Abstract
The zebrafish is an important vertebrate model for the mutational analysis of genes effecting developmental processes. Understanding the relationship between zebrafish genes and mutations with those of humans will require understanding the syntenic correspondence between the zebrafish and human genomes. High throughput gene and EST mapping projects in zebrafish are now facilitating this goal. Map positions for 523 zebrafish genes and ESTs with predicted human orthologs reveal extensive contiguous blocks of synteny between the zebrafish and human genomes. Eighty percent of genes and ESTs analyzed belong to conserved synteny groups (two or more genes linked in both zebrafish and human) and 56% of all genes analyzed fall in 118 homology segments (uninterrupted segments containing two or more contiguous genes or ESTs with conserved map order between the zebrafish and human genomes). This work now provides a syntenic relationship to the human genome for the majority of the zebrafish genome.
Zebrafish is an important model system for analysis of vertebrate development (Kimmel 1989; Driever et al. 1996) and an emerging model system for human disease (Zon 1999). Understanding the relationship between the zebrafish and human genomes will help identify roles for human genes from zebrafish mutations, and help identify zebrafish models for genes identified by human disease (Brownlie et al. 1998). Hundreds of zebrafish genes and thousands of zebrafish ESTs have been identified that provide the basis for comparing the relationship between the human and zebrafish genomes. These can be compared with human genes to identify orthologs. Subsequent mapping can be used to define the extent of conservation between zebrafish and human genomes. Earlier reports identify map locations for 124 zebrafish genes with mapped human orthologs (Postlethwait et al. 1998; Gates et al. 1999). Analysis of this mapping data revealed many instances of conserved synteny, whereby two or more genes that are found on the same chromosome in zebrafish are also found on the same chromosome in humans. In some cases, members of such syntenic groups were contiguous with one another and had conserved map order suggesting no large-scale rearrangements between zebrafish and human genomes in these regions (we call these homology segments). Nevertheless, not enough genes were analyzed to give a global picture of the extent of conserved synteny between zebrafish and human genomes. We have increased the number of analyzed genes and ESTs to 523, allowing a more complete analysis of the syntenic relationship between human and zebrafish genomes.
RESULTS
We used 523 mapped zebrafish genes and ESTs with mapped human orthologs to compare the syntenic relationship of the zebrafish and human genomes. These included 25 genes and 228 ESTs mapped in this study on the LN54 zebrafish radiation hybrid panel (Hukriede et al. 1999) in addition to 270 genes and ESTs with previously reported map positions (Johnson et al. 1996; Postlethwaite et al. 1998; Gates et al. 1999; Geisler et al. 1999; Hukriede et al. 1999). Related gene clusters (such as hox clusters, dlx gene pairs, the major histocompatibility complex, or hemoglobin loci) are represented as single genes in our analysis to prevent an overestimate of the extent of conserved synteny. Orthology was determined by WU-BLAST analysis (W. Gish, unpubl.; http://BLAST.wustl.edu), selecting for highly significant matches (maximum WU-BLASTN probability of e-20, see Materials and Methods). Genes and ESTs positioned with other mapping panels were integrated onto our map with respect to markers shared between each panel (Johnson et al. 1996; Postlethwaite et al. 1998; Gates et al. 1999; Geisler et al. 1999; Hukriede et al. 1999). Approximately 400 additional mapped genes and ESTs were excluded from this analysis because they had no obvious human or mouse ortholog, or map positions of human orthologs were unknown (data not shown). A small subset of ESTs and genes had multiple possible orthologs, which prevented unambiguous orthology assignments (see below).
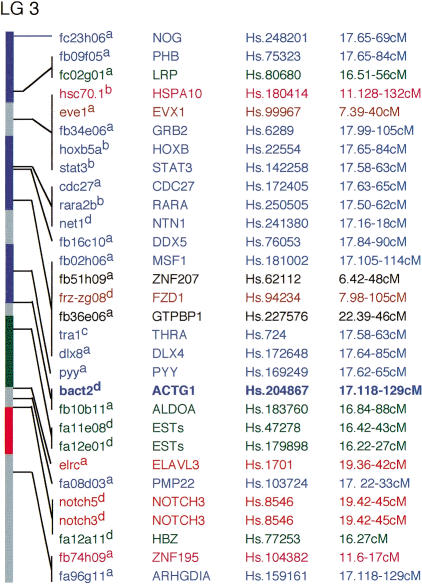
An example of the extent of syntenic correspondence of zebrafish and human genomes is shown in Figure 1. Of the 29 LG3 genes and ESTs with mapped human orthologs, 27 (93%) belong to five conserved synteny groups, corresponding to human chromosomes Hsa7, Hsa11, Hsa16, Hsa17, and Hsa19. The 14 genes of the LG3-Hsa17 conserved synteny group (excluding bact2 for this analysis; see below) are separated into four uninterrupted segments of conserved map order (fc23h06–fb09f05, fb34e06–net1, rara2–fb02h06, and dlx8–pyy) that likely represent homologous segments conserved intact, or nearly intact, between human and zebrafish. An additional two ESTs, fa08d03 and fa96g11 from the LG3–Hsa17 conserved synteny group (that BLAST analyses suggest identify zebrafish orthologs to human PMP22 and ARHGDIA genes) are not contiguous with other genes from the conserved synteny group. However, their membership in the LG3–Hsa17 conserved synteny group adds support to the predicted orthology, and suggests that these ESTs may nucleate additional zebrafish–human homology segments as more genes are analyzed. By similar logic, the other four conserved synteny groups represented on LG3 may identify an additional nine multiple- or single-gene homology segments, increasing the number of homology segments on LG3 to 15. Two ESTs on LG3, fb51h09 and fb36e06, are not identified as members of defined conserved synteny groups and thus lack independent support for the existence of additional homology segments (see below for possible alternatives). We refer to this class of mapped gene as singletons.
Figure 1.
Syntenic relationship between zebrafish linkage group 3 and the human genome. Vertical staff shows map of zebrafish LG3 derived from genes and ESTs (column 1) typed on the LN54 Radiation Hybrid panel 1, or genes and ESTs typed on other panels integrated onto the LN54 map with respect to SSLP markers typed in common. Because gene and EST marker order cannot always be precisely determined when typed on different panels, we show them in high-confidence bins with respect to position of framework markers of the LN54 panel (Hukriede et al. 1999). Order within confidence bins is not established and we have inferred minimal chromosomal rearrangements for our analysis. Superscripts indicate sources of mapping data: (a) the LN54 zebrafish RH panel (Hukriede et al. 1999; this study), (b) the MOP meiotic panel (Johnson et al. 1996;Postlethwait et al. 1998), (c) the GAT meiotic panel (Gates et al. 1999), or (d) the Goodfellow zebrafish RH panel (Geisler et al. 1999). Orthologous human genes (column 2), UniGene reference sequence (http://www.ncbi.nlm.nih.gov/UniGene) (column 3), and Gene Map 98 (Deloukas et al. 1998) position (column 4) are shown to right. Conserved synteny groups are as shown as follows: blue, Hsa17; green, Hsa16; light red, Hsa7; dark red, Hsa19; pink, Hsa11; and singletons, black. Contiguous regions with two or more genes from the same conserved synteny group are shaded the corresponding color on the map staff (left). Bold type shows gene (bact2) where determination of orthology was assisted by syntenic relationships. See http://zfish.wustl.edu, or supplemental information at the Genome Research web site (http://www.genome.org) for maps showing other zebrafish-to-human or human-to-zebrafish relationships.
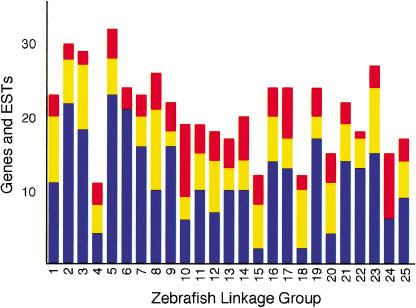
Genome-wide, 421 of 523 mapped genes and ESTs were in 113 conserved synteny groups, averaging 4.5 groups (range 2–7) per zebrafish chromosome (Table 1). As observed above for LG3, genes and ESTs in conserved synteny groups fall into two classes: one class of uninterrupted segments of two or more genes and ESTs with conserved gene order in zebrafish and human that likely represent homology segments conserved intact, or nearly intact, between human and zebrafish; and a second class of single genes and ESTs that belong to conserved synteny groups, but are otherwise isolated from members of their conserved synteny group. Thus, we found 292 genes and ESTs (56% of total) in the first class arranged in 118 multiple-gene homology segments and a further 129 genes and ESTs in the second class separated from other members of their conserved synteny group (presumably by intrachromosomal rearrangements). The fact that this second class of genes are part of conserved synteny groups tends to support their predicted orthology, thus providing evidence for additional homology segments and therefore raising the number of likely zebrafish–human homology segments to 247 (118 + 129). The remaining 102 mapped genes and ESTs (19% of total) that are not currently in conserved synteny groups (thus, singletons, see Figure 2), may reflect the existence of additional conserved synteny groups and homology segments, or instead may reflect errors in determining orthology, errors in mapping, yet unidentified genes in the human (or mouse) data set, or instances where the corresponding orthologous gene has been lost from the human lineage. Putting these possibilities aside and assuming a Poisson distribution of genes and ESTs in synteny groups and singletons suggests the existence of a further 69 synteny groups not yet identified by mapped genes (data not shown). Therefore, the 247 homology segments supported by syntenic relationships provides a lower limit for the number of such segments but there may be upwards of 418 (247 + 102 + 69) homology segments defining the relationship between the zebrafish and human genomes. This compares favorably with the 201 homology segments described between the mouse and human (DeBry and Seldin 1996).
Table 1.
Zebrafish–Human Conserved Syntenies
| Zebrafish linkage group | Human chromosome |
|---|---|
| 1 | 1, 2, 4, 13, 14 |
| 2 | 1, 2, 3, 7, 8, 9, 19 |
| 3 | 7, 11, 16, 17, 19 |
| 4 | 3, 7, 11, 12 |
| 5 | 5, 9, 11, 14, 17, 19, X |
| 6 | 2, 12, 13, 19 |
| 7 | 7, 11, 16, 19 |
| 8 | 1, 3, 4, 5, 7, 8, X |
| 9 | 2, 11, 21, X |
| 10 | 3, 4, 11, 21 |
| 11 | 1, 3, 8, 12, 17 |
| 12 | 2, 10, 17, 22 |
| 13 | 4, 6, 10, 19 |
| 14 | 5, 11, X |
| 15 | 3, 11, 17 |
| 16 | 3, 6, 8, 17, 19 |
| 17 | 2, 4, 14, 20 |
| 18 | 11, 15, 19, 22 |
| 19 | 1, 3, 6, 7 |
| 20 | 2, 4, 6, 20 |
| 21 | 5, 6, 9, 10, 11 |
| 22 | 1, 2, 7, 12, 19 |
| 23 | 1, 3, 6, 7, 12, X |
| 24 | 8, 10 |
| 25 | 5, 11, 15, 22 |
Human chromosomes (right) with two or more orthologous genes or ESTs mapped on corresponding zebrafish linkage groups (left).
Figure 2.
Distribution of genes and ESTs in synteny groups. Bars indicate the distribution of zebrafish genes and ESTs according to class of synteny relationship (Y-axis) for each linkage group (X-axis). Number of genes and ESTs from homology segments with two or more contiguous members where gene order is conserved between zebrafish and human are shown in blue. Additional genes and ESTs in conserved synteny groups but not in contiguous sets are shown in yellow. Genes and ESTs that are not part of conserved synteny groups (singletons) are depicted in red. Together these three classes account for all the mapped genes and ESTS with orthologs predicted unambiguously by WU-BLAST analysis (see Methods).
Previous analyses have suggested that a genome-wide duplication may have occurred in the teleost lineage since its divergence from the tetrapod lineage (Amores et al. 1998; Postlethwaite et al. 1998; Wittbrodt et al. 1998; Gates et al. 1999). Consistent with the notion of genome-wide duplication, we find 38 examples where two or more mapped, unlinked zebrafish genes share a single mammalian ortholog (Table 2). These are distributed on 20 of the 25 zebrafish linkage groups, and 14 of 23 human chromosomes. A further seven pairs of tightly linked zebrafish genes also share a single human ortholog, suggesting that in some cases, tandem duplications may also have played a role in generating extra zebrafish genes. However, paralogous gene pairs are not the rule for the described zebrafish genes. Analysis of ESTs from 12 ribosomal protein genes, an abundantly expressed class of genes that has been sufficiently sampled to draw inferences about gene number, revealed only two with duplicate expressed genes (S. Johnson, unpubl.), raising the possibility that if the entire genome were additionally duplicated, most of the duplicate copies have been lost or inactivated.
Table 2.
Human Genes with Two or More Zebrafish Orthologs
| Human gene | Reference (NCVI unigene) | Human map position | Zebrafish ortholog | Reference (NCBI gi) | Zebrafish map position |
|---|---|---|---|---|---|
| HES5 | no ref | 1.49-52cMa | her2 | 1279391 | 8.472cRc |
| her4 | 1279395 | 23.99cRc | |||
| HFH2 | Hs.166188 | 1.95-102cM | fkd8 | 2982352 | 8.299cRd |
| fkd6 | 2982348 | 6.273cRb | |||
| SOX11 | Hs.32964 | 2.0-32cM | sox11a | NA | 17.234cRd |
| sox11b | NA | 20.499cRd | |||
| RARA | Hs.173205 | 2.51-54cM | rara2a | 704369 | 12.125cRc |
| rara2b | 215025 | 3.161cRd | |||
| SIX3 | Hs.227277 | 2.73-88cM | six6 | 3047418 | 12.188cRf |
| six3 | 304716 | 13.278cRf | |||
| EN1 | II.2019 | 2.127-134cM | eng4 | 4322043 | 1.59cRd |
| eng1 | 62515 | 9.9cRd | |||
| DLX2 | Hs.419 | 2.182-188cM | dlx5 | 1620515 | 1.179cRc |
| dlx2 | 460126 | 9.131cRc | |||
| IHH | Hs.69351 | 2.200-215cM | ehh | 1616584 | 6.115cRd |
| hha | NA | 9.140cRd | |||
| FZD5 | Hs.152251 | 2.211-218 | fz8a | 4164470 | 24.133cRf |
| frz-zg06 | 1245193 | 2.438cRf | |||
| FZD7 | Hs.173859 | 2.200-206cM | frz-zg07 | 1245195 | 9.170cRf |
| fb38g02 | 6.115cRb | ||||
| frz-zg13 | 1245207 | 6.129cRf | |||
| GATA2 | Hs.760 | 3.142-146cM | gata1 | 1132418 | 11.230cRd |
| gata2 | 1132420 | 11.390cRc | |||
| ATP1B3 | Hs.76941 | 3.157-158cM | atp1b | 974773 | 2.150cRf |
| fb13c07 | 15.57cRa | ||||
| EPHA5 | Hs.31092 | 4.68-78cM | fb82e05 | 24.301cRb | |
| rtk7 | 3005904 | 24.301cRb | |||
| NPY1R | Hs.169266 | 4.157-169cM | zya | 3098345 | 17.79cRd |
| zyb | 2739140 | 8.563cRd | |||
| zyc | 3098347 | 10.385cRd | |||
| EFNA5 | Hs.37142 | 5.108-116cM | al1 | 1834430 | 8.10cRc |
| ephra5 | 2462952 | 21.129cRb | |||
| CSX | Hs.54473 | 5.161-163cM | nkx2.7 | 1518150 | 8.505cRe |
| nkx2.5 | 1518148 | 14.341cRd | |||
| MSX2 | Hs.89404 | 5.185-196cM | msxe | 1399516 | 14.27cRc |
| msxa | 608508 | 14.464cRd | |||
| msxd | 62544 | 21.211cRc | |||
| ISL1 | Hs.505 | 5.54-61cM | islet1 | 497897 | 5.143cRc |
| islet2 | 1037165 | 25.406cRc | |||
| islet3 | 1037167 | 25.406cRf | |||
| AHR | Hs.170087 | 7.24-35cM | ahr2 | 4321818 | 22.88cRf |
| ahr | 2764987 | 16.196cRb | |||
| EVX1 | Hs.99967 | 7.38-42cM | eve1 | 475049 | 3.113cRc |
| evx1 | no ref. | 16.175cRd | |||
| HOXA | N/A | 7.39-40cM | hoxa13b | 4322052 | 16.175cRd |
| hoxa4a | 4322059 | 19.170cRc | |||
| EN2 | Hs.134989 | 7.167-175cM | eng2 | 62517 | 7.158cRc |
| eng3 | 62521 | 2.343cRc | |||
| SHH | Hs.121539 | 7.181-184cM | shh | 5714439 | 7.158cRc |
| twhh | 1171139 | 2.346cRd | |||
| SLUG | Hs.93005 | 8.57-68cM | sna2 | 841423 | 23.41cRd |
| sna1 | 468620 | 11.284cRc | |||
| NOTCH1 | II.4851 | 9.136-148cM | notch1b | 2569967 | 5.267cRf |
| notch1 | 433866 | 21.75cRf | |||
| RXRA | Hs.20084 | 9.143-166cM | rxrg | 1046288 | 5.222cRf |
| rxra | 1046294 | 2.309cRc | |||
| FTH1 | Hs.62954 | 11.16-23cM | fb06g09 | 7.45cRb | |
| fb01e08 | 24.144cRb | ||||
| WNT11 | Hs.108219 | 11.80-84cM | wnt11 | 3169686 | 5.125cRe |
| wnt11r | NA | 10.306cRd | |||
| HSPA10 | Hs.180414 | 11.128-132cM | hsc70.1 | 1408566 | 3.113cRd |
| fb01g06 | 10.304cRb | ||||
| SPON1 | Hs.5378 | 11.24-25cM | fspdin2 | 2529226 | 25.70cRf |
| mindin1 | 2529220 | 14.379cRf | |||
| mindin2 | 2529222 | 14.341cRf | |||
| HOXC | N/A | 12.70-72cM | hoxc5a | 414104 | 23.324cRc |
| hoxc13b | 4322091 | 11.459cRd | |||
| ASCL1 | Hs.1619 | 12.106-113cM | zasha | 540237 | 4.149cRc |
| zashb | 540239 | 7.177cRc | |||
| OTX2 | II.5015 | 14.0-1cM | otx2 | 540243 | 17.304cRb |
| otx3 | 633134 | 1.381cRc | |||
| RTN1 | Hs.99947 | 14.54-58cM | deltab | 2772824 | 5.125cRd |
| dla | 2809388 | 1.395cRd | |||
| HOXB | N/A | 17.62-69cM | hoxb4a | 341108 | 3.113cRf |
| hoxb1b | 1127809 | 12.188cRc | |||
| LHX1 | Hs.157449 | 17.58-63cM | lim1 | 577524 | 15.189cRd |
| lim6 | 2155288 | 5.171cRd | |||
| NOTCH3 | Hs.8546 | 19.42-45cM | notch3 | 3153196 | 3.430cRf |
| notch5 | 2569969 | 3.430cRf | |||
| PR65 | Hs.173902 | 19.59-98cM | fa02h04 | 5.171cRf | |
| fb38a08 | 15.138cRb | ||||
| CKM | Hs.118843 | 19.59-98cM | fa28d05 | 5.125cRf | |
| fc14g11 | 13.183cRb | ||||
| MYRL2 | Hs.9615 | 19.59-98cM | fa93e09 | 7.284cRb | |
| fa97a12 | 2.340cRb | ||||
| BMP2 | Hs.73853 | 20.18-27cM | bmp2 | 2804174 | 20.678cRc |
| bmp2a | 2149147 | 17.43cRd | |||
| SNAP25 | Hs.84389 | 20.27-37cM | snap25a | 3703097 | 20.459cRc |
| snap25b | 3703099 | 17.79cRc | |||
| L1CAM | Hs.1757 | X.188-198cM | nadl1.1 | 1065713 | 23.22cRc |
| nadl1.2 | 1065715 | 23.163cRc |
Orthologs predicted with aid of syntenic correspondence (see Table 3) are shown in bold.
Position for human gene is inferred from map position of orthologous mouse gene and the mouse–human syntenic relationship (DeBry and Seldin 1996).
Genes and ESTs mapped in this study.
The described syntenic relationship between the zebrafish and human genomes can be used as a tool for predicting human orthologs for zebrafish genes and ESTs. We found 32 zebrafish genes or ESTs where multiple human homologs were suggested by WU-BLAST analysis. For 20 of these genes (61%), the syntenic relationships revealed by the foregoing analysis allowed us to predict the human orthologs (Table 3). For example, our WU-BLAST analysis failed to distinguish between human ACTB (on Hsa1), ACTC (on Hsa15), and ACTG1 (on Hsa17) as the most likely ortholog for zebrafish bact2 (Kelly and Reversade 1997). The map position for bact2 on LG3 (Geisler et al. 1999) near Pyy (on Hsa17; Lundell et al. 1997) argues that bact2 is the zebrafish ortholog for ACTG1, rather than ACTB or ACTC. Similarly, WU-BLAST analysis fails to unambiguously establish the orthologous relationship between zebrafish msxa, msxb, msxc, msxd, and msxe genes (Ekker et al. 1997) and the human MSX1 and MSX2, and mouse Msx3 (human MSX3 has not yet been identified) genes. Because the regions of the zebrafish linkage groups in which msxa (LG14), msxd (LG21) and msxe (LG14) reside are syntenic to or map near syntenic regions to the region on human chromosome 5 that contains MSX2, syntenic comparison suggests that the zebrafish msxa, msxd, and msxe genes are orthologous to human MSX2. Likewise, synteny analysis suggests that the zebrafish msxb gene (LG1) is orthologous to MSX1 (Hsa4) and zebrafish msxc is orthologous to mouse Msx3. These and other zebrafish–human orthology relationships predicted by synteny are shown in Table 3.
Table 3.
Predicting Orthology Using Synteny Relationship
| Zebrafish gene | Reference NCBI gi | Zebrafish map position | Human synteny predictionsa | Possible human orthologues | Reference NCBI unigene | Human map position |
|---|---|---|---|---|---|---|
| bact | 3044209 | 1.59cRb | 1, 2 | ACTB | Hs.180952 | 1.49-82cM |
| ACTG1 | Hs.204867 | 17.118-129cM | ||||
| ACTC | Hs.118127 | 15.25-32cM | ||||
| bact2 | 2822455 | 3.304cRg | 16, 17 | ACTG1 | Hs.204867 | 17.118-129cM |
| ACTB | Hs.180952 | 1.49-82cM | ||||
| ACTC | Hs.118127 | 15.25-32cM | ||||
| brn1.2 | 222975 | 6.218cRd | 1, 2, 9, 17 | POU3F1 | Hs.1837 | 1.49-82cM |
| POU3F2 | Hs.182505 | 6.91-96cM | ||||
| POU3F3 | Hs.248158 | 3.80-100 | ||||
| POU3F4 | Hs.2229 | X.97-105cM | ||||
| elrd | 608548 | 8.108cRd | 1 | ELAVL4 | Hs.75236 | 1.49-82cM |
| ELAVL2 | Hs.3198 | 9.57-93cM | ||||
| frz-zg01 | 1245183 | 15.272cRg | 2, 3, 11, 17 | FZD4 | II.8322 | 11.84-100cM |
| FZD9 | Hs.158335 | 7.84-91cM | ||||
| glr | 3378595 | 14.433cRb | 5, 11, 12 | GLRA1 | Hs.121490 | 5.153-158cM |
| GLRA3 | Hs.167742 | 4.170cM | ||||
| GLRA2 | Hs.2700 | X.0-42cM | ||||
| groucho1 | 2104717 | 7.119cRb | 11, 15, 16 | TLE3 | Hs.167086 | 15.70-71cM |
| TLE1 | Hs.28935 | 9 | ||||
| TLE4 | Hs.83958 | 9.77.7-82.3cM | ||||
| TLE2 | Hs.173063 | 19.0.0-31.9cM | ||||
| hha | N/A | 9.140cRe | 2 | IHH | Hs.69351 | 2.200-215cM |
| SHH | Hs.121539 | 7.181-184cM | ||||
| Idb4 | 3078004 | 13.278cRg | 2, 6, 10 | LDB1 | Hs.26002 | 10.114-131cM |
| LDB2 | Hs.4980 | 4.0-32cM | ||||
| msxa | 608508 | 14.464cRe | 5 | MSX2 | Hs.89404 | 5.185-199cM |
| MSX1 | Hs.194 | 4.4-28cM | ||||
| MSX3 | Mm.4816 | 10.170-182cMc | ||||
| msxb | 608510 | 1.381cRb | 4, 13, 14 | MSX1 | Hs.194 | 4.4-28cM |
| MSX2 | Hs.89404 | 5.185-196cM | ||||
| MSX3 | Mm.4816 | 10.170-182cMc | ||||
| msxc | 399912 | 13.312cRd | 6, 10 | MSX3 | Mm.4816 | 10.170-182cMc |
| MSX1 | Hs.194 | 4.4-28cM | ||||
| MSX2 | Hs.89404 | 5.185-196cM | ||||
| msxd | 62544 | 21.211cRd | 5, 7, 10 | MSX2 | Hs.89404 | 5.185-196cM |
| MSX2 | Hs.89404 | 5.185-196cM | ||||
| MSX3 | Mm.4816 | 10.170-182cMc | ||||
| msxe | 1399516 | 14.27cRd | 5, 6, 8, 22 | MSX2 | Hs.89404 | 5.185-196cM |
| MSX1 | Hs.194 | 4.4-28cM | ||||
| MSX3 | Mm.4816 | 10.170-182cMc | ||||
| otx3 | 633134 | 1.381cRd | 4, 7, 14 | OTX2 | II.5015 | 14.0-1cM |
| OTX1 | II.5013 | 2.84-88cM | ||||
| plasticin | 1881763 | 11.390cRf | 3, 12, 17 | PRPH | Hs.37044 | 12.53-70cM |
| VIM | Hs.2064 | 10.40-44cM | ||||
| rtk7 | 3005904 | 24.301cRb | 4, 8 | EPHA5 | Hs.31092 | 4.67.7-77.9cM |
| EHK-1 | Hs.194771 | N/A | ||||
| EPHNA4 | Hs.739641 | N/A | ||||
| EPHA7 | Hs.73962 | 6.101-104cM | ||||
| EPHA3 | Hs.123642 | 3.111-113cM | ||||
| zef1 | 4099173 | 14.534cRd | 4, 5, 12, X | ELF4 | Hs.151139 | X.150-184cM |
| ELF1 | Hs.154365 | 13.37-46cM | ||||
| fb38g02 | 6.115cRb | 2, 19 | FZD7 | Hs.173859 | 2.200-212cM | |
| FZD2 | Hs.81217 | 17.74-75cM | ||||
| fb18b11 | 24.388cRb | 1, 8 | UBE2V2 | Hs.79300 | 8.66-67cM | |
| UBE2V1 | Hs.75875 | 20.74-75cM | ||||
| FZD10 | Hs.31664 | 12.160-169cM |
Human genes in bold are orthologues predicted by sytenic correspondence.
Corresponding human synteny group or groups for zebrafish genes in same mapping bin or flanking positions to zebrafish gene in column 1.
Genes and ESTs mapped in this study.
Corresponding human map position inferred from human-mouse syntenic relationship and mouse gene position.
DISCUSSION
Increasing the number of mapped zebrafish genes and ESTs with likely human (or in a few cases, mouse) orthologs to 523 has revealed extensive conserved synteny between the zebrafish and human genomes. We find 80% of genes and ESTs in this analysis fall in conserved synteny groups, averaging 3.7 genes/synteny group. A previous analysis of 124 zebrafish genes and ESTs identified only 64% (79/124) in conserved synteny groups, averaging 2.8 genes/group (Gates et al. 1999). Presumably, as more and more zebrafish genes and ESTs are mapped, the fraction that fall in synteny groups will continue to increase, and may approach 100%. Similarly, Gates et al. (1999) identified 28 synteny groups between zebrafish and human, and our analysis increases this number to 113. The existence of yet unidentified synteny groups is suggested by the 102 genes and ESTs in the singleton class. Singletons may reflect errors in mapping or in orthology determination, or may instead nucleate additional synteny groups as additional genes are mapped. Using the singleton class for Poisson analysis (and assuming no error) predicts a further 69 synteny groups as yet undiscovered. This allows us to predict an upper limit for synteny groups between zebrafish and human of 284 (113 +102 + 69).
The finding that most zebrafish genes in this study are in conserved synteny groups with human genes raises the possibility that significant portions of the zebrafish genome are uninterrupted by rearrangements since the teleost–tetrapod divergence. Indeed, we find that 292 of the genes and ESTs analyzed in this study define 118 homology segments (uninterrupted segments with conserved map order) covering ∼56% of the zebrafish genome (assuming random marker distribution). Taking into account the 1.7 × 109 bp size of the haploid zebrafish genome (Hinegardner 1968), we suggest an average size of 8.1 × 106 bp/homology segment identified in this study. This analysis suggests that zebrafish workers wishing to positionally clone zebrafish mutant genes can profitably use the syntenic comparison between zebrafish and human to identify candidates from the nearly complete human genome sequence.
Comparative biology often utilizes functional analysis of orthologous gene pairs, yet gene orthology is not always solvable by sequence comparison. For instance, members of multigene families may be too similar for BLAST or phylogenetic methods to unambiguously distinguish orthologous pairs of genes. One alternative to sequence-based orthology determination is a synteny-based approach. Such an approach first requires an understanding of the syntenic relationship between species compared. We suggest that the extensive correspondence between the human and zebrafish genomes revealed by this analysis can be used in predicting orthologous gene relationships. Of 32 zebrafish genes or ESTs whose human ortholog could not be unambiguously identified by BLAST analysis (data not shown), we suggest a human ortholog for 20 of these based on the syntenic correspondence of the zebrafish and human genomes (Table 3). Examples of such predictions include members of the zebrafish msx gene family. BLAST analysis fails to confidently predict the orthology relationships between the zebrafish msxa, msxb, msxc, msxd, or msxe genes and the human MSX1 and MSX2 and mouse MSX3 genes. Phylogenetic analysis (data not shown), suggests that zebrafish msxb and msxc are orthologous to mouse Msx3 (the human ortholog has not been identified), and zebrafish msxe is orthologous to human MSX1. We can use synteny as an alternative predictor of orthology, which suggests that msxa, msxd, and msxe are orthologous to MSX2; zebrafish msxb is orthologous to MSX1; and zebrafish msxc is orthologous to mouse MSX3. The addition of more genes to the zebrafish genetic map may further resolve this issue.
Recent observations suggest a whole genome duplication occurred in the teleost lineage since it's divergence from the tetrapod lineage (Amores et al. 1998; Postlethwaite et al. 1998; Wittbrodt et al. 1998; Gates et al. 1999). Consistent with this notion are the 38 examples where two or more mapped, unlinked zebrafish genes share a single mammalian ortholog, distributed among 20 of the 25 zebrafish chromosomes. The alternative hypothesis, that the duplications observed may have accrued individually, rather than in a single, whole-genome event, cannot yet be excluded. Indeed, instances of three zebrafish orthologs for a single human gene may argue for some role of regional duplication in generating duplicate copies of zebrafish genes. For instance, two of the three ISL1 orthologs, islet2 and islet3, map to a similar location on LG 25 (Geissler et al. 1999; Hukriede et al. 1999), and thus may have arisen by a tandem duplication. Identifying the syntenic relationship between the entire zebrafish and human genome may help resolve this issue.
A full understanding of the role of human genes in development and physiology will require models where gene function can be examined readily. Forward mutant screens in zebrafish are performed routinely, resulting in sizable collections of mutations causing a variety of developmental and physiological defects (e.g., Driever et al. 1996; Haffter et al. 1996; Henion et al. 1996). Molecular analysis of these mutations is beginning to reveal their utility as models for human disease (Zon 1999). Furthermore, the zebrafish is being established as a genetic and physiological model for vertebrate-specific processes such as organogenesis (Zhong et al. 2000). Knowledge of the relationship between the zebrafish and human genomes will provide the link to compare zebrafish genes and mutations with their orthologous human genes and diseases.
METHODS
RH Mapping and Map Construction
RH mapping was performed as described (Hukriede et al. 1999) on the LN54 zebrafish RH panel. Briefly, STS primers for genes were designed from 3′ ends of gene sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov), or for representative 3′ EST reads preselected for highly significant WU-BLASTX matches to the nonredundant protein database (http://zfish.wustl.edu). Primer sequences were designed using OSP (Hillier and Green 1991), (see http://zfish.wustl.edu for primer sequences). Each marker was positioned relative to the LN54 framework (Hukriede et al. 1999) using the RHMAPPER radiation hybrid mapping program (http://waldo.wi.mit.edu/ftp/distribution/software/rhmapper/) by web submission of the RH vector to http://mgchd1.nichd.nih.gov:8000/zfrh/beta.cgi, and placed accordingly in the bin following the framework marker, using the position of the framework marker to denote their position on the map.
Orthology Prediction
Each mapped zebrafish EST or gene was subjected to extensive WU-BLASTX and WU-BLASTN (filter = seg, E = 1e−10) (W. Gish, unpubl.; http://blast.wustl.edu) analysis against the comprehensive GenBank EST database, release 113 (http://ncbi.nlm.nih.gov) as well as the nonredundant protein and nucleotide database. The reports were postprocessed to recover the top matching hits from zebrafish, and the top EST, protein, and nucleotide hits from human sequences. All alignments were assessed manually, using a BLASTN cutoff at a maximum p value of e−20(the vast majority of predicted ortholog showed matches with p values < e−40. Zebrafish–human sequence pairs identified as putative orthologs by BLASTN similarity were likewise confirmed by BLASTX similarity. When available, we determined the UniGene reference sequence (http://www.ncbi.nih.nlm.gov/UniGene/) representing the human ortholog and acquired its Gene Map 98 map location (Deloukas et al. 1998; http://www.ncbi.nlm.gov/genemap98). In some cases human mapping data was obtained from Online Mendeliean Inheritance in Man (OMIM) (http://www.ncbi.nlm.nih.gov/Omim). All zebrafish–human orthologous pair BLASTN/BLASTX results, GenBank accession numbers, GenBank records, human reference numbers, and map positions are available at http://www.zfish.wustl.edu.
Acknowledgments
We thank Susan Dutcher, David Parichy, and John Rawls for critical reading of the manuscript, Warren Gish and Sean Eddy for providing additional local computer support, and Jonathon Epstein, Neil Hukriede, and Igor Dawid (NICHD) for providing the RHMAPPER web site. We are especially grateful to Matt Clark, Sandy Clifton, Marco Marra, and the WashU-MPIMG zebrafish EST project for generation of EST sequence used in this study. This work was funded by RO1 DK55379 (S.L.J.). S.L.J. is a Pew Scholar in Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sjohnson@genetics.wustl.edu; FAX (314) 362-7855.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.144700.
REFERENCES
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Brownlie A, Donovan A, Pratt SJ, Paw BH, Oates AC, Brugnara C, Witkowska HE, Sassa S, Zon LI. Positional cloning of the zebrafish sauternes gene: A model for congenital sideroblastic anaemia. Nat Genet. 1998;20:244–250. doi: 10.1038/3049. [DOI] [PubMed] [Google Scholar]
- DeBry RW, Seldin MF. Human/mouse homology relationships. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Ekker M, Akimenko MA, Allende ML, Smith R, Drouin G, Langille RM, Weinberg ES, Westerfield M. Relationships among msx gene structure and function in zebrafish and other vertebrates. Mol Biol Evol. 1997;10:1008–1022. doi: 10.1093/oxfordjournals.molbev.a025707. [DOI] [PubMed] [Google Scholar]
- Gates MA, Kim L, Egan ES, Cardozo T, Sirotkin HI, Dougan ST, Lashkari D, Abagyan R, Schier AF, Talbot WS. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 1999;9:334–347. [PubMed] [Google Scholar]
- Geisler R, Rauch GJ, Baier H, van Bebber F, Brobeta L, Dekens MP, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Henion PD, Raible DW, Beattie CE, Stoesser KL, Weston JA, Eisen JS. Screen for mutations affecting development of Zebrafish neural crest. Dev Genet. 1996;18:11–17. doi: 10.1002/(SICI)1520-6408(1996)18:1<11::AID-DVG2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hillier L, Green P. OSP: A computer program for choosing PCR and DNA sequencing primers. PCR Meth Appl. 1991;1:124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]
- Hinegardner R. Cellular DNA content and the evolution of teleostean fishes. Am Nat. 1968;102:517–523. [Google Scholar]
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Gates MA, Johnson M, Talbot WS, Horne S, Baik K, Rude S, Wong JR, Postlethwait JH. Centromere-linkage analysis and consolidation of the zebrafish genetic map. Genetics. 1996;142:1277–1288. doi: 10.1093/genetics/142.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly GM, Reversade B. Characterization of a cDNA encoding a novel band 4.1-like protein in zebrafish. BiochemCell Biol. 1997;75:623–632. [PubMed] [Google Scholar]
- Kimmel CB. Genetics and early development of zebrafish. Trends Genet. 1989;5:283–288. doi: 10.1016/0168-9525(89)90103-0. [DOI] [PubMed] [Google Scholar]
- Lundell I, Berglund MM, Starback P, Salaneck E, Gehlert DR, Larhammar D. Cloning and characterization of a novel neuropeptide Y receptor subtype in the zebrafish. DNA Cell Biol. 1997;16:1357–1363. doi: 10.1089/dna.1997.16.1357. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Meyer A, Schartl M. More genes in fish? Bioessays. 1998;20:511–515. [Google Scholar]
- Zon LI. Zebrafish: A new model for human disease. Genome Res. 1999;9:99–100. [PubMed] [Google Scholar]
- Zhong TP, Rosenburg M, Mohideen MPK, Weinstein B, Fishman MC. Gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]