Abstract
Hammerhead ribozymes previously were found in satellite RNAs from plant viroids and in repetitive DNA from certain species of newts and schistosomes. To determine if this catalytic RNA motif has a wider distribution, we decided to scrutinize the GenBank database for RNAs that contain hammerhead or hammerhead-like motifs. The search shows a widespread distribution of this kind of RNA motif in different sequences suggesting that they might have a more general role in RNA biology. The frequency of the hammerhead motif is half of that expected from a random distribution, but this fact comes from the low CpG representation in vertebrate sequences and the bias of the GenBank for those sequences. Intriguing motifs include those found in several families of repetitive sequences, in the satellite RNA from the carrot red leaf luteovirus, in plant viruses like the spinach latent virus and the elm mottle virus, in animal viruses like the hepatitis E virus and the caprine encephalitis virus, and in mRNAs such as those coding for cytochrome P450 oxidoreductase in the rat and the hamster.
The hammerhead ribozyme originally was discovered as a self-cleaving motif in viroids and satellite RNAs. These RNAs replicate using the rolling circle mechanism, which generates long multimeric replication intermediates. They use the cleavage reaction to resolve the multimeric intermediates into monomeric forms. The region able to self-cleave has three base paired helices (I–III) connected by two conserved single stranded regions and a bulged nucleotide (Forster and Symons 1987; for reviews see Symons 1992; Bratty et al. 1993; Birikh et al. 1997). The hammerhead ribozyme also seems to function in the generation of unit length sequences from multimeric transcripts of repetitive DNA sequences. Two of these RNAs have been characterized: one in several newt species (Epstein and Gall 1987) and the other one in three Schistosome species (Ferbeyre et al. 1998). Among the repetitive sequences of these two organisms, note that not all contained a bona fide hammerhead ribozyme. Indeed, many mutations also were found creating variants of the original motif. Overall, the rather limited distribution of this motif contrast with the simplicity of its secondary structure in which only a core of 14 nucleotides is absolutely required for cleavage.
We recently have conducted an extensive research of different RNA motifs in the GeneBank database (Bourdeau et al. 1999). The results showed that most of the motifs were distributed randomly among gene sequences suggesting that most RNA motifs originate by random drift. We now wish to extend these observations to the self-cleaving hammerhead ribozyme and its variants in which either an essential nucleotide in the single strand positions is allowed to be random or the identity of a conserved base pair from helices II and III is changed. We found that most of the hammerhead motifs are apparently underrepresented among gene sequences, but this comes from the bias of the GenBank for sequences with low CpG representation. We also report the finding of intriguing motifs in several repetitive sequences and mRNAs.
RESULTS
Searching for Self-cleaving RNA Motifs of the Hammerhead Type in the GenBank
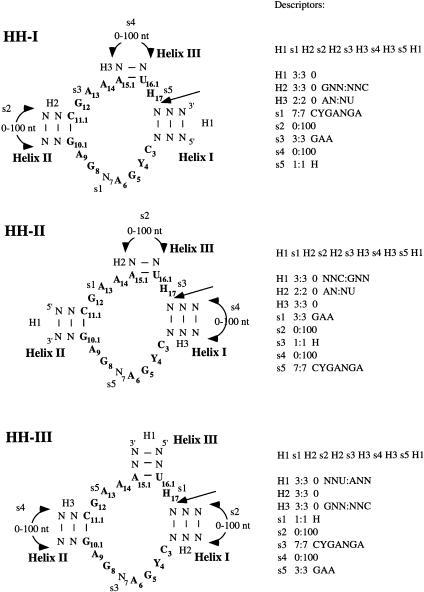
The hammerhead ribozyme can be described by three helices separated by three single stranded regions of conserved nucleotides. There are three equivalent conformations of the self-cleaving hammerhead depending on which helix bears the 5′ and 3′ end of the motif. We named them HH-I, HH-II, and HH-III (Figure 1). The descriptors composed as input for the search program are presented beside each motif and described in the legend of Figure 1 (see also Methods). They were designed to detect any sequence with all the minimal nucleotide requirements to have some catalytic activity and with the possibility to fold like the hammerhead. In this context, it is expected that sequences will be found that combine several nonoptimum features and be inactive for this reason, i.e., a non-GUC cleavage, a C in position 4, short helices, and long loops. It is also possible that they contain all the requirements for being catalytically active but the active conformation is inaccessible because the RNA molecule that bears them folds into an alternative secondary structure.
Figure 1.
Structures and descriptors of the hammerhead self-cleaving ribozyme motifs. The three descriptors, HH-I, HH-II, and HH-III, are defined by which helix is at the 5′ end and named according to the helix number (Hertel et al. 1992). Each descriptor is composed of single stranded (s) and double stranded (H) regions. The regions first are named in order from 5′ to 3′ and then specified for their length (minimum:maximum), number of mismatches (in the case of H only), and presence of specific nucleotides. For example, HH-I consists of the following features: H1 s1 H2 s2 H2 s3 H3 s4 H3 s5 H1 where H1 is an helix of a fixed length of three base pairs with no mismatches and no specific nucleotides; H2 is also of three base pairs with no mismatches but with a starting G-C base pair; H3 is an helix of 2 base pairs beginning with an A-U base pair; s1 is a single stranded region of seven nucleotides exactly with a specific sequence; s2 varies between 0 and 100 undetermined nucleotides; and so on. The hammerhead-like motifs are the same as the three shown but with an “N” replacing one of the nucleotides in boldface or with a different identity of one of the base pairs in boldface. These motifs are named according to the original motif and the position of the mutation, e.g., HH-I-3 motif is as HH-I but with an N instead of a C at position 3; thus, HH-I-3 descriptor has a modified s1 as follows: s1 7:7 NYGANGA, similarly with HH-I-iiAU, which is a HH-I motif with a A:U base pair in the Helix II instead of a G:C; thus, the descriptor HH-I-iiAU has this particularity: H2 3:3 0 ANN:NNU. The cleaving site is after H17. (H) A, C, or U; (N) A, C, G, or U; (Y) C or U. See Methods for the basis of the sequence requirements.
The search for hammerhead self-cleaving motifs through the GenBank database (Benson et al. 1999) was performed using the program RNAMOT (Gautheret et al. 1990; Laferrière et al. 1994). The sequences detected with our descriptors are referred to as occurrences. The ability of the descriptors to identify the hammerhead motifs already characterized is illustrated in Table 1. The program recognizes most of the known plant derived hammerheads (Symons 1997; see also http://callisto.si.usherb.ca/∼jpperra/organisms.html; Bussière et al. 1996; Lafontaine et al. 1999) and all those present in satellite DNA sequences. Note that there is no known natural incidence of a hammerhead of the HH-II type.
Table 1.
Known Hammerhead Motifs Identified in Our Search
| HH-I | HH-III |
|---|---|
| Avocado sunblotch viroid | Schistosoma mansoni DNA for repeated sequences |
| Tobacco Ringspot virus satellite RNA | Barley yellow virus satellite RNA |
| Ambyostoma talpoideum satellite 2 | Chrysanthemum chlorotic mottle viroid |
| Cryptobranchus alleganiensis satellite 2 | Cherry small circular viroid-like RNA |
| Cyrrops pyrrhogaster satellite 2 | Lucerne transient streak virus RNA 2 |
| Eurycea longicauda satellite 2 | Peach latent mosaic viroid |
| Plethodon glutinosus satellite 2 | Subterranean clover mottle virus satellite RNA |
Table 2 presents the frequencies of occurrences of potential hammerhead motifs in the different sections of the GenBank as well as the expected frequencies calculated from the number of occurrences obtained in a database of random sequences. In general, the number of occurrences observed are half of the frequency expected if our motifs were randomly distributed among the sequences of the GenBank. HH-I and HH-II detect twice as many motifs as HH-III because we designed the motifs in a way that Helix III had a 2–base pair requirement in HH-I and HH-II descriptors versus 3 base pairs in the HH-III descriptor (see Methods). This increase was predicted by the number of occurrences obtained in the random database.
Table 2.
Distribution of Hammerhead and Hammerhead-like Motifs in the Different Sections of the GenBank: Mutants of the Single Stranded Regions
| pri | rod | mam | vrt | inv | pln | bct | rna | vrl | phg | syn | una | est | pat | sts | gss | htg | Total | Total over expected | Expected (A = C = G = T) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HH-I | 51 | 21 | 0 | 14 | 176 | 119 | 130 | 0 | 37 | 3 | 5 | 0 | 186 | 45 | 4 | 17 | 108 | 916 | 0.56 | 1635 |
| HH-I-3 | 182 | 72 | 12 | 45 | 997 | 560 | 553 | 2 | 159 | 16 | 6 | 0 | 702 | 125 | 10 | 119 | 486 | 4046 | 0.57 | 7081 |
| HH-I-4 | 270 | 29 | 0 | 32 | 365 | 228 | 226 | 10 | 69 | 3 | 8 | 1 | 358 | 55 | 4 | 33 | 272 | 1963 | 0.55 | 3576 |
| HH-I-5 | 310 | 70 | 14 | 97 | 506 | 400 | 309 | 2 | 77 | 10 | 7 | 1 | 641 | 60 | 13 | 92 | 382 | 2991 | 0.46 | 6488 |
| HH-I-6 | 197 | 60 | 17 | 37 | 516 | 312 | 399 | 3 | 107 | 5 | 8 | 1 | 700 | 93 | 11 | 67 | 288 | 2821 | 0.49 | 6973 |
| HH-I-8 | 171 | 44 | 11 | 22 | 790 | 407 | 455 | 10 | 138 | 11 | 5 | 0 | 521 | 86 | 10 | 61 | 420 | 3162 | 0.46 | 6883 |
| HH-I-9 | 247 | 56 | 12 | 29 | 508 | 260 | 454 | 0 | 107 | 9 | 7 | 3 | 536 | 99 | 9 | 65 | 344 | 2745 | 0.38 | 7314 |
| HH-I-12 | 946 | 224 | 41 | 103 | 754 | 613 | 454 | 0 | 180 | 13 | 8 | 2 | 1856 | 137 | 48 | 263 | 794 | 6436 | 0.94 | 6865 |
| HH-I-13 | 209 | 76 | 16 | 41 | 428 | 308 | 462 | 5 | 93 | 10 | 11 | 0 | 739 | 143 | 12 | 66 | 226 | 2845 | 0.42 | 6775 |
| HH-I-14 | 317 | 94 | 30 | 36 | 404 | 377 | 487 | 7 | 104 | 5 | 30 | 5 | 737 | 102 | 8 | 77 | 327 | 3147 | 0.50 | 6362 |
| HH-I-17 | 65 | 25 | 0 | 18 | 201 | 144 | 162 | 0 | 50 | 4 | 11 | 0 | 225 | 53 | 4 | 19 | 122 | 1103 | 0.l54 | 2031 |
| HH-II | 83 | 33 | 1 | 5 | 175 | 126 | 141 | 1 | 80 | 1 | 1 | 3 | 435 | 30 | 4 | 8 | 91 | 1218 | 0.54 | 2246 |
| HH-II-3 | 180 | 78 | 8 | 29 | 994 | 645 | 715 | 3 | 204 | 14 | 25 | 23 | 1022 | 102 | 11 | 82 | 502 | 4637 | 0.66 | 7063 |
| HH-II-4 | 133 | 56 | 7 | 11 | 387 | 261 | 278 | 1 | 100 | 1 | 2 | 5 | 658 | 43 | 7 | 39 | 188 | 2177 | 0.51 | 4277 |
| HH-II-5 | 244 | 82 | 16 | 37 | 573 | 477 | 372 | 7 | 141 | 10 | 4 | 4 | 910 | 70 | 12 | 71 | 362 | 3392 | 0.39 | 8788 |
| HH-II-6 | 234 | 81 | 23 | 17 | 522 | 348 | 977 | 38 | 153 | 7 | 7 | 9 | 856 | 74 | 8 | 59 | 311 | 3724 | 0.49 | 7674 |
| HH-II-8 | 209 | 64 | 11 | 22 | 921 | 385 | 519 | 16 | 130 | 3 | 10 | 4 | 1265 | 53 | 7 | 65 | 452 | 4136 | 0.54 | 7710 |
| HH-II-9 | 255 | 58 | 8 | 24 | 457 | 319 | 540 | 6 | 140 | 9 | 11 | 5 | 884 | 82 | 10 | 51 | 281 | 3140 | 0.39 | 7979 |
| HH-II-12 | 1290 | 253 | 61 | 60 | 879 | 716 | 534 | 1 | 209 | 14 | 18 | 3 | 2491 | 125 | 56 | 273 | 1027 | 8010 | 0.97 | 8285 |
| HH-II-13 | 214 | 91 | 45 | 26 | 421 | 326 | 534 | 7 | 162 | 8 | 23 | 3 | 1031 | 71 | 7 | 53 | 228 | 3250 | 0.44 | 7404 |
| HH-II-14 | 281 | 81 | 15 | 37 | 414 | 524 | 493 | 6 | 142 | 7 | 2 | 4 | 1038 | 87 | 9 | 67 | 256 | 3463 | 0.43 | 8069 |
| HH-II-17 | 100 | 35 | 1 | 7 | 220 | 152 | 173 | 1 | 97 | 1 | 1 | 4 | 504 | 36 | 7 | 16 | 108 | 1463 | 0.53 | 2786 |
| HH-III | 42 | 6 | 0 | 2 | 93 | 67 | 65 | 0 | 21 | 1 | 2 | 0 | 96 | 57 | 3 | 8 | 64 | 527 | 0.55 | 952 |
| HH-III-3 | 96 | 32 | 8 | 18 | 625 | 305 | 245 | 0 | 74 | 6 | 3 | 0 | 310 | 72 | 5 | 74 | 291 | 2164 | 0.64 | 3397 |
| HH-III-4 | 295 | 12 | 1 | 21 | 192 | 128 | 98 | 0 | 36 | 3 | 5 | 0 | 204 | 63 | 3 | 23 | 229 | 1313 | 0.76 | 1725 |
| HH-III-5 | 144 | 38 | 16 | 35 | 301 | 205 | 231 | 0 | 34 | 2 | 2 | 0 | 388 | 76 | 11 | 32 | 219 | 1734 | 0.48 | 3612 |
| HH-III-6 | 134 | 29 | 5 | 18 | 290 | 183 | 197 | 9 | 58 | 2 | 3 | 0 | 426 | 66 | 10 | 33 | 180 | 1643 | 0.46 | 3540 |
| HH-III-8 | 92 | 26 | 4 | 6 | 528 | 203 | 260 | 0 | 722 | 1 | 2 | 0 | 294 | 154 | 4 | 29 | 255 | 2580 | 0.68 | 3774 |
| HH-III-9 | 109 | 22 | 0 | 12 | 239 | 143 | 196 | 0 | 87 | 3 | 4 | 0 | 462 | 63 | 7 | 38 | 164 | 1549 | 0.49 | 3918 |
| HH-III-12 | 658 | 97 | 40 | 32 | 369 | 318 | 217 | 0 | 98 | 3 | 6 | 0 | 1071 | 135 | 20 | 121 | 477 | 3662 | 1.03 | 3540 |
| HH-III-13 | 119 | 29 | 8 | 14 | 226 | 176 | 246 | 3 | 53 | 6 | 2 | 0 | 345 | 70 | 6 | 28 | 142 | 1473 | 0.41 | 3576 |
| HH-III-14 | 133 | 33 | 10 | 11 | 242 | 184 | 195 | 1 | 65 | 2 | 21 | 0 | 408 | 86 | 5 | 39 | 146 | 1581 | 0.45 | 3504 |
| HH-III-17 | 49 | 6 | 0 | 3 | 123 | 81 | 86 | 0 | 23 | 2 | 2 | 0 | 154 | 67 | 4 | 11 | 74 | 685 | 0.49 | 1402 |
| GenBank relative size | 0.14 | 0.03 | 0.01 | 0.01 | 0.07 | 0.07 | 0.07 | <0.01 | 0.03 | <0.01 | <0.01 | <0.01 | 0.37 | 0.02 | 0.01 | 0.06 | 0.10 | 1.00 |
The motifs are named as explained in Methods and in Figure 1. The “expected” number of occurrences was obtained by searching the different motifs in a database of 1000 random sequences of 100,000 nucleotides (equal representations of A, C, G, and T) and correcting the frequency to the relative size of the GenBank. The “total over expected” shows the ratio of occurrences obtained in the GenBank versus the expected ones according to the search performed in a random database with the size of the GenBank.
(pri) Primate sequence entries (from the two GenBank files); (rod) rodent sequence entries; (mam) other mammalian sequence entries; (vrt) other vertebrate sequence entries; (inv) invertebrate sequence entries; (pln) plant sequence entries (including fungi and algae); (bct) bacterial sequence entries; (rna) structural RNA sequence entries; (vrl) viral sequence entries; (phg) phage sequence entries; (syn) synthetic and chimeric sequence entries; (una) unannotated sequence entries; (est) EST (expressed sequence tag) (from 23 GenBank files); (pat) patent sequence entries; (sts) STS (sequence tagged site) sequence entries; (gss) GSS (genome survery sequence) sequence entries; (htg) HTGS (high throughput genomic sequencing) sequence entries.
The Frequency of Mutated Versions of the Hammerhead Self-cleaving RNAs
We also composed descriptors for variants of the hammerhead ribozyme motif. Substitutions were made by replacing, one at a time, each of the essential nucleotides located in the single stranded regions of the ribozyme core by N (boldface in Fig. 1) or by changing the identity of each one of the 2 conserved base pairs of the hammerhead motif (also boldface in Fig. 1).
Table 2 presents the data on the distribution of the mutated variants of HH-I, HH-II, and HH-III from the single stranded region. It is expected that every mutant will increase the frequency of occurrences by a factor of four because we changed the requirements in every position from only one to all four nucleotides except in position 4 where C and U already were allowed and in the cleavage site where only G originally was excluded. Thus, in position 4 we expected to double the frequency, and in the cleavage site we expected a 25% increase. The results are mostly those anticipated based on these calculations. However the mutants of position 12 doubled the expected increase in all the orientations. This effect was not uniformly observed in the different subdivisions of the GenBank. Actually, most of the extra occurrences are located in the files containing ESTs (Expressed Sequence Tags) and mammalian sequences. These preferences were not observed in the random database in which the mutants showed the anticipated increase in their frequency in comparison with the original motif. The number of occurrences obtained in the virus section of the GenBank for the HH-III-8 variant was 722 instead of the 113 expected (HH-III has 3774 expected occurrences and viruses represent 3% of the GenBank). However, a quick analysis of the occurrences obtained with this descriptor revealed that most of them are the same motif repeated in 679 hepatitis C sequences.
Table 3 presents the frequencies obtained with the mutant hammerhead ribozymes using a different identity for the conserved base pair of helices II or III (positions 10.1:11.1 and 15.1:16.1, respectively). One striking observation is that all the mutants in Helix II (iiNN) have total occurrences two to six times higher than expected whereas the mutants in Helix III (iiiNN) have half the expected frequency. One more interesting point is the high number of occurrences obtained with the three orientations of the hammerhead ribozyme having a A:U base pair in Helix II (10.1:11.1) instead of the usual G:C.
Table 3.
Distribution of Hammerhead and Hammerhead-like Motifs in the Different Sections of the GenBank: Mutants of Helices II and III
| pri | rod | mam | vrt | inv | pln | bct | rna | vrl | phg | syn | una | est | pat | sts | gss | htg | Total | Total over expected | Expected (A = C = G = T) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HH-I | 51 | 21 | 0 | 14 | 176 | 119 | 130 | 0 | 37 | 3 | 5 | 0 | 186 | 45 | 4 | 17 | 108 | 916 | 0.56 | 1635 |
| HH-I-iiAU | 569 | 100 | 30 | 36 | 822 | 545 | 426 | 3 | 58 | 7 | 8 | 0 | 1113 | 93 | 32 | 341 | 544 | 4727 | 2.56 | 1851 |
| HH-I-iiCG | 343 | 59 | 12 | 20 | 203 | 117 | 512 | 11 | 24 | 2 | 1 | 11 | 669 | 69 | 15 | 66 | 221 | 2355 | 1.27 | 1851 |
| HH-I-iiGU | 452 | 65 | 9 | 38 | 361 | 227 | 203 | 0 | 73 | 3 | 49 | 2 | 794 | 58 | 15 | 93 | 332 | 2774 | 1.77 | 1564 |
| HH-I-iiUA | 606 | 124 | 21 | 82 | 406 | 364 | 260 | 2 | 165 | 10 | 2 | 0 | 941 | 43 | 15 | 175 | 436 | 3652 | 2.03 | 1797 |
| HH-I-iiUG | 455 | 69 | 25 | 38 | 354 | 268 | 287 | 4 | 57 | 6 | 24 | 2 | 664 | 47 | 9 | 92 | 268 | 2669 | 1.43 | 1869 |
| HH-I-iiiCG | 51 | 19 | 5 | 6 | 91 | 63 | 163 | 0 | 43 | 2 | 2 | 0 | 355 | 77 | 1 | 10 | 27 | 915 | 0.48 | 1887 |
| HH-I-iiiGC | 121 | 53 | 13 | 13 | 103 | 116 | 189 | 1 | 48 | 1 | 6 | 0 | 494 | 60 | 2 | 28 | 52 | 1300 | 0.74 | 1761 |
| HH-I-iiiGU | 77 | 31 | 18 | 12 | 98 | 116 | 130 | 0 | 21 | 0 | 0 | 1 | 284 | 44 | 1 | 17 | 64 | 914 | 0.58 | 1564 |
| HH-I-iiiUA | 52 | 23 | 2 | 8 | 118 | 89 | 79 | 2 | 13 | 0 | 8 | 0 | 177 | 28 | 2 | 18 | 38 | 657 | 0.39 | 1707 |
| HH-I-iiiUG | 36 | 19 | 4 | 6 | 100 | 63 | 155 | 6 | 21 | 2 | 152 | 0 | 154 | 73 | 1 | 19 | 90 | 901 | 0.50 | 1797 |
| HH-II | 83 | 33 | 1 | 5 | 175 | 126 | 141 | 1 | 80 | 1 | 1 | 3 | 435 | 30 | 4 | 8 | 91 | 1218 | 0.54 | 2246 |
| HH-II-iiAU | 719 | 117 | 40 | 39 | 860 | 572 | 453 | 0 | 118 | 13 | 2 | 0 | 1021 | 87 | 24 | 190 | 663 | 4918 | 2.76 | 1779 |
| HH-II-iiCG | 404 | 97 | 21 | 25 | 183 | 113 | 148 | 1 | 50 | 1 | 7 | 0 | 856 | 38 | 20 | 83 | 284 | 2331 | 1.34 | 1743 |
| HH-II-iiGU | 560 | 96 | 36 | 39 | 352 | 270 | 237 | 1 | 192 | 5 | 1 | 0 | 783 | 35 | 15 | 124 | 367 | 3113 | 1.77 | 1761 |
| HH-II-iiUA | 668 | 126 | 133 | 39 | 465 | 382 | 308 | 4 | 85 | 10 | 22 | 1 | 843 | 66 | 21 | 248 | 462 | 3883 | 2.12 | 1833 |
| HH-II-iiUG | 694 | 123 | 36 | 24 | 335 | 239 | 260 | 1 | 71 | 0 | 5 | 0 | 749 | 67 | 17 | 212 | 319 | 3152 | 1.81 | 1743 |
| HH-II-iiiCG | 65 | 28 | 3 | 14 | 96 | 45 | 215 | 2 | 15 | 4 | 1 | 0 | 159 | 10 | 1 | 20 | 49 | 727 | 0.40 | 1833 |
| HH-II-iiiGC | 136 | 67 | 16 | 21 | 92 | 103 | 232 | 0 | 20 | 7 | 1 | 2 | 498 | 51 | 2 | 19 | 72 | 1339 | 0.71 | 1887 |
| HH-II-iiiGU | 69 | 42 | 3 | 12 | 101 | 88 | 182 | 0 | 33 | 6 | 2 | 0 | 365 | 30 | 1 | 19 | 65 | 1018 | 0.48 | 2103 |
| HH-II-iiiUA | 60 | 16 | 3 | 11 | 107 | 78 | 102 | 1 | 27 | 0 | 0 | 0 | 136 | 8 | 1 | 21 | 65 | 636 | 0.35 | 1833 |
| HH-II-iiiUG | 50 | 22 | 4 | 27 | 85 | 77 | 129 | 0 | 39 | 0 | 0 | 0 | 154 | 23 | 0 | 17 | 63 | 690 | 0.30 | 2282 |
| HH-III | 42 | 6 | 0 | 2 | 93 | 67 | 65 | 0 | 21 | 1 | 2 | 0 | 96 | 57 | 3 | 8 | 64 | 527 | 0.55 | 952 |
| HH-III-iiAU | 360 | 59 | 17 | 11 | 482 | 324 | 241 | 0 | 35 | 7 | 11 | 1 | 723 | 31 | 6 | 354 | 328 | 2990 | 5.05 | 593 |
| HH-III-iiCG | 203 | 37 | 4 | 9 | 98 | 58 | 101 | 0 | 9 | 0 | 0 | 0 | 469 | 8 | 6 | 42 | 132 | 1176 | 1.17 | 1006 |
| HH-III-iiGU | 251 | 29 | 3 | 19 | 194 | 126 | 121 | 0 | 43 | 4 | 0 | 0 | 440 | 40 | 13 | 52 | 217 | 1552 | 1.73 | 899 |
| HH-III-iiUA | 333 | 48 | 10 | 23 | 246 | 224 | 153 | 13 | 39 | 1 | 0 | 0 | 486 | 22 | 5 | 70 | 257 | 1930 | 2.24 | 863 |
| HH-III-iiUG | 285 | 56 | 11 | 31 | 212 | 139 | 113 | 0 | 29 | 0 | 1 | 0 | 399 | 27 | 8 | 63 | 148 | 1522 | 2.02 | 755 |
| HH-III-iiiCG | 23 | 9 | 2 | 3 | 45 | 22 | 108 | 0 | 8 | 1 | 0 | 0 | 158 | 52 | 1 | 7 | 19 | 458 | 0.50 | 917 |
| HH-III-iiiGC | 78 | 14 | 24 | 15 | 38 | 66 | 100 | 0 | 12 | 2 | 1 | 0 | 173 | 27 | 2 | 18 | 38 | 608 | 0.57 | 1060 |
| HH-III-iiiGU | 52 | 18 | 10 | 13 | 47 | 51 | 83 | 1 | 18 | 1 | 11 | 1 | 235 | 7 | 4 | 9 | 43 | 604 | 0.53 | 1150 |
| HH-III-iiiUA | 43 | 15 | 7 | 3 | 72 | 64 | 137 | 2 | 8 | 3 | 304 | 0 | 98 | 133 | 3 | 18 | 126 | 1036 | 1.56 | 665 |
| HH-III-iiiUG | 26 | 17 | 6 | 5 | 40 | 33 | 82 | 3 | 17 | 1 | 152 | 0 | 97 | 64 | 4 | 8 | 58 | 613 | 0.67 | 917 |
| GenBank relative size | 0.14 | 0.03 | 0.01 | 0.01 | 0.07 | 0.07 | 0.07 | <0.01 | 0.03 | <0.01 | <0.01 | <0.01 | 0.37 | 0.02 | 0.01 | 0.06 | 0.10 | 1.00 |
See legend of Table 2 for explanation of abbreviations.
The mutants in position 12 and the mutants of the conserved base pair of Helix II have in common that they disrupt the presence of a dinucleotide CpG in the resulting sequence. It is well known that CpG is underrepresented in vertebrate sequences (Karlin and Mrazek 1997). The GenBank is biased for those sequences mainly owing to human and rodent entries. In those files, the mutants that disrupt the CpG requirement have a higher frequency. To confirm that the overall frequency of the hammerhead motifs containing CpG dinucleotides is half of the expected one because of the low CpG content of vertebrate sequences, we built a new random database in which the frequency of CpG was reduced by half in favor of either AG, CA, CC, CT, GG, or TG to simulate the frequencies observed by Karlin and Mrazek (1997; see Methods). In this database, we observed an overall doubling of the original expected frequencies for all the motifs needing a CpG but not for the others (data not shown).
Still, the mutants with a A:U base pair in position 10.1:11.1 of the Helix II have a very high frequency in all three conformations of the motif: two to three times higher than expected even considering the CpG effect discussed above. So far, we have no explanation for this intriguing observation.
Finally, we made three more searches by changing the cleavage site from NUH to NHH based on the report of Kore et al. (1998) that such hammerheads were still active. We obtained for these new mutants a number of occurrences corresponding to half of what we expected according to the search in an equal A-C-G-T random database. Moreover, as for the previous motifs, the number of occurrences in the GenBank is comparable to the expected frequency according to the search in the reduced for CpG database. All the occurrences found in the GenBank are available in our web site at http://www.centrcn.umontreal.ca/∼bourdeav/HH.
Some Intriguing Hammerhead Motifs that Might Have Functional Significance
This section presents a sample of motifs considered interesting either because of their location or because their structure is optimal for self cleavage. The hammerhead ribozyme occurs naturally in satellite RNAs, viroids, and transcripts from repetitive sequences. The probability of finding an active hammerhead should be higher among these genetic elements. Several potential hammerhead motifs were found in distinct families of repetitive DNA.
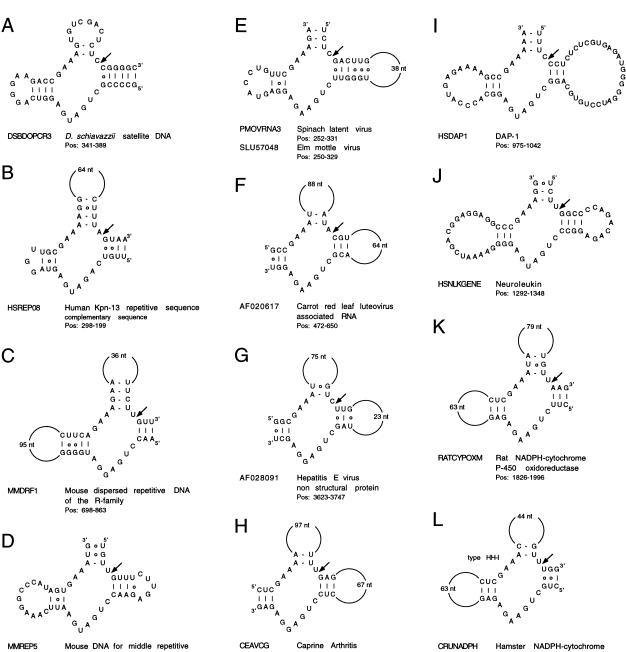
Hammerhead ribozymes were found in the satellite DNA from Dolichopoda schiavazzii (cricket) by using the HH-I descriptor (example in Figure 2A). Fourteen have a conserved HH-I motif and two have a HH-I-iiGU motif (G:U in position 10.1:11.1 instead of G:C). This ribozyme cleaves after CUA (A.A. Rojas, A. Vazques-Tello, G. Ferbeyre, F. Venanzetti, L. Bachmann, B. Paquin, and R. Cedergren, in prep.). Helix I has the GG:CC base pairs and the internal loop common to the hammerhead motifs in schistosomes (Ferbeyre et al. 1998) and newts (Pabon-Peña et al. 1991). It is noteworthy that among the 20 similar sequences submitted to GenBank, the four sequences not found through the search contained either mismatches in one of the helices or combined two point mutations.
Figure 2.
A–L show putative hammerhead motifs.
A hammerhead-like motif was detected in the Kpn-13 family of human repetitive DNA by using the descriptor HH-I-4 (Fig. 2B). The motif is found in several ESTs containing Kpn-repetitive sequences (also known as L1-repetitive elements) indicating its expression at the RNA level. All the occurrences contain a disabling A at position 4, but one (AA564135) possesses a C. The latter motif is inactivated by a G per A substitution at position 12. Variants of this motif also are found in genomic clones containing Kpn repetitive sequences. Intriguingly, the L1 motif interrupting the dystrophin gene of a muscular dystrophy patient (accession number HSU09115) also has a disruption in Helix I. Four additional hammerhead-like motifs were found in the satellite DNA array from the rodent Microtus chrotorrhinus (accession number MICSATB, position 921–1079, not shown), in the repetitive DNA from the protozoan parasite Theileria parva (accession number S37077, position 84–223, not shown) with the descriptor HH-I-7 and in mouse repetitive DNA with descriptors for the HH-I-iiUA and HH-III-iiAU motifs (Fig. 2C,D). The first two motifs are predicted to be inactive because they contain A instead of G in position 12.
Viruses are good candidates for using catalytic RNA motifs. We have found several new intriguing hammerhead motifs in different viruses (Fig. 2E). Two similar hammerhead ribozyme motifs were found in the 5′ untranslated region of two viruses of the Ilarvirus genus, family of Bromoviridae, which are single stranded positive RNA viruses. One motif is in the spinach latent virus (accession number PMOVRNA3, position 252–331) and the other in the Elm mottle virus (accession number SLU57048, position 250–329) (Fig. 2E). Both motifs were found using the HH-III descriptor. The region containing the hammerhead is highly conserved among these viruses. The hammerhead motif found with HH-II in an RNA associated to carrot red luteovirus that is also very interesting because satellite RNAs were the first molecules found to contain hammerhead ribozymes (Fig. 2F). This motif is predicted to cleave after AUA. Mammalian viruses also contain potential hammerhead ribozymes, and two of them found with HH-II are illustrated in Figure 2G,H, one in the hepatitis E virus, and the other in the caprine encephalitis virus.
Two hammerhead motifs in human mRNAs also are presented in Figure 2I,J. Self-cleaving motifs in mRNA might regulate gene expression by promoting RNA decay. The genes coding for the interferon-induced DAP1 and the neuroleukin gene possess potentially active hammerhead motifs found with HH-III that are predicted to cleave after UUC and CUC, respectively. Perhaps even more remarkable are the conserved hammerhead motifs found in the genes coding for NADPH-cytochrome P450 oxidoreductase both in the rat and the hamster (Fig. 2K,L). All together, the motifs presented here suggest that the hammerhead ribozyme might have functions other than those previously suggested for satellite RNA and transcripts for repetitive sequences.
DISCUSSION
We have used the search engine RNAMOT to scrutinize the GenBank for potential self-cleaving hammerhead ribozyme motifs. Our search extends earlier efforts to find a subset of potential hammerheads in Escherichia coli sequences (Ruffner et al. 1990). Because this motif has relatively few structural constraints, we designed an extensive set of descriptors for both the wild-type motif and variants of its essential nucleotides. The results show a wide distribution of potential hammerhead-like motifs in all regions of the GenBank with a higher frequency for the variants that do not require the presence of a CpG dinucleotide in the final sequence of the motifs. This CpG dinucleotide in positions 11.1 and 12 is not absolutely required for self-cleavage because other base pairs are acceptable in positions 10.1:11.1. We conclude that the reduction we observed in the frequency of most hammerhead motifs in this search is fortuitous.
We expect that most of the motifs found here are inactive because we designed descriptors that include mutations or nonoptimal features of the hammerhead self-cleaving motif (Ruffner et al. 1990). However, our results illustrate the possibility that natural sequences might end up forming self-cleaving motifs by random drift. In other words, it would be sufficient to mutate one or two residues to activate the potential hammerhead ribozymes described here. This is not only true for the hammerhead ribozyme motif because other RNA motifs can be found randomly in natural sequences (Fontana et al. 1993; Reidys et al. 1997; Bourdeau et al. 1999).
The use of variants of the hammerhead ribozyme was stimulated by previous work that showed that satellite DNA encoding hammerhead ribozymes is enriched with mutated variants of the motif (Zhang and Epstein 1996; Ferbeyre et al. 1998). The ribozyme motif found in the cricket satellite DNA follows this rule because 14 of the 20 sequences deposited until now in the GenBank contains an active motif. Other mutant hammerheads were found in different families of repetitive DNA by using descriptors for hammerhead-like motifs, raising the possibility that other members of these families, not yet sequenced, contain the active motifs. The occurrence of hammerhead ribozymes in transcripts of repetitive DNA from different species suggests a functional role for the self-cleavage reaction in the propagation and/or the metabolism of these transcripts. We previously have proposed that self-cleavage might limit the expansion of repetitive sequences through the genome by retrotransposition (Ferbeyre et al. 1998). This model predicts that recent insertions of these elements will contain disabling mutations in the hammerhead motif. The family of L1 repetitive elements for example contains mutated versions of the hammerhead and members of this family still retrotranspose in humans, sometimes causing genetic diseases (Holmes et al. 1994). Another intriguing possibility is that viroids and satellite RNAs originated from transcripts of repetitive sequences when these transcripts parasitizes a viral replication machinery. Subsequently, they might jump from one organism to another using the virus as a vector, and as a result their distribution will cross phylogenetic barriers.
Many ESTs and mRNAs were found here to possess hammerhead-like motifs. To test any role of the hammerhead motifs identified in this work, we need a combination of biochemical and genetic analysis. Our group has finished the characterization of hammerhead motifs in repetitive DNA of Schistosome (Ferbeyre et al. 1998) and the cricket (A.A. Rojas, A. Vazques-Tello, G. Ferbeyre, F. Venanzetti, L. Bachmann, B. Paquin, and R. Cedergren, in prep.). All the occurrences we found in the GenBank are available at our web site (URL: http://www.centrcn.umontreal.ca/∼bourdeav/HH) for those interested in finding where “hammers” can cut.
METHODS
The pattern searching for RNA secondary structures was performed by RNAMOT (Gautheret et al. 1990; Laferrière et al. 1994). The inputs for this program are nucleotide sequences, and a descriptor file defining the structural motif to be searched. RNAMOT reports all the occurrences of the motif as well as its positions along the sequence. Two of the three helices defining the hammerhead self-cleaving motif are closed by loops. The remaining helix connects the motif to the rest of the RNA molecule. As a result, there are three ways of defining a self-cleaving hammerhead ribozyme motif. We have built descriptors for these three different orientations of the motif taking into account the following constraints (Fig. 1):
Three nucleotides in Helix I. Helix I has no specific nucleotide requirements although the hammerhead motif found in the newt and in Schistosome possess a conserved GG:CC base pairing, three nucleotides downstream from the cleavage site as well as an internal loop farther downstream (Pabon-Peña et al. 1991; Ferbeyre et al. 1998).
The conserved sequence CYGANGA. This sequence is part of the catalytic core of the ribozyme and is entirely conserved with the exception of position 7. In the latter, although all nucleotides are accepted, the preferred ones are U then G or A and finally C. More recently, position 4 was reported to accept also U, so we have included this feature in our search (Ambros and Flores 1998).
Three nucleotides in Helix II. There is a strong preference for a R:Y base pair in positions 10.1:11.1, but the pair G:C confers the better activity and was the only one allowed in our original descriptors.
The conserved sequence GAA is absolutely required for catalysis. In the X-ray model of the hammerhead, nucleotides G12 and A13 form two reverse Hoogsteen G-A base pairs with nucleotides A9 and G8, respectively, whereas A14 form a non-Watson Crick base pair with N7 (Scott et al. 1995).
Helix III requires an A:U base pair which is also of non Watson Crick type and a minimum of one more pair in two of the orientations (HH-I and HH-II). When the helix is open as in HH-III, two more pairs are required.
The cleavage site was defined as NUH (H is any nucleotide but G). However, natural ribozymes contain GUC, GUA, AUA, and AUC because they allow the highest reaction rates (Shimayama et al. 1995; Ferbeyre et al. 1998).
The loops closing the helices were allowed to have from 0 to 100 nucleotides.
Sixty-three additional mutants also were included in the study. These were derived from the original motifs shown in Figure 1 by changing either one base in the conserved single stranded regions for an N (any nucleotide; 30 mutants), the identity of one of the constrained base pair (positions 10.1:11.1 and 15.1:16.1; 30 mutants), or by changing the cleavage site from NUH to NHH (three more motifs; Kore et al. 1998).
The search was performed in the July 15, 1998 release of the GenBank sequence database (National Center for Biotechnology Information-GenBank flat file release 108.0). Searches were performed on both strands and all occurrences of motifs involving unidentified bases denoted by N in the database were disregarded. A Power Challenge XL with 32 CPUs IP 19, R4400, 150-MHz processor (3072 Mbytes) running UNIX IRIX 6.2 was used.
To help establish the significance of their presence, frequencies of each motif in the database were compared with frequencies in a random sequence database generated by a uniform pseudo-random number generator (L'Écuyer and Andres 1997) with a period length near 2121. The random sequence databases contained 1000 sequences of 100,000 nucleotides each; the four nucleotides A, C, G, and T were used with equal probabilities. An “expected” frequency N in GenBank was calculated from the number M of occurrences of each motif in the random databases as follows: N = (a ×M)/(104 × 105), where a is the number of nucleotides in GenBank (1.797 × 109 in the release 108.0).
The random database reduced in CpG dinucleotides was generated using the same procedure, but each time a CpG dinucleotide was created a second generator (evolving in parallel) would enter in function to decide if yes or no (50% frequency) the dinucleotide would be changed. If a change had to take place, a third generator (also evolving in parallel) would be able to choose among six replacing dinucleotides: AG, CA, CC, CT, GG, or TG (choices made according to the dinucleotide frequencies reported by Karlin and Mrazek 1997). The expected frequency was evaluated as before.
Acknowledgments
We thank Bruno Paquin for valuable comments, and to NSERC of Canada which financed this project. V.B. holds a doctoral fellowship from NSERC of Canada. The late R.C. was Richard Ivey Scholar of the Canadian Institute for Advanced Research (CIAR) program in Evolutionary Biology. We acknowledge previous efforts from Dr. Daniel Gautheret to search for hammerhead sequences with RNAMOT in our laboratory. In addition, we thank Bernard Lorazo, Daniel Raymond and André Fourrier of the DITER (Direction des infrastructures technologiques d'enseignement et de recherche) at the Université de Montréal for their assistance. P.M. wishes to thanks the hospitality of the Université de Montréal and the Institute of Physics, UNAM.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ferbeyre@cshl.org; FAX (516) 367 8454.
REFERENCES
- Ambros S, Flores R. In vitro and in vivo self-cleavage of a viroid RNA with a mutation in the hammerhead catalytic pocket. Nucleic Acids Res. 1998;26:1877–1883. doi: 10.1093/nar/26.8.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birikh KR, Heaton PA, Eckstein F. The structure, function and application of the hammerhead ribozyme. Eur J Biochem. 1997;245:1–16. doi: 10.1111/j.1432-1033.1997.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Ferbeyre G, Pageau M, Paquin B, Cedergren R. The distribution of RNA motifs in natural sequences. Nucleic Acids Res. 1999;27:4457–4467. doi: 10.1093/nar/27.22.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratty J, Chartrand P, Ferbeyre G, Cedergren R. The hammerhead RNA domain, a model ribozyme. Biochim Biophys Acta. 1993;1216:345–359. doi: 10.1016/0167-4781(93)90001-t. [DOI] [PubMed] [Google Scholar]
- Bussière F, Lafontaine D, Perreault J-P. Compilation and analysis of viroid and viroid-like RNA sequences. Nucleic Acids Res. 1996;24:1793–1798. doi: 10.1093/nar/24.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LM, Gall JG. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana W, Konings DA, Stadler PF, Schuster P. Statistics of RNA secondary structures. Biopolymers. 1993;33:1389–1404. doi: 10.1002/bip.360330909. [DOI] [PubMed] [Google Scholar]
- Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Gautheret D, Major F, Cedergren R. Pattern searching/alignment with RNA primary and secondary structures: an effective descriptor for tRNA. Comput Appl Biosci. 1990;6:325–331. doi: 10.1093/bioinformatics/6.4.325. [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Pardi A, Uhlenbeck OC, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach WL, Hodgson R, et al. Numbering system for the hammerhead. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrazek J. Compositional differences within and between eukaryotic genomes. Proc Natl Acad Sci. 1997;94:10227–10232. doi: 10.1073/pnas.94.19.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kore AR, Vaish NK, Kutzke U, Eckstein F. Sequence specificity of the hammerhead ribozyme revisited; the NHH rule. Nucleic Acids Res. 1998;26:4116–4120. doi: 10.1093/nar/26.18.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferrière A, Gautheret D, Cedergren R. An RNA pattern matching program with enhanced performance and portability. Comput Appl Biosci. 1994;10:211–212. doi: 10.1093/bioinformatics/10.2.211. [DOI] [PubMed] [Google Scholar]
- Lafontaine DA, Deschênes P, Bussière F, Poisson V, Perreault J-P. The viroid and viroid-like RNA database. Nucleic Acids Res. 1999;27:186–187. doi: 10.1093/nar/27.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Écuyer P, Andres TH. A random number generator based on the combination of four LCGs. Math Comput Simulation. 1997;44:99–107. [Google Scholar]
- Pabon-Peña LM, Zhang Y, Epstein LM. Newt satellite 2 transcripts self-cleave by using an extended hammerhead structure. Mol Cell Biol. 1991;11:6109–6115. doi: 10.1128/mcb.11.12.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidys C, Stadler PF, Schuster P. Generic properties of combinatory maps: Neutral networks of RNA secondary structures. Bull Math Biol. 1997;59:339–397. doi: 10.1007/BF02462007. [DOI] [PubMed] [Google Scholar]
- Ruffner DE, Stormo GD, Uhlenbeck OC. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Shimayama T, Nishikawa S, Taira K. Generality of the NUX rule: Kinetic analysis of the results of systematic mutations in the trinucleotide at the cleavage site of hammerhead ribozymes. Biochemistry. 1995;34:3649–3654. doi: 10.1021/bi00011a020. [DOI] [PubMed] [Google Scholar]
- Symons R. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Symons R. Plant pathogenic RNAs and RNA catalysis. Nucleic Acid Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Epstein LM. Cloning and characterization of extended hammerheads from a diverse set of caudate amphibians. Gene. 1996;172:183–190. doi: 10.1016/0378-1119(96)00126-6. [DOI] [PubMed] [Google Scholar]