Abstract
Se-dependent glutathione peroxidase-1 (GPX1) and Cu,Zn-superoxide dismutase (SOD1) are two major intracellular antioxidant enzymes. This study was to elucidate biochemical mechanisms for the 40% loss of hepatic GPX1 activity in SOD1−/− mice. Compared with the wild-type (WT), the SOD1−/− mice showed no change in the total amount of GPX1 protein. However, their total enzyme protein exhibited a 31 and 38% decrease (P < 0.05) in the apparent kcat for hydrogen peroxide and tert-butyl peroxide (at 2 mM GSH), respectively. Most striking, mass spectrometry revealed two chemical forms of the 47th residue of GPX1: the projected native selenocysteine (Sec) and the Se-lost dehydroalanine (DHA). The hepatic GPX1 protein of the SOD1−/− mice contained 38% less Sec and 77% more DHA than that of WT, respectively, and showed aggravated dissociation of the tetramer structure. In conclusion, knockout of SOD1 elevated the conversion of Sec to DHA in the active site of hepatic GPX1, leading to proportional decreases in the apparent kcat and activity of the enzyme protein as a whole. Our data reveal a structural and kinetic mechanism for the in vivo functional dependence of GPX1 on SOD1 in mammals, and provide a novel mass spectrometric method for the assay of oxidative modification of the GPX1 protein.
Keywords: Dehydroalanine, glutathione peroxidase, liver, mass spectrometry, mouse, selenocysteine, superoxide dismutase
Se-dependent cellular glutathione peroxidase-1 (GPX1, EC 1.11.19) and Cu,Zn-superoxide dismutase (SOD1, EC 1.15.1.1) are widely considered to be the two major intracellular antioxidant enzymes in mammals. While SOD1 converts superoxide anion into H2O2, GPX1 catalyzes the reduction of H2O2 and lipid peroxides by glutathione [1, 2]. Although the product of SOD1 is a major substrate of GPX1, catalytic coordination and(or) mutual functional regulation between these two enzymes under physiological conditions remain unclear. Intriguingly, several groups have shown a 40% decrease in hepatic GPX1 activity [3–6] in SOD1 knockout (SOD1−/−) mice compared with the wild-type (WT).
Presumably, tissue GPX1 activity is affected by the stead-state of GPX1 protein (amount) and post-translational modifications of the enzyme. As the most abundant member in liver among the 25 identified mammalian selenoproteins [7, 8], GPX1 incorporates Se into its catalytic site as selenocysteine (Sec) via a co-translational mechanism[9]. Deemed as a selective advantage during evolution [10, 11], Sec is encoded by a TGA codon [12], and requires a unique consensus Sec insertion sequence (SECIS) in the 3′ untranslated region of the selenoprotein mRNA for its synthesis [13]. Numerous studies have shown that Se is the most important regulator of GPX1 activity because its deprivation causes drastic losses of GPX1 mRNA, protein, and activity in tissues or cultured cells [14, 15]. The primary regulation is attributed to a nonsense codon-mediated decay of GPX1 mRNA [16–19]. In addition, transcriptional regulations of GPX1 may also account for its activity changes [20–24]. Notably, an antibiotic aminoglycoside decreases GPX1 activity by interfering the Sec incorporation in Se deficiency [25].
Post-translational modification of GPX1 protein may serve as a physiologically-relevant regulation of GPX1 function in Se adequacy. Indeed, c-Abl and Arg tyrosine kinases [26, 27] can directly bind to a proline-rich site of GPX1 protein and activate it by phosphorylation of Tyr96 [28]. In many cases, however, GPX1 protein is susceptible to oxidative inactivation despite its antioxidant catalysis [29, 30]. A NO donor, S-nitroso-N-acetyl-D,L-penicillamine (SNAP), decreases GPX1 activity via oxidizing Sec to form a selenenyl sulfide (Se-S) with a free thiol [31]. Recently, Rhee and Cho (2010) have reported that incubating GPX1 protein purified from human red blood cells with 1 mM H2O2 for 1 h at 37°C resulted in a substantial loss of peroxidase activity and a conversion of Sec at the active site into dehydroalanine (DHA) [32, 33]. This conversion is likely mediated by an oxidation of Sec by H2O2 or related derivatives, resulting in a subsequent loss of Se oxide. They have suggested DHA-GPX1 in red blood cells as a surrogate biomarker of body oxidative stress status, and developed a blot-based assay for DHA-containing GPX1 using biotin-conjugated cysteamine. However, there is no report for such conversion of the Sec residue of GPX1 into DHA under physiological conditions, in particular by changes in intracellular superoxide status. Despite convenience, the blot-based assay of DHA-GPX1 does not give accurate quantification or detect any other modifications of the peptide [34, 35].
The 40% decrease of GPX1 activity in the SOD1−/− mouse liver prompted us to determine if the activity loss was due to a lowered GPX1 expression and (or) an inactivation of the produced protein by deficiency of SOD1 [36]. After we verified that the SOD1 knockout caused no change in the total amount of hepatic GPX1 protein, but a ~ 40% decrease in its apparent kcat, we applied both qualitative and quantitative mass spectrometry to characterize modifications of the hepatic GPX1 protein in the SOD1−/− mice. Strikingly, the relative decrease in the apparent kcat of the hepatic GPX1 enzyme in the SOD1−/− mice corroborated with a similarly proportional increase in the conversion of Sec into DHA in the target peptide of the GPX1.
Materials and methods
Animals and diet
The SOD1−/− and WT mice were generated from the 129SVJ × C57BL/6 strain [37]. Deletion of sod1 gene was verified by PCR and by enzyme activity assay in various tissues. Hepatic SOD activity in SOD1−/− mice was <1% of the WT. All mice were fed a Se-adequate diet (0.4 mg/kg) [38], given free access to feed and distilled water, and housed in shoebox cages in a constant temperature (22°C) animal room with a 12-h light/dark cycle. All animals used in this study were bred in our mouse facility and were 8-wk old males (n = 3–5). Our experiments were approved by the Institutional Animal Care and Use Committee at Cornell University and conducted in accordance with National Institutes of Health guidelines for animal care.
SDS-PAGE, Western blot and immune-precipitation
Liver samples used for Western blot analysis were homogenized in buffer A [100 mM Tris, pH 7.4, containing 250 mM sucrose/protease inhibitor mixture (1 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 10 μg of leupeptin/mL,10 μg of aprotinin/mL, 1 μM microcystin, 1 mM PMSF, 10 mM sodium fluoride)]. Samples used for co-immunoprecipitation were homogenized in buffer B (50 mM Hepes, pH 7.4, containing 100 mM sodium chloride, 0.5% NP40, 5 mM EDTA, and protease inhibitor mixture as in buffer A). Liver homogenates were centrifuged at 14,000×g for 10 min at 4°C. Western blot analysis was performed as described previously [38]. For co-immunoprecipitation, supernatant (0.1 mg of protein) of liver homogenates was pre-cleared by incubation with 5 μL of rabbit serum and 30 μL of Protein A/G PLUS-Agarose beads (50% slurry) for 30 min at 4°C. After centrifugation at 3,000×g for 1 min, the supernatant was incubated with the antibody against SOD1 (Chemicon, Temecula, CA) or GPX1 (Lab Frontier, Seoul, Korea) overnight at 4°C with rotation. After the preparation was incubated with 50 μL of protein A/G beads for 60 min at 4°C with rotation, the beads were washed four times with 1 mL of buffer B on ice. Thereafter, Buffer B and 5×SDS buffer (20 μL each) were added to the washed beads to elute the precipitant for respective Western blot analysis. For mass spectrometry analysis, the immunoprecipitant of liver homogenate supernatant (500 ug) by the GPX1 antibody was subject to SDS-PAGE (12% gel). After the gel was stained with Sypro Ruby (Invitrogen, Carlsbad, CA), the GPX1 band was excised and submitted to the Cornell Proteomic and Mass Spectrometry Core facility. For determining the GPX1 protein integrity in vivo, a non-denaturing gel was run at 4°C without reducing agent, SDS, or heating.
GPX1 activity and kinetics
Fresh liver samples were homogenized in sucrose buffer (20 mM Tris-HCl, pH 7.6, containing 0.25 M sucrose and 1.1 mM EDTA) and centrifuged (10,000×g, 45 min, 4°C). The supernatant was used for the GPX1 activity assay [10]. For kinetic studies, the hepatic GPX1 protein was purified as described below from both genotypes. Because the enzyme reaction of GPX1 represents a ter-uni ping pong mechanism, only apparent kinetic parameters (appKm, appVmax, appkcat, appkcat/Km) against substrates hydrogen peroxide (H2O2) and tert-butyl peroxide (TBHP) were measured at 25°C using the coupled assay in 0.1 M potassium phosphate, pH 7.0[39, 40]. Accordingly, a fixed GSH concentration (2 mM) was maintained in all assays [41], while 9 substrate concentrations were tested ranging from 0.1 to 10 times of predicted Km. The reaction mixture contained 10 μg protein as determined by the BCA proteinassay (Thermo Fisher Scientific, Rockford, IL). The Michaelis-Menten equation was fit to the plot of initial velocity vs. substrate concentration for determination of appKm and appVmax using Kaleidagraph (v. 3.5, Synergy Software, Reading, PA). A monomer molecular weight of 22,198 was used for appkcat calculation. Statistical analysis was done using the Student’s t-test (n = 4).
Purification of hepatic GPX1 protein
Liver samples were homogenized in sucrose buffer as described above, and applied to a 100 × 2.5 cm column of Sephadex G-100 (Pharmacia Fine Chemicals, Piscataway, NJ). The GPX1 protein was eluted with 20 mM Tris-HCl, 1 mM GSH, pH 7.6. All procedures were performed at 4°C. The fractions collected were subjected to Western blot with anti-GPX1 antibody while the same membrane was also probed with anti-SOD1 and anti-JNK1 (Cell Signaling, Danvers, MA) antibodies as controls.
Mass spectrometry analysis
The excised GPX1 protein gel band was subjected to reduction and alkylation with iodoacetamide followed by in-gel digestion and extraction [42]. The extracted tryptic peptides were reconstituted in 15 μL of 0.5% formic acid (FA), and 5 μL was injected in nanoLC-MS/MS analysis using 4000 Q Trap as described previously [43]. The nanoLC was carried out by a LC Packings Ultimate Plus system (Dionex, Sunnyvale, CA). The tryptic peptides were loaded on a Dionex’s trapping column and separated on a PepMap C-18 RP nano column (3 μm, 75 μm id × 150 mm, Dionex), eluted in a 60-min gradient of 5% to 45% acetonitrile in 0.1% FA at 275 nL/min. The MS data acquisition was performed using Analyst 1.4.2 software (Applied Biosystems, Framingham, MA) for information dependent acquisition (IDA) analysis in which, after each survey and an enhanced resolution scan, the three highest intensity ions with multiple charge states were selected for tandem MS (MS/MS).
The MS/MS data generated from IDA analysis were submitted to Mascot 2.2 (Matrix Science, London, UK) for database searching against the Mouse RefSeq database (downloaded in July 2007), with one missed cleavage site by trypsin allowed. Carbamidomethyl modification of cysteine or selenocysteine and a methionine oxidation were set as variable modifications. Only significant scores for the peptides defined by Mascot probability analysis greater than “identity” were considered for the peptide identification and modification site determinations. All acquired MS and MS/MS spectra for identified peptides in GPX1 protein from WT and SOD1−/− samples were manually inspected and interpreted with the Analyst 1.4.2 software along with its integrated BioAnalysis 1.4 software (Applied Biosystems).
For relatively quantitative analysis on the two target peptide isoforms (Sec47 and DHA47) detected in WT and SOD1−/− samples, the peak areas of detected precursor ions were determined by extracted ion chromatogram (XIC) at each specific m/z corresponding Se-peptide and DHA-peptide, along with the rest of identified peptides of GPX1 serving as internal controls. An alternative quantitation approach, selected reaction monitoring (SRM or often called MRM) for selected target peptides was also applied for the same gel-extracted samples using the same nanoLC-4000 Q Trap platform. The IDA data on all detected GPX1 peptides were used to select precursor → fragment ion m/z values for MRM assays using MRMPilot 2.0 software (Applied Biosystems). The selection for MRM analysis was based on three main criteria: (i) the peptides chosen were detected by IDA with no missed cleavages and 99% confidence in identification; (ii) the m/z value of fragment ions for monitoring transition ion pairs was always larger than that of their precursor ions for enhancing the selectivity; (iii) for each peptide two or more MS/MS fragments with relatively high signal intensity were chosen to be monitored. As a result, 8 GPX1 tryptic peptides with 20 transition ion pairs were monitored with dwell time at 100 ms for each pair in the nanoLC-MRM analysis. The acquired MRM data for WT and SOD1−/− samples were processed and analyzed using MultiQuant 1.2 software (Applied Biosystems).
Results
No genotype difference was detected in hepatic GPX1 protein expression
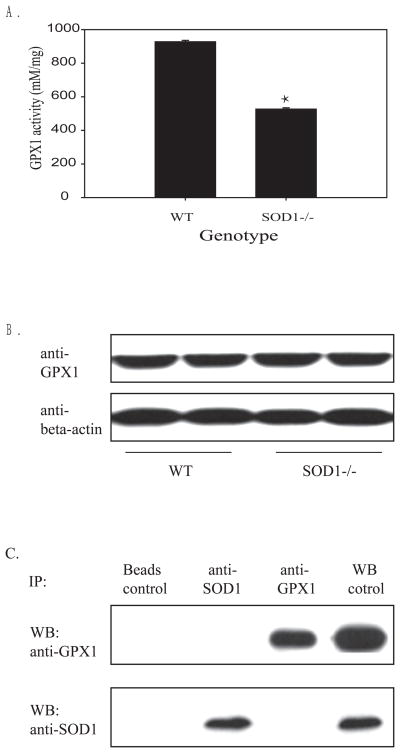
While hepatic GPX1 activity in the SOD1−/− mice was 39% (P < 0.01) lower than that of WT mice (Fig. 1A), there was no genotype difference in total amount of hepatic GPX1 protein (Fig.1B). Thus, the loss of GPX1 activity in the SOD1−/− mouse liver was caused by post-translational modification or inactivation of the produced protein. In addition, reciprocal co-immunoprecipitation analyses of liver homogenate supernatant against the GPX1 and SOD1 antibodies depicted no protein-protein interaction between these two enzymes (Fig.1C). Likewise, loss of GPX1 activity in the SOD1−/− mouse liver was unlikely due to a missing of the SOD1 protein for a physical interaction.
Fig. 1.
Impact of SOD1 knockout on hepatic GPX1 activity (A), protein amount (B) and co-immunopreciptation of hepatic GPX1 and SOD1 (C). A. Hepatic GPX1 activity was expressed as mean ± SE (n = 5). B. Western blot analysis of hepatic GPX1 protein (10 μg protein/lane). The image is a representative of three independent experiments. C. Co-immunoprecipation of GPX1 and SOD1 in liver homogenates. After 100 μg of total liver homogenate protein was incubated with 1 μg anti-GPX1 or anti-SOD1 antibody overnight at 4°C. The precipitates were loaded to SDS-PAGE (12% gel) and transferred to membranes that were blotted against the designated antibodies. WT liver homogenate was used as the positive control for Western blot. The image is a representative of three independent experiments.
Knockout of SOD1decreased the apparent catalytic efficiency of total hepatic GPX1 protein for hydroperoxides
Compared with that of the WT, the hepatic GPX1 enzyme protein, including both native and inactive forms, from the SOD1−/− mice had a 31% lower appkcat [H2O2] [GSH=2mM] (8.1 vs. 1.18 ×104/sec, P < 0.0001) and 38% lower appkcat [TBHP] [GSH=2mM] (4.2 vs. 6.7 ×104/sec, P < 0.0001), respectively (Table 1). Similar genotype differences were also observed in the enzyme appVmax and appkcat/Km (P < 0.0001). The apparent substrate affinity (appKm) of the GPX1 enzyme from the SOD1−/− mice was slightly better for TBHP (24 vs. 29 μM, P < 0.01), but somewhat worse for H2O2 (22 vs. 14 μM, P < 0.05) than that of WT (at 2 mM GSH).
Table 1.
Kinetics of hepatic GPX1 from WT and SOD−/− mice*
| Substrate | Genotype | appKm (μM) | appVmax (mM min−1 mg−1) | appkcat (×103 sec−1) | appkcat/Km (×109 M−1 sec−1) |
|---|---|---|---|---|---|
| TBHP | WT | 29a±1 | 182a±2 | 67a±1 | 2.4a±0.03 |
| SOD−/− | 24b±1 | 113b±2 | 42b±1 | 1.7b±0.03 | |
|
| |||||
| H2O2 | WT | 14b±1 | 319a±3 | 118a±1 | 8.6a±0.29 |
| SOD−/− | 22a±2 | 220b±9 | 81b±3 | 3.7b±0.25 | |
Data are presented as the mean ± standard error (n = 4). Values within a given substrate and column differ (P < 0.05) without sharing a common superscript letter (a vs. b). A fixed GSH concentration was maintained constant at 2 mM in the reaction mixture.
Knockout of SOD1caused simultaneous decrease of Sec and increase of DHA in the hepatic GPX1 protein
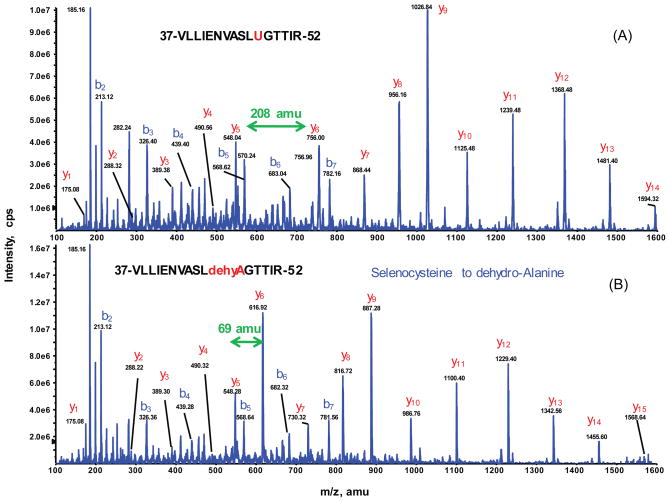
The peptides identified by nanoLC-MS/MS yielded 90% of GPX1 protein sequence coverage (data not shown) for both genotype samples, including the active site Sec-containing tryptic peptide. Strikingly, the 47th residue in the GPX1 tryptic peptide (residues 37–52) existed in two isoforms: 1) the projected native Sec47 peptide eluted at 52.4 min with both doubly-charged precursor ion at m/z 903.97 and triply-charged ion at m/z 603.4; and 2) a new isoform with a mass decreased by 139 amu. The latter isoform, with only 69 amu locating at the 47th residue, matched exactly the resultant DHA from the loss of the Se group. The selective MS/MS spectra of the doubly charged peptides ions: [903.97]2+ for the native form of Sec (with additional carbamidomethyl modification) and [834.46]2+ for the Se-lost form of DHA were identified in both of WT and SOD−/− samples (Fig. 2).
Fig. 2.
Identification of seleno-peptide for GPX1 by nanoLC-MS/MS. Representative MS/MS spectra of the doubly charged peptides ions at m/z [903.97]2+ for VLLIENVASLUGTTIR (A) and [834.46]2+ for VLLIENVASLdehydroAGTTIR (B), acquired from nanoLC-MS/MS analysis of tryptic digest of SOD1−/− gel band. All y-ion series including y6 and higher match to the GPX1 tryptic peptide covering residues #37–52 confirm that the residue 47 is Sec (selenocycteine) which is carbamidomethylated (208 amu) as shown in the top panel. The bottom panel shows that the same peptide sequence is unambiguously identified with the peptide mass decrease by 139amu. The MS/MS spectrum clearly indicates that the shift of −139amu begins at y6 and higher y-ion series, suggesting Sec at residue 47 be converted to DHA (dehydroalanine) (69 amu).
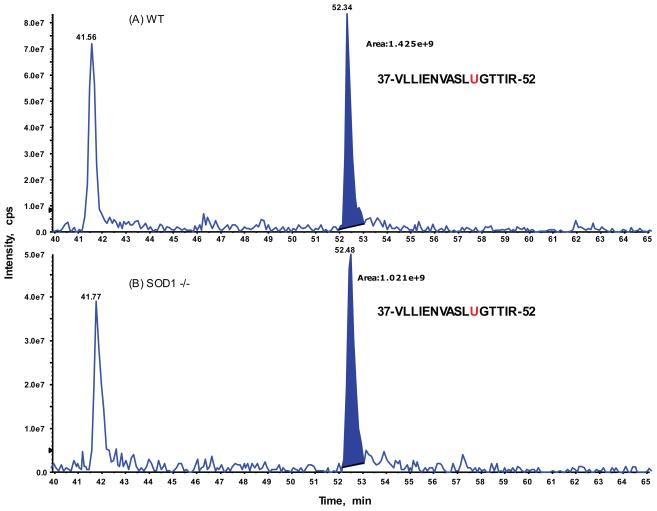
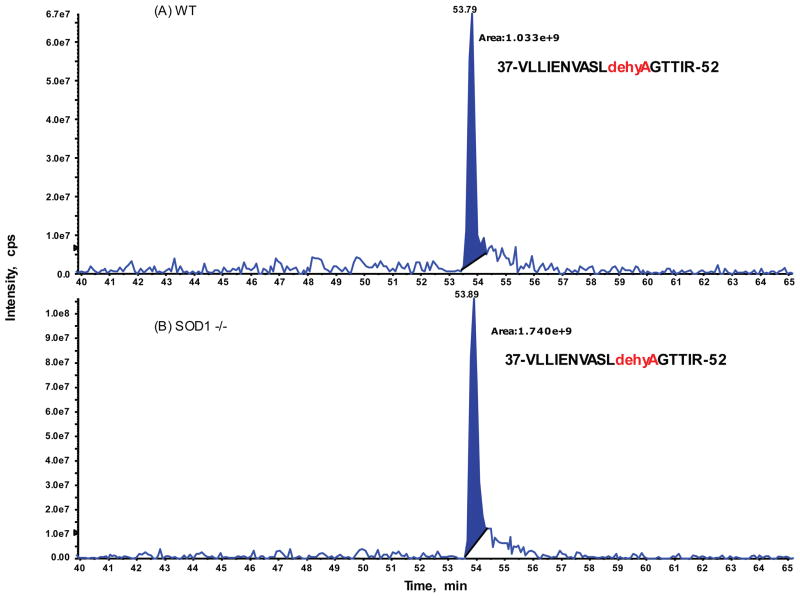
To explore if relative distributions of these two isoforms of Sec and DHA in the hepatic GPX1 accounted for the genotype difference in hepatic GPX1 activity, we conducted intensive analysis of the extracted ion chromatogram (XIC) for precursor ions on the targeted peptide with two isoforms identified by previous IDA analysis on both genotype samples (Fig. 3 and 4). The ratios of peak areas corresponding to the native Sec- and the Se-lost DHA-containing peptides from the XIC on the doubly-charged ion only between the SOD1−/− and WT samples were 0.71 and 1.68, respectively (Table 2). These ratios were 0.62 and 1.77, respectively, when both doubly and triply-charged ions were combined. Meanwhile, relative ratios for eight additional GPX1 peptides identified from IDA analysis were also compared and showed a range from 0.96 to 1.15, with an average of 1.01 and RSD of 6.3%. Thus, there were no significant changes in the GPX1 protein abundances between the WT and SOD1−/− gel extracted samples.
Fig. 3.
Relative quantification of the identified selenocysteine peptide (residue 37 to 52 of GPX1) between SOD1−/− and WT by extracted ion chromatogram (XIC) analysis. Expanded view of XIC on a doubly-charged ion: [903.97]2+ for tryptic GPX1 peptide, VLLIENVASLUGTTIR acquired from WT sample at top panel (A) and from SOD1−/− mutant sample at bottom panel (B). The integrated peak areas at retention of 52 min plus those for the triply-charged ion of the same peptide (data not shown here) were used to determine the relative ratio changes between two samples. The same XIC approach was applied for the rest detected GPX1 tryptic peptides (see Table 2).
Fig. 4.
Relative quantification of the identified Se-lost DHA peptide (residue 37 to 52 of GPX1) between SOD1−/− and WT by extracted ion chromatogram (XIC) analysis. Expended view of XIC on a doubly-charged ion: [834.46]2+ for VLLIENVASLdehydroAGTTIR acquired from WT sample at top panel (A) and from SOD1−/− mutant sample at bottom panel (B). The integrated peak areas at retention of 54 min plus those for the triply-charged ion of the same peptide (data not shown here) were used to determine the relative ratio changes between two samples (see Table 2).
Table 2.
Relative quantification of GPX1 tryptic peptides between the two genotypes by XIC and MRM analyses*
| Peptide sequence | m/z (z) | Ratio of SOD−/− to WT
|
|
|---|---|---|---|
| XIC peak | MRM | ||
| 37-VLLIENVASLUGTTIR-52 | 904.0 (2) | 0.71 | 0.65 |
| 603.4 (3) | 0.49 | ND | |
| Sum +2 & +3 | 0.62 | NA | |
| 37-VLLIENVASLdehydroAGTTIR-52 | 834.5 (2) | 1.68 | 1.30 |
| 556.7 (3) | 2.04 | ND | |
| Sum +2 & +3 | 1.77 | NA | |
| 3-AARLSAAAQSTVYAFSARPLTG GEPVSLGSLR-34 | 812.6 (4) | 0.99 | 1.0 |
| 68-GLVVLGFPCNQFGHQENGKKE EILNSLK-95 | 1045.1 (3) | 1.15 | 0.93 |
| 96-YVRPGGGFEPNFTLFEK-112 | 979.6 (2) | 0.96 | ND |
| 120-AHPLFTFLR-128 | 551.3 (2) | 0.96 | 1.0 |
| 147-YIIoxiWSPVCR-155 | 605.3 (2) | 1.04 | 1.08 |
| 147-YIIWSPVCR-155 | 597.3 (2) | 1.0 | 0.93 |
| 165-FLVGPDGVPVR-175 | 578.4 (2) | 0.96 | 1.05 |
| 183-TIDIEPDIETLLSQQSGNS-201 | 1030.6 (2) | 1.01 | ND |
The values were calculated based on the integrated peak areas at the specific retention time for doubly-charged ions plus those for the triply-charged ion of the same peptide by XIC. MRM-based quantification was conducted with the same gel-extracted samples under the same nanoLC-4000 Q Trap platform and elution conditions. R = acetyl arginine; C = cysteic acid.
To further validate the XIC-based relative quantitation results, we applied a more selective and accurate technique MRM approach. Again, six non-targeted control peptides from the GPX1 protein were included in MRM analysis serving as normalization between two samples. The relative ratios of the SOD1−/− to the WT samples for Sec and DHA-containing peptides were 0.65 and 1.30, respectively for the doubly-charged ion pairs. Likewise, the relative ratios for six control peptides of GPX1 by the MRM analysis ranged from 0.93 to 1.08 with an average of 0.998 and RSD of 6.0% (Table 2). The MRM results were consistent with the initial XIC data for all tryptic peptides, indicating that partial Sec47 isoform in the target peptide were converted to DHA47 isoform in response to the SOD1 knockout.
Meanwhile, other modifications in several peptides of GPX1 were identified. These modifications included acetylation of the several Arg residues, conversion of the residue Cys76 and Cys154 into cysteic acid, and oxidation of the residue Trp150. However, neither XIC nor MRM analysis showed a difference between the two genotypes (Table 2), suggesting that these modifications were not directly related to the knockout of SOD1.
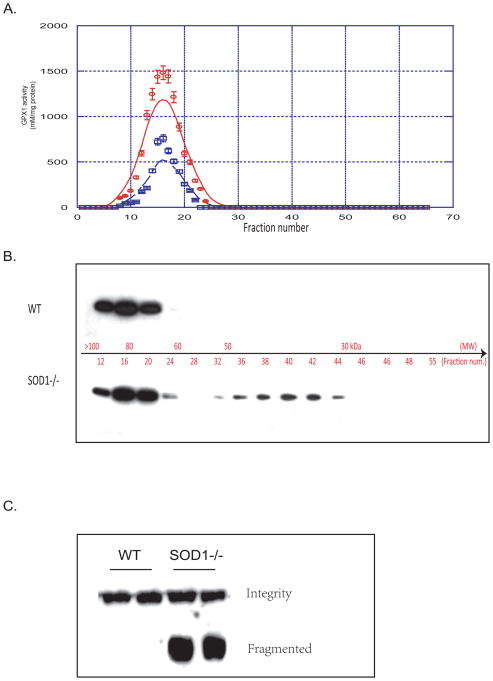
Dissociation of hepatic GPX1 protein tetramer was elevated in the SOD1−/− mice
The SOD1−/− and the WT mice shared similar GPX1 activity distribution profiles for size exclusion chromatography fractions of liver homogenates, although the peak activity in the former was lower than the latter (Fig. 5A). Western blot analysis indicated the GPX1 tetramer peak was centered on the fraction 16 in both genotype samples, which matched the activity distribution. Another GPX1 protein fragment that was close to the molecular weight of the GPX1 dimer was centered on the fraction 40 only in the SOD1−/− mice (Fig. 5B). Similarly dissociated GPX1 protein, smaller than the native tetramer, was also seen in the SOD1−/− mouse liver homogenates by Western blot analysis after a non-denaturing gel separation (Fig. 5C).
Fig. 5.
Distribution of GPX1 activity and protein in size exclusion chromatography fractions and native gel electrophoresis. A. GPX1 activity of fractions of liver homogenates separated on a 100 × 2.5 cm Sephadex G-100 column (flow rate = 0.1 ml/min; 1.5 ml/fraction). B. SDS-PAGE followed by Anti-GPX1 Western blot of selected fractions (5μl). C. Native gel (7.5%) electrophoresis followed by Anti-GPX1 Western blot of liver homogenates showing dissociated GPX1.
Discussion
The most significant finding of this study is that the knockout of SOD1 in mice resulted in an accelerated conversion of Sec in the active site of hepatic GPX1protein into Se-eliminated DHA. Remarkably, our qualitative and quantitative mass spectrometry illustrated a nearly perfect match for the reciprocal changes in DHA and Sec of the GPX1 peptide to the decrease in the enzyme appkcat in the SOD1−/− mice. Thus, we herein illustrate a structural and kinetics mechanism to explain the previously-reported activity loss of hepatic GPX1 in these mice [3–5]. The lack of genotype difference in total amount of hepatic GPX1 protein excludes potential effects of SOD1 knockout on GPX1 gene expression or protein production via altering body Se status. Likewise, the lack of co-immunoprecipitation of GPX1 and SOD1 by each other eliminated a possible missing of their protein-protein interaction in the SOD1−/− mice as the reason for the loss of GPX1 activity. Likely, oxidative modification of Sec in the active site of GPX1 was attributed to this SOD1 knockout-mediated decrease of hepatic GPX1 activity. Although inactivation of GPX1 protein by its own peroxide substrates and conversion of Sec to DHA in the active site of GPX1 have been previously shown in in vitro experiments [29, 30, 32, 33], findings from the present study signify at least three novel aspects. First, the elevated DHA in the hepatic GPX1 protein of the SOD1−/− mice, to our best knowledge, represents the first in vivo evidence for the oxidative modification of Sec under physiological conditions. Second, it is also the first illustration for the elevated superoxide anion-associated conversion of Sec of GPX1 into DHA [36]. Third, the qualitative and quantitative mass spectrometry provides an accurate estimate of the relative portion of the intact (Sec) and the modified (DHA) forms for the Sec residue in the hepatic GPX1 protein.
Our study was conducted to rule out several concerns that may cause variable conversions of Sec to DHA during the assay. All sample preparations and purifications were carried out in parallel without bias for both of WT and SOD1−/− proteins. Prior to MS analysis, the gel discs were reduced and alkylated with iodoacetamide to avoid potential Se-Se bond formation during the electrospray oxidation process. Chemically, the alkylation of Sec (carbamidomethylated Sec) should be more stable and resistant to possible subsequent oxidation in MS analysis than its unalkylated native form. Therefore, the loss of Se could not result from the alkylation. Because the in vitro experiments by Rhee and Cho have provided a biochemical possibility for the oxidative modification of Sec into DHA in the GPX1 protein by elevated H2O2 [32, 33], the escalated intracellular superoxide anion and the resultant oxidative stress in the SOD1−/− mice might induce the same oxidative conversion.
Located in the catalytic center of GPX1, the Sec residue contains one selenyl group [10]. During catalysis, the selenolate (GPX1-Se−) reacts with peroxide substrates to produce selenenic acid (GPX1-SeOH). Subsequently, glutathionylated intermediate (GPX1-Se-S-G) is formed with GSH, and it can react with another GSH molecule to re-generate GPX1-Se− along with GSSG. The H2O2-mediated conversion of Sec to DHA in the GPX1 protein purified from human red blood cells was achieved via the initial oxidation of Sec and then the loss of Se oxide [32, 33]. Because the catalytic intermediate GPX1-SeOH can serve as a source of the conversion of Sec to DHA [32, 33], it may partially explain the baseline of DHA detected in the hepatic GPX1 protein of the WT. Obviously, elevated intracellular superoxide anion in the SOD1−/− mice could further oxidize GPX1-SeOH into GPX1-SeO2H that is a better leaving group for the β-elimination to produce DHA [36]. Consequently, these mice exhibited a greater level of irreversible inactivation of the GPX1 than the WT. Despite fairly sensitive mass spectrometry used for this study, we did not detect any isoforms in the target peptide covering residues 37–52 other than Sec47 and DHA47. We also observed no appearance of the previously-reported modifications of GPX1 such as oxidization of Trp150 and phosphorylation of Tyr96 [26–28, 34, 35]. Seemingly, the conversion of Sec to DHA of the GPX1 protein represents a specific biomarker for in vivo oxidative stress induced by the knockout of SOD1. It would be fascinating to determine if the SOD1 knockout mediates similar modifications of other GPX enzymes. Meanwhile, the MRM-based quantitative MS described herein may be used not only for relative estimate but also for absolute quantification of the DHA isoform in peptides.
Our second most interesting discovery is the association of the Se-deleted modification of Sec with compromised apparent enzyme kinetics and accelerated dissociation of the GPX1 tetramer. Replacements of Sec by cysteine or arginine in selenoenzymes including GPX1, type III iodothyronine deiodinase, and thioredoxin reductase attenuate their activities [25, 31, 44–47]. In the present study, the relative decrease in appkcat (31–38%) of hepatic GPX1 enzyme in the SOD1−/− mice was proportional to the relative loss of Sec (38%) in the peptide. This remarkable match verifies the importance of an intact Sec for catalytic function of GPX1, and implicates that this decreased turnover number was due to the active center loss in a portion of the enzyme protein, rather than a uniform decline in the kinetic constant of the active enzyme per se. Because the GPX1 activity was measured at the H2O2 concentration (120 μM) that was more than 5 times of its appKm, the observed genotype difference in the GPX1 apparent substrate affinity (appKm) was unlikely to account for the ~ 40% loss of activity in the SOD1−/− mice compared with the WT. We found a number of GPX1 bands smaller than the tetramer in size exclusion chromatography fractions only in the SOD1−/− mice. This type of dissociation produced inactive protein fragments around the dimer size, as confirmed by non-denaturing gel electrophoresis and Western blot analysis. Unlike thioredoxin reductase 1 [48], the GPX1 enzyme requires integrity of the homotetramer for its activity. Although there is no clear-cut evidence that the tetramer structure of GPX1 depends on Sec, this residue (at 47) is located in the functional loop (residues 42 to 50) connecting β1 and α1, and is known to be important for the interface formation between the two subunits [49, 50]. Compared with the monomeric or dimeric GPX proteins, GPX1 tetrameric structure is deemed to be stabilized, at least partly by a mixture of specific hydrophobic interactions and hydrogen bonds in the functional loops around the active center [51]. Therefore, it is reasonable for us to postulate that the conversion of Sec to DHA may attenuate the interactions of the tetramers and promote their dissociation, although other unknown factors could not be excluded. In fact, Sec participates in the formation of a selenyl sulfide bridge with Cys91 and hydrogen bond network with its neighboring residues. Because those forces are known to stabilize the protein structure [31, 49], loss of Se in the Sec may destabilize the GPX1 tetramer by destroying or decreasing these forces.
The partial in vivo inactivation of hepatic GPX1 enzyme by the SOD1 knockout via the modification of Sec depicts a unique coordination of redox enzymes in mammals. This down regulation seems to be specific for GPX1 because SOD1 deficiency did not decrease functional expression of other closely-related antioxidant enzymes including catalase, peroredoxin, and another Se-dependent thioredoxin reductase [3, 5, 52]. However, knockout of SOD1 resulted in a 50% decrease in activity of hepatic P450 enzyme 2E1, without impact on its protein level [5, 53]. While P450 2E1 is an enzyme upstream of SOD1 in catalyzing superoxide generation, GPX1 functions downstream of SOD1 in removing its catalysis product of superoxide. Down regulation of both GPX1 and P450 2E1 activities in the SOD1−/− mice may serve as a coordinated adaptation to control intracellular superoxide tone in the absence of SOD1. On the other hand, superoxide generator and hydrogen peroxide, in the presence of SOD1, induce or activate GPX1 [54, 55]. In contrast, a reciprocal response of SOD1 activity to the GPX1 knockout or GPX1 overexpression was not seen [38, 56]. This asymmetric scenario underscores the complexity of redox enzymes and reactive oxygen species in metabolism [57–59].
Acknowledgments
This work was supported in part by National Institutes of Health Grant DK53108 to X.G.L.
Abbreviations
- App
apparent
- DHA
dehydroalanine
- FA
formic acid
- GPX1
glutathione peroxidase-1
- GSH
glutathione
- GSSG
glutathione disulfide
- IDA
information dependent acquisition
- JNK1
c-Jun NH(2)-terminal protein kinase 1
- LC
liquid chromatography
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- Sec
selenocysteine
- SOD1
copper, zinc-superoxide dismutase
- SRM
selected reaction monitoring
- TBHP
tert-butyl peroxide
- WT
wild type
- XIC
extracted ion chromatogram
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, Hoekstra WG. Selenium: biochemical role as a component of glutathione peroxidase. Science. 1973;179:588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- 2.Lei XG, Cheng WH, McClung JP. Metabolic regulation and function of glutathione peroxidase-1. Annu Rev Nutr. 2007;27:41–61. doi: 10.1146/annurev.nutr.27.061406.093716. [DOI] [PubMed] [Google Scholar]
- 3.Iuchi Y, Okada F, Onuma K, Onoda T, Asao H, Kobayashi M, Fujii J. Elevated oxidative stress in erythrocytes due to a SOD1 deficiency causes anaemia and triggers autoantibody production. Biochem J. 2007;402:219–227. doi: 10.1042/BJ20061386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lei XG. In vivo antioxidant role of glutathione peroxidase: evidence from knockout mice. Methods Enzymol. 2002;347:213–225. doi: 10.1016/s0076-6879(02)47021-8. [DOI] [PubMed] [Google Scholar]
- 5.Lei XG, Zhu JH, McClung JP, Aregullin M, Roneker CA. Mice deficient in Cu,Zn- superoxide dismutase are resistant to acetaminophen toxicity. Biochem J. 2006;399:455–461. doi: 10.1042/BJ20060784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy MA, Tsai YH, Reaume A, Bray TM. Cellular response of antioxidant metalloproteins in Cu/Zn SOD transgenic mice exposed to hyperoxia. Am J Physiol Lung Cell Mol Physiol. 2001;281:L172–182. doi: 10.1152/ajplung.2001.281.1.L172. [DOI] [PubMed] [Google Scholar]
- 7.Cheng WH, Ho YS, Valentine BA, Ross DA, Combs GF, Jr, Lei XG. Cellular glutathione peroxidase is the mediator of body selenium to protect against paraquat lethality in transgenic mice. J Nutr. 1998;128:1070–1076. doi: 10.1093/jn/128.7.1070. [DOI] [PubMed] [Google Scholar]
- 8.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 9.Flohe L, Gunzler WA, Schock HH. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32:132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 10.Cheng WH, Ho YS, Ross DA, Han Y, Combs GF, Jr, Lei XG. Overexpression of cellular glutathione peroxidase does not affect expression of plasma glutathione peroxidase or phospholipid hydroperoxide glutathione peroxidase in mice offered diets adequate or deficient in selenium. J Nutr. 1997;127:675–680. doi: 10.1093/jn/127.5.675. [DOI] [PubMed] [Google Scholar]
- 11.Gromer S, Johansson L, Bauer H, Arscott LD, Rauch S, Ballou DP, Williams CH, Jr, Schirmer RH, Arner ES. Active sites of thioredoxin reductases: why selenoproteins? Proc Natl Acad Sci USA. 2003;100:12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. EMBO J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 14.Saedi MS, Smith CG, Frampton J, Chambers I, Harrison PR, Sunde RA. Effect of selenium status on mRNA levels for glutathione peroxidase in rat liver. Biochem Biophys Res Commun. 1988;153:855–861. doi: 10.1016/s0006-291x(88)81174-4. [DOI] [PubMed] [Google Scholar]
- 15.Baker RD, Baker SS, LaRosa K, Whitney C, Newburger PE. Selenium regulation of glutathione peroxidase in human hepatoma cell line Hep3B. Arch Biochem Biophys. 1993;304:53–57. doi: 10.1006/abbi.1993.1320. [DOI] [PubMed] [Google Scholar]
- 16.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, Moriarty PM, Maquat LE. Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J. 2000;19:4734–4744. doi: 10.1093/emboj/19.17.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, Burk RF, Berry MJ, Diamond AM, Lee BJ, Gladyshev VN, Hatfield DL. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burk RF, Hill KE. Regulation of selenoproteins. Annu Rev Nutr. 1993;13:65–81. doi: 10.1146/annurev.nu.13.070193.000433. [DOI] [PubMed] [Google Scholar]
- 20.Cowan DB, Weisel RD, Williams WG, Mickle DA. Identification of oxygen responsive elements in the 5′-flanking region of the human glutathione peroxidase gene. J Biol Chem. 1993;268:26904–26910. [PubMed] [Google Scholar]
- 21.Merante F, Altamentova SM, Mickle DA, Weisel RD, Thatcher BJ, Martin BM, Marshall JG, Tumiati LC, Cowan DB, Li RK. The characterization and purification of a human transcription factor modulating the glutathione peroxidase gene in response to oxygen tension. Mol Cell Biochem. 2002;229:73–83. doi: 10.1023/a:1017921110363. [DOI] [PubMed] [Google Scholar]
- 22.Cowan DB, Weisel RD, Williams WG, Mickle DA. The regulation of glutathione peroxidase gene expression by oxygen tension in cultured human cardiomyocytes. J Mol Cell Cardiol. 1992;24:423–433. doi: 10.1016/0022-2828(92)93196-q. [DOI] [PubMed] [Google Scholar]
- 23.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 24.Dong J, Sulik KK, Chen SY. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: implications for the prevention of fetal alcohol spectrum disorders. Antioxid Redox Signal. 2008;10:2023–2033. doi: 10.1089/ars.2007.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handy DE, Hang G, Scolaro J, Metes N, Razaq N, Yang Y, Loscalzo J. Aminoglycosides decrease glutathione peroxidase-1 activity by interfering with selenocysteine incorporation. J Biol Chem. 2006;281:3382–3388. doi: 10.1074/jbc.M511295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao C, Ren X, Kharbanda S, Koleske A, Prasad KV, Kufe D. The ARG tyrosine kinase interacts with Siva-1 in the apoptotic response to oxidative stress. J Biol Chem. 2001;276:11465–11468. doi: 10.1074/jbc.C100050200. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Majumder P, Shioya H, Wu F, Kumar S, Weichselbaum R, Kharbanda S, Kufe D. Activation of the cytoplasmic c-Abl tyrosine kinase by reactive oxygen species. J Biol Chem. 2000;275:17237–17240. doi: 10.1074/jbc.C000099200. [DOI] [PubMed] [Google Scholar]
- 28.Cao C, Leng Y, Huang W, Liu X, Kufe D. Glutathione peroxidase 1 is regulated by the c- Abl and Arg tyrosine kinases. J Biol Chem. 2003;278:39609–39614. doi: 10.1074/jbc.M305770200. [DOI] [PubMed] [Google Scholar]
- 29.Blum J, Fridovich I. Inactivation of glutathione peroxidase by superoxide radical. Arch Biochem Biophys. 1985;240:500–508. doi: 10.1016/0003-9861(85)90056-6. [DOI] [PubMed] [Google Scholar]
- 30.Pigeolet E, Corbisier P, Houbion A, Lambert D, Michiels C, Raes M, Zachary MD, Remacle J. Glutathione peroxidase, superoxide dismutase, and catalase inactivation by peroxides and oxygen derived free radicals. Mech Ageing Dev. 1990;51:283–297. doi: 10.1016/0047-6374(90)90078-t. [DOI] [PubMed] [Google Scholar]
- 31.Asahi M, Fujii J, Takao T, Kuzuya T, Hori M, Shimonishi Y, Taniguchi N. The oxidation of selenocysteine is involved in the inactivation of glutathione peroxidase by nitric oxide donor. J Biol Chem. 1997;272:19152–19157. doi: 10.1074/jbc.272.31.19152. [DOI] [PubMed] [Google Scholar]
- 32.Rhee SG, Cho CS. Blot-based detection of dehydroalanine-containing glutathione peroxidase with the use of biotin-conjugated cysteamine. Methods Enzymol. 2010;474:23–34. doi: 10.1016/S0076-6879(10)74002-7. [DOI] [PubMed] [Google Scholar]
- 33.Cho CS, Lee S, Lee GT, Woo HA, Choi EJ, Rhee SG. Irreversible inactivation of glutathione peroxidase 1 and reversible inactivation of peroxiredoxin II by H2O2 in red blood cells. Antioxid Redox Signal. 2010;12:1235–1246. doi: 10.1089/ars.2009.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai J, Jin WH, Sheng QH, Shieh CH, Wu JR, Zeng R. Protein phosphorylation and expression profiling by Yin-yang multidimensional liquid chromatography (Yin-yang MDLC) mass spectrometry. J Proteome Res. 2007;6:250–262. doi: 10.1021/pr0604155. [DOI] [PubMed] [Google Scholar]
- 35.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Vatamaniuk M, Roneker C, Pepper MP, Hu L, Simmons R, Lei X. Knockouts of SOD1 and GPX1 exert different impacts on murine islet function and pancreatic integrity. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho YS, Gargano M, Cao J, Bronson RT, Heimler I, Hutz RJ. Reduced fertility in female mice lacking copper-zinc superoxide dismutase. J Biol Chem. 1998;273:7765–7769. doi: 10.1074/jbc.273.13.7765. [DOI] [PubMed] [Google Scholar]
- 38.McClung JP, Roneker CA, Mu W, Lisk DJ, Langlais P, Liu F, Lei XG. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc Natl Acad Sci USA. 2004;101:8852–8857. doi: 10.1073/pnas.0308096101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flohe L, Loschen G, Gunzler WA, Eichele E. Glutathione peroxidase, V. The kinetic mechanism. Hoppe Seylers Z Physiol Chem. 1972;353:987–999. doi: 10.1515/bchm2.1972.353.1.987. [DOI] [PubMed] [Google Scholar]
- 40.Ursini F, Maiorino M, Gregolin C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim Biophys Acta. 1985;839:62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- 41.Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein P. J Biol Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- 42.Zhu JH, Zhang X, Roneker CA, McClung JP, Zhang S, Thannhauser TW, Ripoll DR, Sun Q, Lei XG. Role of copper,zinc-superoxide dismutase in catalyzing nitrotyrosine formation in murine liver. Free Radic Biol Med. 2008;45:611–618. doi: 10.1016/j.freeradbiomed.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, Zinder SH, Richardson RE. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl Environ Microbiol. 2007;73:320–326. doi: 10.1128/AEM.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuiper GG, Klootwijk W, Visser TJ. Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase reduces catalytic efficiency and alters substrate preference. Endocrinology. 2003;144:2505–2513. doi: 10.1210/en.2003-0084. [DOI] [PubMed] [Google Scholar]
- 45.Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 46.Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci USA. 2000;97:2521–2526. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian t hioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci USA. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rengby O, Cheng Q, Vahter M, Jornvall H, Arner ES. Highly active dimeric and low- activity tetrameric forms of selenium-containing rat thioredoxin reductase 1. Free Radic Biol Med. 2009;46:893–904. doi: 10.1016/j.freeradbiomed.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Ren B, Huang W, Akesson B, Ladenstein R. The crystal structure of seleno-glutathione peroxidase from human plasma at 2.9 A resolution. J Mol Biol. 1997;268:869–885. doi: 10.1006/jmbi.1997.1005. [DOI] [PubMed] [Google Scholar]
- 50.Ren B, Huang W, Akesson B, Ladenstien R. Crystallization and preliminary X-ray diffraction analysis of glutathione peroxidase from human plasma. Acta Crystallogr D Biol Crystallogr. 1995;51:824–826. doi: 10.1107/S0907444994011765. [DOI] [PubMed] [Google Scholar]
- 51.Toppo S, Vanin S, Bosello V, Tosatto SC. Evolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid Redox Signal. 2008;10:1501–1514. doi: 10.1089/ars.2008.2057. [DOI] [PubMed] [Google Scholar]
- 52.Sentman ML, Granstrom M, Jakobson H, Reaume A, Basu S, Marklund SL. Phenotypes of mice lacking extracellular superoxide dismutase and copper- and zinc- containing superoxide dismutase. J Biol Chem. 2006;281:6904–6909. doi: 10.1074/jbc.M510764200. [DOI] [PubMed] [Google Scholar]
- 53.Zhu JH, McClung JP, Zhang X, Aregullin M, Chen C, Gonzalez FJ, Kim TW, Lei XG. Comparative impacts of knockouts of two antioxidant enzymes on acetaminophen-induced hepatotoxicity in mice. Exp Biol Med (Maywood) 2009;234:1477–1483. doi: 10.3181/0904-RM-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhee SG, Yang KS, Kang SW, Woo HA, Chang TS. Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post- translational modification. Antioxid Redox Signal. 2005;7:619–626. doi: 10.1089/ars.2005.7.619. [DOI] [PubMed] [Google Scholar]
- 55.de Haan JB, Bladier C, Griffiths P, Kelner M, O’Shea RD, Cheung NS, Bronson RT, Silvestro MJ, Wild S, Zheng SS, Beart PM, Hertzog PJ, Kola I. Mice with a homozygous null mutation for the most abundant glutathione peroxidase, Gpx1, show increased susceptibility to the oxidative stress-inducing agents paraquat and hydrogen peroxide. J Biol Chem. 1998;273:22528–22536. doi: 10.1074/jbc.273.35.22528. [DOI] [PubMed] [Google Scholar]
- 56.Cheng W, Fu YX, Porres JM, Ross DA, Lei XG. Selenium-dependent cellular glutathione peroxidase protects mice against a pro-oxidant-induced oxidation of NADPH, NADH, lipids, and protein. FASEB J. 1999;13:1467–1475. doi: 10.1096/fasebj.13.11.1467. [DOI] [PubMed] [Google Scholar]
- 57.Imamura Y, Noda S, Hashizume K, Shinoda K, Yamaguchi M, Uchiyama S, Shimizu T, Mizushima Y, Shirasawa T, Tsubota K. Drusen, choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: a model of age-related macular degeneration. Proc Natl Acad Sci USA. 2006;103:11282–11287. doi: 10.1073/pnas.0602131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han ES, Muller FL, Perez VI, Qi W, Liang H, Xi L, Fu C, Doyle E, Hickey M, Cornell J, Epstein CJ, Roberts LJ, Van Remmen H, Richardson A. The in vivo gene expression signature of oxidative stress. Physiol Genomics. 2008;34:112–126. doi: 10.1152/physiolgenomics.00239.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blander G, de Oliveira RM, Conboy CM, Haigis M, Guarente L. Superoxide dismutase 1 knock-down induces senescence in human fibroblasts. J Biol Chem. 2003;278:38966–38969. doi: 10.1074/jbc.M307146200. [DOI] [PubMed] [Google Scholar]