on the reverse strand and originally was detected as an anonymous cDNA clone (D17Wsu15e) mapped to the t-complex region (Ko et al. 1998) and was used to isolate this BAC clone. This gene encodes the transcript for the recently identified JSAP1 protein, a novel JNK (Jun N-Terminal Protein Kinase) binding protein (Ito et al. 1999). Homology to the reported mRNA sequence reveals 30 exons. These 30 exons define an mRNA length of 6 kb. Before publication of Jsap1, 26 exons had been identified by homology to mouse, rat, and human ESTs as belonging to this gene. Of these 26 exons, 20 exons matched those present in published mRNA. The remaining six exons are also probably true, bringing the total number of exons for this gene to 36. Three of these exons (defined by ESTs AV039876, AU051436, AA146208, and AA198480) likely extend the reported 5′ untranslated region by 850 nucleotides, and three (defined by ESTs AA414323, AI115545, and AI608397) likely indicate at least one alternative isoform of this gene (Fig. 2). However, a portion (20 bp) of the Jsap1 coding sequence could not be resolved in the genomic sequence. A putative 12-bp exon (one of the five exons demonstrated by EST AI608397) is localized in the region of the expected location of this 20-bp omission (Fig. 2). The 3′ untranslated region is now better defined with the 3′ sequence of D17Wsu15e (AA407329), demonstrating an additional 1266 nucleotides and a canonical polyadenylation signal. ESTs for this gene can be found in embryo and adult libraries. Based on the amino acid sequences of the hypothetical translation product, the putative protein is similar to a 168.6 kDa protein (1139 amino acids) predicted by the complete sequence of C. elegans chromosome III (P34609), with 45% identity and 64% positive identity over a stretch of 130 amino acids that may represent an evolutionarily conserved domain.
D17Ertd441e
This gene was identified originally as an anonymous cDNA clone recovered from a fertilized egg cDNA library (Ko et al. 2000). The existence of a gene in the region is attested firmly by numerous GenBank EST entries from mouse libraries (and one from a human library). Seven exons spanning 21 kb of genomic sequence define the putative gene. Based on the matches to ESTs, exons 3 to 7 can be spliced together. No EST currently reported links exons 1–2 to exons 3–7. However, based on the position of CpG island 6 (predicted by GRAIL to extend across 741 bp at its 5′ end) a single gene is speculatively transcribed from the region.
t-complex Expressed Gene 3 (Tce3)
Putative gene Tce3 is defined by six exons. Exons 1–3 are defined by homology to human EST AA239845, and exons 4–6 are defined by homology to human EST W78724. In the absence of expression data for other exons or interstitial CpG islands, the simplest structure for the region would combine these exons into the single putative gene. Neither 5′ nor 3′ untranslated regions have been identified for this gene. All exons have open reading frames, and when spliced together, they elucidate 816 nucleotides of the open reading frame in the primary transcript. Once again, EST sequences (in this instance, W78724, with the three 3′ exons) are homologous to regions of human chromosome 16 (see Discussion).
t-complex Expressed Gene 4 (Tce4)
The putative transcript of Tce4 is defined by three exons and the 5′ end CpG island 7 (662 bp in length predicted by GRAIL). Mouse ESTs AA302050 and AI119548 define two exons at the 5′ end, where mouse EST AI119731 demonstrates exon 3. These exons include 5′ and 3′ untranslated sequence, canonical splice sites, and an open reading frame of 461 nucleotides.
t-complex Expressed Gene 5 (Tce5)
Currently, this putative gene is inferred with four exons starting from CpG island 8 (1696 bp in length according to the GRAIL prediction) and transcribed from the forward strand of the BAC. Matches to two mouse ESTs (AI118204 and AI118602) predict the proximal two exons. The distal two exons, also predicted strongly by GRAIL, can be assembled from homology to human EST AA379972 (derived from a tumor cell line), and they show canonical splice junction sequences. The first two exons have no open reading frame and may be part of a 5′ untranslated region. The subsequent exons are open for translation, though the predicted translation products show no similarity to any entries in protein databases.
DISCUSSION
Tight Clustering of Embryonic-related Genes in the t-complex
Four unique cDNAs that had been mapped to the same 1 cM bin in previous studies [Igfals, Nubp2, D17Wsu15e (Jsap1), and D17Ertd441e] all fall into the 176-kb BAC clone analyzed here. Thus, this BAC sequence is inferred to represent one of the unduplicated regions of the t-complex.
The unambiguous determination of the gene content in genomic sequence depends on the progressive enrichment of cDNA and EST collections. In regions where no cDNAs are as yet recovered, gene prediction programs provide suggestions that can interact with hunts for transcripts. The programs remain probabilistic in their identification of genes. However, the recognition of CpG islands as telltales for the 5′ ends of genes and the scoring of putative exons as excellent by the GRAIL program have, thus far, been substantiated by cDNA findings in every case examined.
The overall high GC content of the region is a general index of high gene content and is confirmed by the census of one gene/16.0 kb (Saccone et al. 1997). The extent of transcription provides a more striking confirmation of the region's coding capacity. As schematized (Fig. 1), more than 75% transcription is inferred, and part of the remaining sequence is involved in promoter elements. As one might expect, the large extent of transcription in the region is correlated with a relatively low content of repetitive elements (about 20%). In other instances where comparable stretches of sequence have been annotated, very gene-rich, GC-rich regions also contain about 20% repetitive elements, whereas less gene-rich, lower GC-content zones may be less than 40% transcribed and contain up to 50% repetitive sequence (Chen et al. 1996).
The tight packing of genes leads to several instances in which genes lie very close to one another or even overlap. For example, the transcripts of Tce2 and Ndk3, in tandem on the forward strand, are within several hundred nucleotides of each other. In a more extreme instance, Ndk3 and Jsap1 end no more than 6 bp from each other. Even more striking is the case of Tce1 and Nubp2, which are transcribed in opposite directions from the same CpG island. The same sequence tract includes exon 1 of both genes from opposite strands. Such instances of near or direct overlap are rare, but they are certainly not unique. For example, TCP1 and ACAT2 are transcribed tail-to-tail with overlapping 3′ untranslated regions (Shintani et al. 1999). Furthermore, two genes in the HTF9 locus are transcribed in reverse orientation from a shared CpG island (Bressan et al. 1991); and divergently transcribed overlapping genes have been analyzed in the 11p15.5 imprinted domain (Cooper et al. 1998). In all such cases, it is likely that the genes reside in a region that is closely packed with transcription units.
Coexpression and Possible Coregulation
Notably, all the genes detected here, except Igfals, have been found as ESTs in placental or embryonic libraries. Thus, RNA polymerases may be transcribing the entire region during early tissue development.
Such genes become candidates for the many embryonic lethal mutations mapped to the region. For example, tw73, which is mapped to this region, is a mutant that fails to implant because of a defect in trophoblasts (Spiegelman et al. 1976; Schweifer and Barlow 1992). One then might speculate that the high incidence of developmental mutants in the region correlates with two features: the high local density of genes and their expression in embryonic life. The placement of many other early transcripts in the same interval of the t-complex, supports these inferences strongly (Fig. 1 and work in progress).
Simultaneous expression of many genes from the region could result if genes were regulated by independent mechanisms (e.g., tissue-specific transcription factors) that have convergent kinetics during development. However, coexpression also could result, at least partly, from a true coregulation by a common mechanism. For example, chromatin remodeling might open up large subregions of the t-complex early in embryonic life.
The concentration in the region of dosage-sensitive genes also provides a further hint of possible long-range regulatory features. Two imprinted genes in the t-complex, Igf2r (Barlow et al. 1991) and Mas1 (Villar and Pedersen 1994), have been detected earlier, and we have found recently that one of the genes in the segment analyzed here, Nubp2, shows hemizygous methylation patterns at two MspI sites (CCGG) (Y. Sano et al. work in progress), which suggests monoallelic expression.
The results also hint at features of the content and regulation of syntenic regions in human. As detailed previously, fragmentary sequencing of human DNA indicates that this segment of mouse chromosome 17 DNA has its homologous region in 16p. For example, Ndk3 and Tce2 are juxtaposed in both, and one of the ESTs used to infer D17Ertd441e (AA476517) is derived from human and is found on a PAC clone from chromosome 16. More refined determination of the conservation of gene content and possible imprinting center(s) will, of course, be possible as genome sequencing of mouse and human progresses.
METHODS
BAC Selection and Sequencing Strategy
BAC 126c8 was screened from the Caltech mouse BAC library (129SV mouse strain; Shizuya et al. 1992; purchased from Research Genetics) using STSs and ESTs previously mapped to the t-complex as probes. ESTs D17Wsu11e (Nubp2) and D17Wsu15e (Jsap1) recovered six BAC clones (Ko et al. 1998). The overlap among these clones was verified by generating additional STSs from the ends of the BAC and from inter-B1 products (Cox et al. 1991; Detter et al. 1998). The random shotgun sequencing approach was used to obtain the complete sequence without gaps (Chen et al. 1996).
Sequence Analysis
Repetitive elements were found, classified, and masked by the University of Washington's Web-based program, RepeatMasker (Smit and Green 1999). The masked genomic sequence was compared mainly against the GenBank nr, dbEST, Swissprot, and PDB databases using BLAST 2.0 (Altschul et al. 1997). The results were analyzed to ascertain homology and the structure of known genes and ESTs. Consensus sequences of ESTs matching the BAC were used to reconstruct unknown genes. GRAIL (v. 1.3c) exon predictions were studied to verify the utility of this exon detection method (Uberbacher et al. 1996). GRAIL predicted polyadenylation signals and promoter signatures were considered to give us an initial hint at the overall gene structure. In cases where the gene was known in rat or human but not in mouse (e.g., Rsp29 and Ndk3), the translated sequence was used to reconstruct the gene. Because many transcripts contain repetitive sequences in their untranslated regions, and, therefore, would be masked by RepeatMasker, the unmasked genomic sequence was compared against the GenBank dbEST database using BLAST to recover expression data for such genes (e.g., Ertd441e). CpG islands were sought as telltales for the 5′ end of genes using GRAIL, as well as an in-house program that calculated the CpG dinucleotide content across the region with a 50-bp moving window (script available upon request). The data was plotted using Gnuplot (see http://www.geocities.com/SiliconValley/Foothills/6647/), and obvious peaks were identified by visual inspection (Fig. 1). DNASIS (Hitachi Software Engineering, v. 2.6) software was used to determine ORFs across reconstructed genes, which, in turn, aided in determining the putative gene structure. ABI Prism AutoAssembler (Perkin Elmer, v. 2.0) was used in an attempt to build consensus sequences from ESTs collected from serial BLASTs seeded by ESTs showing moderate to high homology to the BAC sequence in localized regions. The resulting consensus sequences were compared to the BAC sequence to determine whether they could be transcripts. This technique was used to analyze Ndk3. Transcription direction and expression data showing untranslated regions flanking long, open reading frames aided in distinguishing neighboring genes, which, in turn, allowed the estimate of gene content.
Acknowledgments
We thank Massimo Cocchia, Yuri Sano, Saied Jaradat, and Reid Huber for their consultation and advice. Further, we thank Yong Qian, Dawood Dudekula, and Choor Chau Yu for contributing their computer expertise toward this project. Finally, we thank Serafino Pantano for the use of his Tce1 Northern blot data, Shoshana Stern for assistance in the analysis, and Kristen Keener for editorial consultation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kom@grc.nia.nih.gov; FAX (410) 558-8331.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Bennett D. The T-locus of the mouse. Cell. 1975;6:441–454. [Google Scholar]
- Boisclair YR, Seto D, Hsieh S, Hurst KR, Ooi GT. Organization and chromosomal localization of the gene encoding the mouse acid labile subunit of the insulin-like growth factor binding complex. Proc Natl Acad Sci USA. 1996;93:10028–10033. doi: 10.1073/pnas.93.19.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan A, Somma MP, Lewis J, Santolamazza C, Copeland NG, Gilbert DJ, Jenkins NA, Lavia P. Characterization of the opposite-strand genes from the mouse bidirectionally transcribed HTF9 locus. Gene. 1991;103:201–209. doi: 10.1016/0378-1119(91)90274-f. [DOI] [PubMed] [Google Scholar]
- Chen CN, Su Y, Baybayan P, Siruno A, Nagaraja R, Mazzarella R, Schlessinger D, Chen E. Ordered shotgun sequencing of a 135 kb Xq25 YAC containing ANT2 and four possible genes, including three confirmed by EST matches. Nucleic Acids Res. 1996;24:4034–4041. doi: 10.1093/nar/24.20.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Zollo M, Mazzarella R, Ciccodicola A, Chen CN, Zuo L, Heiner C, Burough F, Repetto M, Schlessinger D, D́Urso M. Long-range sequence analysis in Xq28: thirteen known and six candidate genes in 219.4 kb of high GC DNA between the RCP/GCP and G6PD loci. Hum Mol Genet. 1996;5:659–668. doi: 10.1093/hmg/5.5.659. [DOI] [PubMed] [Google Scholar]
- Cooper PR, Smilinich NJ, Day CD, Nowak NJ, Reid LH, Pearsall RS, Reece M, Prawitt D, Landers J, Housman DE, Winterpacht A, Zabel BU, Pelletier J, Weissman BE, Shows TB, Higgins MJ. Divergently transcribed overlapping genes expressed in liver and kidney and located in the 11p15.5 imprinted domain. Genomics. 1998;49:38–51. doi: 10.1006/geno.1998.5221. [DOI] [PubMed] [Google Scholar]
- Cox RD, Copeland NG, Jenkins NA, Lehrach H. Interspersed repetitive element polymerase chain reaction product mapping using a mouse interspecific backcross. Genomics. 1991;10:375–384. doi: 10.1016/0888-7543(91)90322-6. [DOI] [PubMed] [Google Scholar]
- Detter JC, Nguyen QA, Kingsmore SF. Identification of novel simple sequence length polymorphisms (SSLPs) in mouse by interspersed repetitive element (IRE)-PCR. Nucleic Acids Res. 1998;26:4091–4092. doi: 10.1093/nar/26.17.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MJ, Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Forejt J, Trachtulec Z, Hamvas R. Mouse Chromosome 17. Mamm Genome. 1999;10:958. doi: 10.1007/s003359901136. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Koschorz B, Wertz K, McLaughlin K, Kispert A. A protein kinase encoded by the t-complex responder gene causes non-Mendelian inheritance. Nature. 1999;402:141–146. doi: 10.1038/45970. [DOI] [PubMed] [Google Scholar]
- Ito M, Yoshioka K, Akechi M, Yamashita S, Takamatsu N, Sugiyama K, Hibi M, Nakabeppu Y, Shiba T, Yamamoto KI. JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a Scaffold factor in the JNK signaling pathway. Mol Cell Biol. 1999;19:7539–7548. doi: 10.1128/mcb.19.11.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Moore HD, Russell RG, Watts DJ. cDNA cloning and characterization of a rat spermatogenesis-associated protein Rsp29. Biochem Biophys Res Commun. 1997;241:714–719. doi: 10.1006/bbrc.1997.7880. [DOI] [PubMed] [Google Scholar]
- Ko MSH, Threat TA, Wang X, Horton JH, Cui Y, Wang X, Pryor E, Paris J, Wells-Smith J, Kitchen JR, Rowe LB, Eppig J, Satoh T, Brant L, Fujiwara H, Yotsumoto S, Nakashima H. Genome-wide mapping of unselected transcripts from extraembryonic tissue of 7.5-day mouse embryos reveals enrichment in the t-complex and under-representation on the X chromosome. Hum Mol Genet. 1998;7:1967–1978. doi: 10.1093/hmg/7.12.1967. [DOI] [PubMed] [Google Scholar]
- Ko MSH, Kitchen JR, Wang X, Threat TA, Wang X, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, Cui Y, Sano Y, Tanaka T, Liang Y, Mason S, Paonessa PD, Saulus AD, DePalma GE, Sharara R, Rowe LB, Eppig J, Morrell C, Doi H. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development. 2000;127:1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- Martinez R, Venturelli D, Perrotti D, Veronese ML, Kastury K, Druck T, Huebner K, Calabretta B. Gene structure, promoter activity, and chromosomal location of the DR- nm23 gene, a related member of the nm23 gene family. Cancer Res. 1997;57:1180–1187. [PubMed] [Google Scholar]
- Nakashima H, Grahovac MJ, Mazzarella R, Fujiwara H, Kitchen JR, Threat TA, Ko MSH. Two novel mouse genes-Nubp2, mapped to the t-complex on chromosome 17, and Nubp1, mapped to chromosome 16-establish a new gene family of nucleotide-binding proteins in eukaryotes. Genomics. 1999;60:152–160. doi: 10.1006/geno.1999.5898. [DOI] [PubMed] [Google Scholar]
- Saccone S, Caccio S, Perani P, Andreozzi L, Rapisarda A, Motta S, Bernardi G. Compositional mapping of mouse chromosomes and identification of the gene-rich regions. Chromosome Res. 1997;5:293–300. doi: 10.1023/B:CHRO.0000038759.09018.a7. [DOI] [PubMed] [Google Scholar]
- Schimenti J, Vold L, Socolow D, Silver LM. An unstable family of large DNA elements in the center of the mouse t complex. J Mol Biol. 1987;194:583–594. doi: 10.1016/0022-2836(87)90235-x. [DOI] [PubMed] [Google Scholar]
- Schweifer N, Barlow DP. The mouse plasminogen locus maps to the recombination breakpoints of the tLub2 and TtOrl partial t haplotypes but is not at the tw73 locus. Mamm Genome. 1992;2:260–268. doi: 10.1007/BF00355436. [DOI] [PubMed] [Google Scholar]
- Shintani S, ÓhUigin C, Toyosawa S, Michalova V, Klein J. Origin of gene overlap: the case of TCP1 and ACAT2. Genetics. 1999;152:743–754. doi: 10.1093/genetics/152.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LM. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends Genet. 1993;9:250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- Smit, A.F.A. and P. Green. 1999. RepeatMasker at: http://ftp.genome.washington.edu/RM/RepeatMasker.html.
- Spiegelman M, Artzt K, Bennett D. Embryological study of a T/t locus mutation (tw73) affecting trophectoderm development. J Embryol Exp Morphol. 1976;36:373–381. [PubMed] [Google Scholar]
- Uberbacher EC, Xu Y, Mural RJ. Discovering and understanding genes in human DNA sequence using GRAIL. Methods Enzymol. 1996;266:259–281. doi: 10.1016/s0076-6879(96)66018-2. [DOI] [PubMed] [Google Scholar]
- Venturelli D, Martinez R, Melotti P, Casella I, Peschle C, Cucco C, Spampinato G, Darzynkiewicz Z, Calabretta B. Overexpression of DR-nm23, a protein encoded by a member of the nm23 gene family, inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells. Proc Natl Acad Sci USA. 1995;92:7435–7439. doi: 10.1073/pnas.92.16.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar AJ, Pedersen RA. Parental imprinting of the Mas protooncogene in mouse. Nat Genet. 1994;8:373–379. doi: 10.1038/ng1294-373. [DOI] [PubMed] [Google Scholar]