Abstract
BACKGROUND/AIMS
Bile acid (BA) malabsorption of moderate severity is reported in 32% of patients with chronic unexplained diarrhea including diarrhea-predominant IBS (IBS-D). We hypothesize that variants of genes regulating hepatic BA synthesis play a role in IBS-D.
METHODS
In 435 IBS and 279 healthy subjects, we tested individual associations of 15 common single-nucleotide polymorphisms (SNPs) from 7 genes critical to BA homeostasis with symptom-based subgroups using dominant genetic models. In a subset of 238 participants, we tested association with colonic transit. SNP-SNP interactions were investigated based on known protein interactions in BA homeostasis. The function of SNP rs17618244 in Klothoβ (KLB) was evaluated using a protein stability assay in HEK293 cells.
RESULTS
SNP rs17618244 (Arg728Gln in KLB) is associated with colonic transit at 24 h. G allele (Arg728) compared to A allele (Gln728) is associated with accelerated colonic transit (p=0.0007) in the overall cohort; this association was restricted to IBS-D (p=0.0018). Interaction tests of KLB rs17618244 with 3 non-synonymous coding SNPs of fibroblast growth factor receptor 4 (FGFR4) revealed that rs1966265 (Val10Ile) and rs351855 (Gly388Arg) modulate KLB’s association with colonic transit in IBS-D (p=0.0025 and p=0.0023, respectively). The KLB Arg728 variant significantly reduced protein stability compared to KLB Gln728, demonstrating KLB rs17618244’s functional significance. No significant associations with symptom-based subgroups of IBS were detected.
CONCLUSIONS
A functional KLB gene variant mediating protein stability associates with colonic transit in IBS-D. This association is modulated by 2 genetic variants in FGFR4. The FGF19-FGFR4-KLB pathway links regulation of BA synthesis to colonic transit in IBS-D.
Keywords: bile acid, KLB, FGFR4, FGF19
INTRODUCTION
Irritable bowel syndrome (IBS) is a common disorder characterized by recurrent abdominal pain or discomfort and altered bowel function that are not explained by overt structural or tissue abnormalities.1 The diarrhea phenotype in IBS-D may have specific etiologies, including bile acid (BA) malabsorption or gluten intolerance. A genetic contribution to IBS is suggested by studies of familial aggregation, twin concordance, candidate gene association, and pharmacogenomics, but there has been no genome wide association study (GWAS) performed and no definitive genetic susceptibility factor(s) identified in IBS to date.2 The heterogeneity of IBS symptoms is a confounder in detecting genotype-phenotype associations, necessitating studies far larger than those performed to date.2 Intermediate phenotypes, particularly measurement of colonic transit, have proven more informative in candidate gene association studies.3
Increased colonic exposure to BA, as in idiopathic bile acid malabsorption (BAM), is associated with functional diarrhea and IBS-D.4-8 A meta-analysis reported BAM in 32% of patients with unexplained chronic diarrhea; patients with BAM and diarrhea respond to treatment with the BA sequestrant, cholestyramine.7 In addition, reducing colonic exposure to BA by another sequestrant, colesevelam, may lead to constipation in diabetics9 and delays ascending colon emptying in IBS-D patients with evidence of increased BA synthesis.10
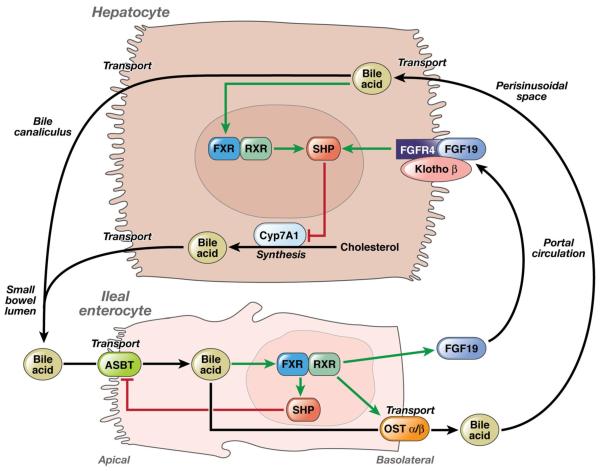
The factors controlling hepatic BA synthesis are well established (Fig. 1). Fibroblast growth factor 19 (FGF19) from the ileal enterocyte binds to fibroblast growth factor receptor 4 (FGFR4) on the hepatocyte cell membrane (Fig. 1). In a manner that is dependent on biochemical interaction between KLB and FGFR4, FGF19 triggers intracellular signaling to downregulate cytochrome P450 7A1 (CYP7A1) activity and suppress BA synthesis.11 Defective FGF19 release from the ileum is reported in IBS-D responsive to cholestyramine, consistent with excessive hepatic BA synthesis due to defective FGF19 signaling leading to BA diarrhea.12 Impaired apical uptake of BA into ileal enterocytes can lead to BA diarrhea. However, loss-of-function mutations in the apical sodium-dependent bile acid transporter (ASBT) gene that cause congenital BAM13 are rare.14 Knockout mice with loss of function of KLB or small heterodimer partner (SHP) result in elevated BA synthesis and diarrhea.15-17 In a pharmacogenomics study, we showed that genetic variants in KLB and FGFR4 influence the degree of acceleration of colonic transit in IBS-C patients mediated by treatment with chenodeoxycholate, a primary bile salt.18
Figure 1.
Molecular mechanisms in homeostatic control of bile acid synthesis. FGF19 plays an important role in BA homeostasis by binding to FGFR4 on the hepatocyte cell membrane, triggering intracellular signaling in a KLB-dependent manner to downregulate CYP7A1 expression and thereby suppress BA synthesis. Defective FGF19 release from the ileum is reported in IBS-D patients whose symptoms improve with the BA sequestrant cholestyramine, suggesting that excessive hepatic BA synthesis due to defective FGF19 signaling is associated with BA diarrhea (adapted from Rao et al.)24.
Our hypothesis is that common single nucleotide polymorphisms (SNPs) in genes critical for BA synthesis play a role in expression of IBS symptoms and colonic transit.
METHODS
Subjects
Our study assessed a total of 717 subjects: 435 patients with functional gastrointestinal disorder (FGID) [Rome II criteria positive consisting of 177 constipation-predominant IBS (IBS-C), 174 IBS-D, 84 alternating-type IBS (IBS-A)] and 282 healthy volunteers (HV). Of these, 239 participants had undergone scintigraphic colonic transit measurement. Baseline characteristics of the full cohort of 714 participants and the subset of 238 participants with colonic transit and genotyping information are shown in Table Ia and Ib, respectively. The demographics of subjects with colonic transit data do not differ from those of the full cohort. Genomic DNA and phenotype data were collected with written informed consent in previous studies from 2000 to 2010.10, 19-24
Table Ia. Baseline Characteristics of All Study Subjects.
| Total | Healthy | IBS-C | IBS-D | IBS-A | |
|---|---|---|---|---|---|
| N† | 717 | 282 | 177 | 174 | 84 |
| Age (Mean ± SEM) | 41.4 ± 0.5 | 37.0 ± 0.7 | 44.9 ± 1.0 | 45.3 ± 1.2 | 40.7 ± 1.4 |
| Gender (% Females) | 82% | 70% | 96% | 82% | 92% |
Note that not all of the 717 had complete genotype data: 3 healthy subjects were missing rs17618244 genotype, and two IBS patients were missing 2 individual genotype results. All missing results were caused by technical problems with the assays for those individual DNA samples.
Table Ib. Baseline Characteristics of Study Subjects Who Underwent Scintigraphic Colonic Transit Measurement.
| Total | Healthy | IBS-C | IBS-D | IBS-A | |
|---|---|---|---|---|---|
| N†† | 238 | 55 | 78 | 80 | 25 |
| Age (Mean ± SEM) | 38.4±0.8 | 33.7 ± 1.3 | 38.6 ± 1.1 | 41.9 ± 1.6 | 37.2 ± 2.3 |
| Gender (% Females) | 87% | 78% | 99% | 78% | 100% |
| BMI (Mean ± SEM) | 26.8±0.3 | 26.2 ± 0.7 | 26.1 ± 0.5 | 27.5 ± 0.6 | 28.3 ± 0.9 |
Note that there were 239 participants with transit data, but one of the healthy volunteer did not have genotype information
Use of the collected data for this study was reviewed and approved by the Mayo Clinic Institutional Review Board. All participants had authorized use of medical records and genomic DNA for research.
Gastrointestinal and Colonic Transit
An established, validated scintigraphic method was used to measure gastrointestinal and colonic transit as previously described.25 Colonic transit by scintigraphy is a valid biomarker of colonic dysmotility.26 The performance characteristics of this test are summarized elsewhere.25-28 During transit studies, participants continued estrogen replacement, birth control pills or depot injection, stable doses of thyroid replacement, low-dose aspirin, and selective serotonergic antidepressants. Exclusion criteria included organic diseases that might explain the patients’ symptoms, and use of medication for IBS or bowel dysfunction within 7 days before or during the study.
The primary endpoint of colonic transit is the geometric center at 24 hours (GC24). Secondary endpoints were the colonic GC at 8 and 48 hours, gastric emptying and small bowel transit. The GC is the weighted average of count percentages in the different colonic regions: ascending (AC), transverse (TC), descending (DC), rectosigmoid (RS), and stool; the weighting factors are 1 to 5 respectively.
A change in colonic GC24 of 1 unit is associated with a discernible (~0.65 point) change in stool consistency on a validated 7-point stool form scale.25
Genotyping
We identified candidate genes critical for regulating absorption, transport, and synthesis of BA (Table II) using a pathway-based approach based on current understanding of these molecular mechanisms.11, 29, 30 We focused on proteins whose change in function can plausibly lead to changes in the amount of bile acids reaching the colon. We selected non-synonymous SNPs with minor allele frequencies (MAF) >9% according to the HapMap-CEU population (Table II). These coding SNPs lead to changes in amino acid sequence and potential changes in function of the synthesized protein. Two candidate genes, OSTβ and CYP7A1, do not have coding SNPs with MAF >9%; therefore, tag SNPs in these genes with r2 >0.8 and MAF >9% were used. In total, 15 SNPs or tag SNPs in 7 candidate genes were genotyped.
Table II. Candidate SNPs with p-values for Hardy-Weinberg Equilibrium and Minor Allele Frequencies.
(SNP=single nucleotide polymorphism; HWE=Hardy-Weinberg Equilibrium; MAF=minor allele frequency) Predicted MAF is based on the HapMap-CEU population. Observed MAF is derived from genotypes of the healthy volunteers (HV) in the full study cohort of 714 subjects (N=282) and the HV subset (N=56) of the 238 subjects with transit measurements. HWE p-value >0.05 denotes that the study sample is in HWE for the corresponding SNP; the two p-values not satisfying HWE are marked with *.
| Gene | SNP rs ID | Major Allele |
Minor Allele |
Predicted MAF |
HV in Full cohort N=282 |
HV in Transit cohort N=56 |
Function | Amino Acid Position |
Amino Acid Change |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed MAF |
HWE p-value |
Observed MAF |
HWE p-value |
||||||||
| SHP | rs6659176 | C | G | 0.092 | 0.061 | 0.266 | 0.055 | 1.000 | missense | 171 | Ala ⇒ Gly |
| ASBT | rs188096 | G | T | 0.192 | 0.111 | 0.759 | 0.164 | 1.000 | missense | 171 | Ala ⇒ Ser |
| KLB | rs17618244 | G | A | 0.155 | 0.181 | 0.109 | 0.164 | 0.325 | missense | 728 | Arg ⇒ Gln |
| KLB | rs4975017 | C | A | 0.325 | 0.355 | 0.513 | 0.409 | 0.100 | missense | 1020 | Gln ⇒ Lys |
| FGFR4 | rs1966265 | G | A | 0.224 | 0.226 | 0.309 | 0.200 | 0.670 | missense | 10 | Val ⇒ Ile |
| FGFR4 | rs376618 | T | C | 0.225 | 0.204 | 0.001 * | 0.273 | 0.047 * | missense | 136 | Leu ⇒ Pro |
| FGFR4 | rs351855 | G | A | 0.283 | 0.333 | 0.079 | 0.318 | 0.210 | missense | 388 | Gly ⇒ Arg |
| OSTα | rs939885 | G | A | 0.458 | 0.470 | 1.000 | 0.518 | 0.279 | missense | 202 | Val ⇒ Ile |
| OSTβ | rs2946676 | A | G | 0.14 | 0.142 | 0.087 | 0.100 | 1.000 | near-gene-3 (dbSNP) |
||
| OSTβ | rs4238399 | C | T | 0.37 | 0.407 | 0.324 | 0.445 | 1.000 | near-gene-3 (dbSNP) |
||
| CYP7A1 | rs8192879 | C | T | 0.37 | 0.407 | 0.535 | 0.336 | 0.362 | utr-3(dbSNP) | ||
| CYP7A1 | rs8192877 | A | G | 0.18 | 0.147 | 0.473 | 0.173 | 0.648 | intron(dbSNP) | ||
| CYP7A1 | rs11786580 | C | T | 0.24 | 0.197 | 0.347 | 0.227 | 0.711 | intron(dbSNP) | ||
| CYP7A1 | rs7833904 | T | A | 0.41 | 0.394 | 0.079 | 0.373 | 0.563 | intergenic(GVS) | ||
| CYP7A1 | rs10957057 | C | T | 0.14 | 0.127 | 0.277 | 0.100 | 1.000 | intergenic(GVS) | ||
Genomic DNA was isolated from whole blood using standard methods shortly after blood draw and stored at −80°C until genotyping. Genotyping was performed using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA) per manufacturer’s instructions. Following PCR amplification, end reactions were analyzed using ABI 7300FAST Real-Time PCR System by Sequence Detection Software (Applied Biosystems).
Protein Stability Assay
We used full-length human KLB cDNA fused to C-terminal Myc and DDK tags cloned into the pCMV6-Entry mammalian plasmid expression vector (OriGene, Rockville, MD). This vector contains the major allele G of SNP rs17618244 at nucleotide position 2280 corresponding to KLB Arg728. Mutagenesis of this single nucleotide into the minor allele A of rs17618244 was performed using QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) to create an expression vector for KLB Gln728. DNA sequencing with the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) was used to confirm sequences of plasmid vectors. HEK293 cells (ATCC, Manassas, VA) transiently transfected with expression vectors corresponding to each of the 2 allelic variants of KLB were treated with 20 μg/ml of cycloheximide (CHX) (Sigma-Aldrich, St. Louis, MO), a protein translation inhibitor, for 0, 2, 4, 6, or 10 hours. Three independent experiments were performed up to 6 hours, with one experiment extended to 10 hours. Cell lysates were prepared from adherent and floating cells using Complete Lysis-M Reagent (Roche Applied Science, Branford, CT). Western blots were performed by standard techniques using mouse anti-c-Myc (Santa Cruz Biotechnology, Santa Cruz, CA) and goat anti-actin (Abcam Inc., Cambridge, MA) primary antibodies and appropriate secondary antibodies. Signal intensities of Western blots were quantified by optical densitometry using an optical scanner and Macintosh computer running NIH Image software version 1.62. Protein half-lives were determined using the formula t½ = ln(2)/λ where λ is the decay constant.
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) for the genotype distribution of each SNP was assessed using Weir’s exact test.31
Dominant genetic models were used for all analyses in order to provide sufficient numbers of subjects in genotype subgroups given the small samples for the minor allele homozygous subgroups for some of the SNPs. The Wilcoxon rank-sum test was used to test for univariate associations between genotypes of each SNP and colonic transit in the overall study cohort. Analysis of covariance (ANCOVA) was used to test for associations between genotypes of a selected SNP and transit within each of the 4 symptom subgroups (three IBS subgroups and HV). An adjustment for population stratification was not used due to the low likelihood of confounding by ethnicity or race in our highly homogenous patient population.32, 33 Among those who volunteer race and ethnicity information at Mayo Clinic in Rochester, MN, >95% are Caucasians
Given that prior cholecystectomy may predispose to post-cholecystectomy diarrhea and potentially confound any association found between genotype and colonic transit, we conducted a separate analysis using prior cholecystectomy status as a covariate in the ANCOVA models.
KLB interacts biochemically with FGFR4 to promote FGF19-mediated suppression of BA synthesis. Therefore, within each symptom subgroup, the potential interactions between KLB rs17618244 and each of 3 SNPs within the FGFR4 gene coding region were assessed. Therefore, ANCOVA was also used to test for differential associations (“interactions”) among selected SNP-SNP pair-wise combinations: KLB rs17618244 and each of 3 SNPs in FGFR4 (rs1966265, rs351855, and rs376618) within each of the 4 symptom subgroups.
The Bonferroni method was used to adjust significance levels for multiple comparisons: Specifically, we correction for 15 tests when assessing individual SNPs (i.e. 0.05/15 SNPs or tagSNPs tested=0.0033). After demonstrating significant association in the overall cohort with SNP KLB rs17618244, the association of this single SNP with colon transit within each of the 4 symptom subgroups (three IBS subgroups and HV) was corrected for 4 tests (i.e. 0.05/4 symptom subgroups =0.0125). Finally, correction for 12 tests was included in the assessment of the 3 SNP-SNP combinations (KLB rs17618244 with each of 3 SNPs in FGFR4) for the 4 symptom subgroups (i.e. 0.05/12=0.0042).
The stability of protein products from the 2 allelic variants of KLB SNP rs17618244 was examined using both a first-order kinetics model (exponential decay with constant t½) and a zero-order kinetics model (linear decay with constant reaction rate). Two-sample t-tests were used to compare the decay parameter estimates from the exponential model, and separately, to compare the slopes from the linear model.
RESULTS
Hardy-Weinberg Equilibrium for Candidate SNPs
Table II lists the 15 SNPs analyzed, p-values for Hardy-Weinberg equilibrium (HWE), and predicted MAF and observed MAF separately for the healthy participants (N=279) within the entire cohort of 714 subjects (with genotype data) and the healthy participants (N=55) within the subset of 238 subjects with colonic transit measurements.
In the healthy participants, FGFR4 rs376618 is the only SNP out of the 15 candidate SNPs to differ significantly from HWE, both in the full cohort and in the colonic transit cohort. This SNP did not show significant genotypic association with any of the phenotypes evaluated, so inclusion of this SNP did not alter the results or interpretation of the reported associations with symptom subgroups or colonic transit.
Association in Overall Cohort of Individual Candidate SNP with Symptom Phenotypes and Transit Measurements
KLB rs17618244 was significantly associated with colonic transit at 24h (primary endpoint) in the overall cohort after correction for multiple comparisons (GC24 2.78±0.09 for GG vs. 2.36±0.11 for AA/AG genotype; p=0.0007, Bonferroni cutoff α=0.0033). The strength of the association detected between KLB rs17618244 genotype and GC24 was virtually the same after adjusting for prior cholecystectomy status.
KLB rs17618244 was also associated with numerically faster secondary endpoints of colonic transit at 8h (GC8 1.63±0.09 for GG vs. 1.32±0.09 for AA/AG, p=0.023) and 48h (GC48 for GG 3.84±0.09 vs. 3.47±0.13 for AA/AG, p=0.025). No other significant associations were found between individual SNPs and gastric and small bowel transit.
No significant associations with symptom-based subgroup status of subjects (IBS subtypes and healthy volunteers) were detected.
Association of KLB SNP rs17618244 with Colonic Transit in IBS Subtypes
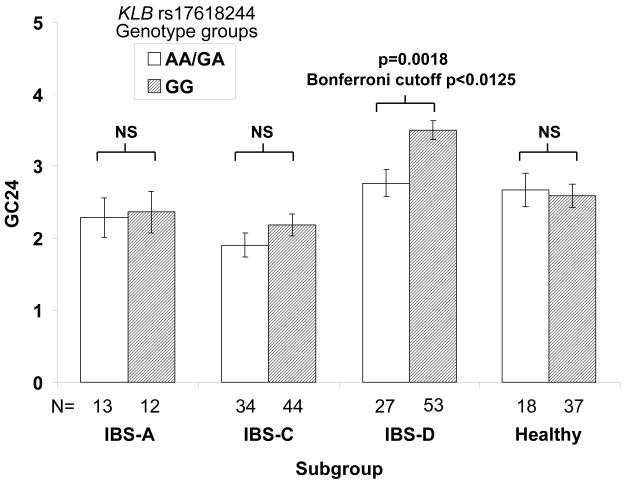
The IBS-D subgroup exhibited the strongest association of KLB rs17618244 with faster colonic transit at 24 and 48 hours (GC24h: 3.50±0.14 for GG vs. 2.76±0.19 for AA/AG, p=0.0018; GC48 4.57±0.15 vs. 3.94±0.20, p=0.0113; Bonferroni cutoff α=0.0125) (Fig. 2). The strength of the associations detected between KLB rs17618244 genotype and either GC24 or GC48 in the IBS-D subgroup alone was not appreciably altered after adjusting for prior cholecystectomy status. No significant associations were found with colonic transit (24 or 48 hours) in the other 3 groups (all p>0.21, Fig. 2).
Figure 2.
KLB SNP rs17618244 is associated with colonic transit in IBS-D. Colonic transit expressed as mean GC24 (y-axis) is shown by KLB rs17618244 genotype groups across the 4 subgroups (x-axis). Error bars denote standard error of the mean (SEM).
Interaction between KLB SNP rs17618244 and 3 FGFR4 SNPs in Individual IBS Subtypes
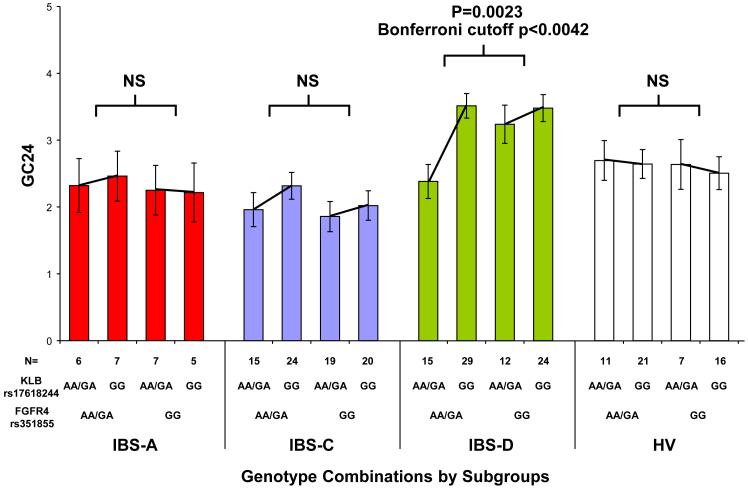
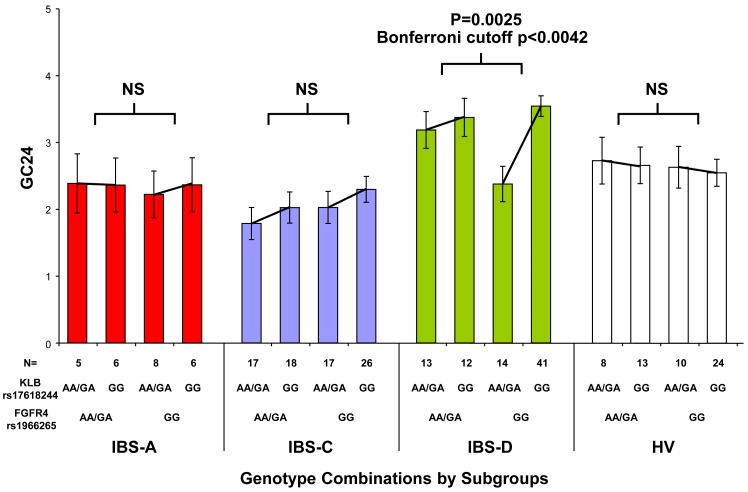
KLB interacts biochemically with FGFR4 to promote FGF19-mediated suppression of BA synthesis. Therefore, within each symptom subgroup, the potential interactions between KLB rs17618244 and each of 3 SNPs within the FGFR4 gene coding region were assessed. Significant interactions between combinations of KLB rs17618244 with FGFR4 rs351855, and separately, of KLB rs17618244 with FGFR4 rs1966265, were observed for GC24 in the IBS-D subgroup (p=0.0023 and p=0.0025, respectively; Bonferroni cutoff α=0.0042) (Fig. 3a and 3b).
Figure 3.
KLB rs17618244’s association with colonic transit in IBS-D is modulated separately by two FGFR4 variants. Colonic transit expressed as mean GC24 (y-axis) is shown by combinations of 2 genotype groups: KLB rs17618244 with either FGFR4 rs351855 (panel A) or FGFR4 rs1966265 (panel B). IBS-D patients generally have accelerated colonic transit. The GG genotype of KLB SNP rs17618244 is associated with accelerated colonic transit, but this association is significantly modulated separately by the genotypes of FGFR4 SNPs rs351855 and rs1966265, with rs351855 GG and rs1966265 AA/GA blunting this acceleration whereas rs351855 AA/GA and rs1966265 GG enhancing this acceleration.
Functional Analysis of KLB SNP rs17618244
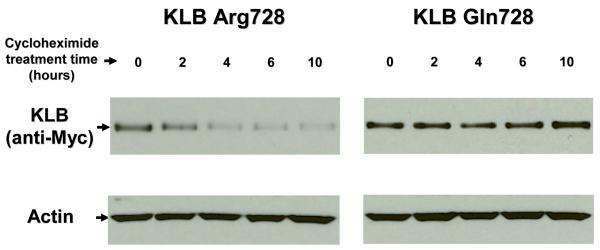
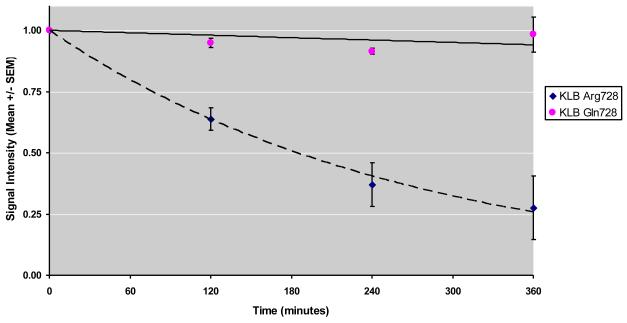
A protein stability assay for protein products of the 2 alleles of KLB SNP rs17618244 (Fig. 4a) showed that KLB Arg728 has a half-life of about 3 hours using an exponential decay best-fit curve (Fig. 4b). In contrast, KLB Gln728 showed signal intensity over 90% of the starting signal both at 6 hours (Fig. 4b) and at 10 hours of CHX treatment (Fig. 4a), suggesting that KLB Gln728’s half-life is >10 hours. The parameter estimates for the KLB Arg728 and KLB Gln728 curves were significantly different using an exponential decay model (p=0.026), as well as the linear model (p=0.008).
Figure 4.
KLB rs17618244 alters protein stability. Cycloheximide protein stability assay was performed using transient transfection into HEK293 cells with vectors expressing Myc-tagged KLB rs17618244 major allele G (KLB Arg728) or minor allele A (KLB Gln728). Cycloheximide (CHX, 20 μg/ml) was added to inhibit protein synthesis for a total of 0, 2, 4, 6, and 10 hours. Panel A shows representative Western blot analysis using anti-Myc. Anti-actin was used to verify equal protein loading. Panel B shows exponential best-fit curves generated from signal intensity measurements by densitometry of three independent experiments. Myc-KLB:actin ratios were calculated and normalized to time 0.
DISCUSSION
In this study, we demonstrate that a functional gene variant mediating stability of KLB, a protein regulating BA synthesis, associates with colonic transit in IBS-D. This association is modulated by two independent genetic variants in FGFR4, whose protein product directly interacts with KLB.34 This study links common polymorphisms in human genes regulating BA synthesis to colonic transit at 24 h, a well-characterized intermediate phenotype, in IBS-D. These data support the hypothesis that genetic differences in regulation of endogenous BA synthesis play a role in the pathophysiology of accelerated colonic transit and diarrhea in IBS-D. This hypothesis is consistent with previous pharmacogenomics observations suggesting that genetic variants in KLB and FGFR4 influence the acceleration of colonic transit by treatment with exogenous BA in IBS-C patients.18
There is a high prevalence of IBS (~10% average) in most western countries. While rare genetic variants may also play important roles in susceptibility to common diseases such as colon cancer35, no such rare variants have been described to date in IBS. Under the “common disease, common variant” paradigm, it is postulated that genetic variations that may contribute to this disorder may be relatively common, partially justifying our choice to study SNPs with MAF ≥9%. Our choice of MAF threshold was also dictated by statistical power considerations in view of the 238 participants with colonic transit measurement. We focused on non-synonymous SNPs in coding regions of candidate genes as these variants have higher likelihood of altering function of the translated protein products.
We did not detect any significant associations between the symptom-based subgroups of IBS and any of the 15 SNPs studied, including KLB rs17618244. We chose the primary endpoint of colonic transit at 24 hours because this is an objective, quantitative intermediate phenotype of IBS, with relatively small coefficient of variation and strong link to the pathophysiology of IBS compared to patient-reported symptoms. When assessing genotype association with IBS symptoms, the possibility of a type II error is magnified, and the sample size required for sufficient power would be much higher than with the quantitative phenotype of colonic transit.3
Differences in protein stability between the 2 alleles of KLB rs17618244 provide a biologically plausible mechanism by which this SNP may affect colonic transit. The difference in KLB protein stability (G allele t½ ~3 h, A allele t½ >10 h) may result in different receptor signaling strength in FGF19-mediated inhibition of hepatic BA synthesis. The diminished stability of KLB Arg728 (G allele) is expected to weaken the strength of intracellular signaling cascades triggered when FGF19 binds to and activates the FGFR4-KLB receptor complex on the hepatocyte plasma membrane. Weakened FGF19 signaling leads to increased CYP7A1 expression and increased hepatic BA synthesis. Excess BAs reaching the small bowel may overwhelm the ileal capacity for BA absorption, spill over into the colonic lumen, and accelerate colonic transit by stimulating motility36 and secretion.37 A recent study suggests that KLB may promote FGFR4’s activation by specifically binding to dominant negative, glycosylated forms of FGFR4 in the endoplasmic reticulum, preferentially mediating their proteosomal degradation to permit FGF19-mediated signaling through active forms of FGFR4.38 The current study does not identify the exact mechanism by which KLB promotes FGF19 signaling; however, our model is consistent with this newly described intracellular effect, as well as with knockout mouse data showing that loss of KLB function leads to BA malabsorption and diarrhea.39
We detected significant interactions in determination of colonic transit between KLB rs17618244 and 2 of the 3 non-synonymous coding SNPs in FGFR4 included in this study (Fig. 3). Such interactions are biologically plausible because of well-documented biochemical interactions between the KLB and FGFR4 proteins in the literature.34, 38, 40 These interactions achieved statistical significance even after correction for multiple comparisons. We chose not to assess for other genetic interactions between multiple loci in the absence of biochemical plausibility. Searching for such interactions that did not have a biochemical basis would be vulnerable to lack of statistical power due to relatively smaller sample sizes that are reduced further by multiple genotype combinations. Confirmation of the biological significance of the specific interactions detected between KLB and FGFR4 gene variants may require future replication in an independent sample population and demonstration of functional changes in KLB-FGFR4 protein interactions by the specific genotype combinations.
IBS-D is the clinical phenotype primarily responsible for the association between KLB rs17618244 and colonic transit in the overall cohort. Walters et al. provided evidence that impaired FGF19 feedback inhibition of BA synthesis plays a role in BA diarrhea.12 Our study builds on these observations, further highlighting the FGF19 pathway’s importance by identification of specific genetic variants in the FGFR4-KLB complex that influence colonic transit in IBS-D.
Our study has several strengths. First, we used scintigraphic colonic transit, a well-established and validated intermediate phenotype in IBS, for evaluating genotype effects. Our prior studies in 287 patients with IBS show that colonic transit is accelerated at 24 hours in ~30% of patients with IBS-D23, 41 and an additional 16% of IBS-D patients had accelerated colonic transit at 48 hours.41 Second, we corrected our statistical analysis for multiple comparisons with the conservative Bonferroni method to minimize the probability of type I errors. Third, the difference in colonic transit between genotype groups is of a magnitude likely to be of clinical significance. Specifically, the GG genotype in IBS-D confers a mean acceleration in GC24 of 0.74 which is large enough to be associated with discernibly looser stool form and increased stool frequency.25 Fourth, we established functional significance of KLB Arg728Gln by showing difference in protein stability between the 2 variants. Lastly, KLB Arg728’s association with accelerated colonic transit and decreased protein stability is biologically plausible, based on current paradigms of BA homeostasis and effects of BA on colonic function. Thus, the G allele results in reduced KLB protein stability, reducing FGF19 feedback inhibition of BA synthesis, and thus resulting in increased BA delivery to the gut and exposure of the colon to the effects of BA, resulting in accelerated transit.
There are some limitations in this study. First, we assessed genotype-phenotype associations using dominant genetic models and did not appraise relationships based on copy numbers of minor A allele, in order to preserve adequate statistical power with the available cohort sample size. Second, our novel results require replication, ideally in different populations. However, since scintigraphic colonic transit studies are only conducted at few centers, this replication would require significant time and resources. Third, we have not directly ascertained whether association of the KLB and FGFR4 gene variants with colonic transit is mediated by colonic exposure to different levels of BAs in IBS-D. Fourth, we did not measure BA absorptive capacity in our cohort. Future studies are needed to explore the role of these gene polymorphisms in BAM in the context of the overlap of BAM with IBS-D.
We conclude that genetic variants in proteins important for regulating BA synthesis may influence colonic transit in IBS-D. The change in protein stability between the two alleles of KLB rs17618244 establishes this SNP’s function and suggests a potential molecular mechanism by which this genetic variant may regulate colonic transit via changes in rate of bile acid synthesis. KLB rs17618244’s effects on colonic transit in IBS-D are modulated by two genetic variants in FGFR4, suggesting that the relationship between genetics of BA synthesis and regulation of colonic transit may be complex and involve the interaction of multiple loci in distinct genes. In summary, a relatively simple genotyping assay for KLB SNP rs17618244 may uncover potential derangements in bile acid homeostasis and facilitate identification of patients with IBS-D for whom more specific therapy with bile acid sequestrants would be indicated.
Acknowledgments
Grant support: Dr. Camilleri’s research in IBS is supported by NIH grants R01-DK079866 and 1RC1-DK086182.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have no disclosures.
REFERENCES
- 1.Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology. 2006;130:1377–90. doi: 10.1053/j.gastro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Saito YA, Mitra N, Mayer EA. Genetic approaches to functional gastrointestinal disorders. Gastroenterology. 2010;138:1276–85. doi: 10.1053/j.gastro.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camilleri M. Genetics and irritable bowel syndrome: from genomics to intermediate phenotype and pharmacogenetics. Dig Dis Sci. 2009;54:2318–24. doi: 10.1007/s10620-009-0903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eusufzai S. Bile acid malabsorption in patients with chronic diarrhoea. Scand J Gastroenterol. 1993;28:865–8. doi: 10.3109/00365529309103126. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Banares F, Esteve M, Salas A, Alsina M, Farre C, Gonzalez C, Buxeda M, Forne M, Rosinach M, Espinos JC, Maria Viver J. Systematic evaluation of the causes of chronic watery diarrhea with functional characteristics. Am J Gastroenterol. 2007;102:2520–8. doi: 10.1111/j.1572-0241.2007.01438.x. [DOI] [PubMed] [Google Scholar]
- 6.Sciarretta G, Furno A, Morrone B, Malaguti P. Absence of histopathological changes of ileum and colon in functional chronic diarrhea associated with bile acid malabsorption, assessed by SeHCAT test: a prospective study. Am J Gastroenterol. 1994;89:1058–61. [PubMed] [Google Scholar]
- 7.Wedlake L, A’Hern R, Russell D, Thomas K, Walters JR, Andreyev HJ. Systematic review: the prevalence of idiopathic bile acid malabsorption as diagnosed by SeHCAT scanning in patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2009;30:707–17. doi: 10.1111/j.1365-2036.2009.04081.x. [DOI] [PubMed] [Google Scholar]
- 8.Wildt S, Norby Rasmussen S, Lysgard Madsen J, Rumessen JJ. Bile acid malabsorption in patients with chronic diarrhoea: clinical value of SeHCAT test. Scand J Gastroenterol. 2003;38:826–30. doi: 10.1080/00365520310004461. [DOI] [PubMed] [Google Scholar]
- 9.Zieve FJ, Kalin MF, Schwartz SL, Jones MR, Bailey WL. Results of the glucose-lowering effect of WelChol study (GLOWS): a randomized, double-blind, placebo-controlled pilot study evaluating the effect of colesevelam hydrochloride on glycemic control in subjects with type 2 diabetes. Clin Ther. 2007;29:74–83. doi: 10.1016/j.clinthera.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Odunsi-Shiyanbade ST, Camilleri M, McKinzie S, Burton D, Carlson P, Busciglio IA, Lamsam J, Singh R, Zinsmeister AR. Effects of chenodeoxycholate and a bile acid sequestrant, colesevelam, on intestinal transit and bowel function. Clin Gastroenterol Hepatol. 2009;8:159–65. doi: 10.1016/j.cgh.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009 doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters JR, Tasleem AM, Omer OS, Brydon WG, Dew T, le Roux CW. A new mechanism for bile acid diarrhea: defective feedback inhibition of bile acid biosynthesis. Clin Gastroenterol Hepatol. 2009;7:1189–94. doi: 10.1016/j.cgh.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 13.Oelkers P, Kirby LC, Heubi JE, Dawson PA. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2) J Clin Invest. 1997;99:1880–7. doi: 10.1172/JCI119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montagnani M, Love MW, Rossel P, Dawson PA, Qvist P. Absence of dysfunctional ileal sodium-bile acid cotransporter gene mutations in patients with adult-onset idiopathic bile acid malabsorption. Scand J Gastroenterol. 2001;36:1077–80. doi: 10.1080/003655201750422693. [DOI] [PubMed] [Google Scholar]
- 15.Ito S, Kinoshita S, Shiraishi N, Nakagawa S, Sekine S, Fujimori T, Nabeshima YI. Molecular cloning and expression analyses of mouse betaklotho, which encodes a novel Klotho family protein. Mech Dev. 2000;98:115–9. doi: 10.1016/s0925-4773(00)00439-1. [DOI] [PubMed] [Google Scholar]
- 16.Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–20. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–31. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 18.Delgado-Aros S, Camilleri M, Garcia MA, Burton D, Busciglio I. High body mass alters colonic sensory-motor function and transit in humans. Am J Physiol Gastrointest Liver Physiol. 2008;295:G382–8. doi: 10.1152/ajpgi.90286.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andresen V, Camilleri M, Kim HJ, Stephens DA, Carlson PJ, Talley NJ, Saito YA, Urrutia R, Zinsmeister AR. Is there an association between GNbeta3-C825T genotype and lower functional gastrointestinal disorders? Gastroenterology. 2006;130:1985–94. doi: 10.1053/j.gastro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Camilleri CE, Carlson PJ, Camilleri M, Castillo EJ, Locke GR, 3rd, Geno DM, Stephens DA, Zinsmeister AR, Urrutia R. A study of candidate genotypes associated with dyspepsia in a U.S. community. Am J Gastroenterol. 2006;101:581–92. doi: 10.1111/j.1572-0241.2006.00481.x. [DOI] [PubMed] [Google Scholar]
- 21.Camilleri M, Carlson P, Zinsmeister AR, McKinzie S, Busciglio I, Burton D, Zaki EA, Boles RG. Mitochondrial DNA and gastrointestinal motor and sensory functions in health and functional gastrointestinal disorders. Am J Physiol Gastrointest Liver Physiol. 2009;296:G510–6. doi: 10.1152/ajpgi.90650.2008. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, Camilleri M, Carlson PJ, Cremonini F, Ferber I, Stephens D, McKinzie S, Zinsmeister AR, Urrutia R. Association of distinct alpha(2) adrenoceptor and serotonin transporter polymorphisms with constipation and somatic symptoms in functional gastrointestinal disorders. Gut. 2004;53:829–37. doi: 10.1136/gut.2003.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manabe N, Wong BS, Camilleri M, Burton D, McKinzie S, Zinsmeister AR. Lower functional gastrointestinal disorders: evidence of abnormal colonic transit in a 287 patient cohort. Neurogastroenterol Motil. 2010;22:293–e82. doi: 10.1111/j.1365-2982.2009.01442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao AS, Wong BS, Camilleri M, Odunsi-Shiyanbade ST, McKinzie S, Ryks M, Burton D, Carlson P, Lamsam J, Singh R, Zinsmeister AR. Chenodeoxycholate in Females With Irritable Bowel Syndrome-Constipation: A Pharmacodynamic and Pharmacogenetic Analysis. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deiteren A, Camilleri M, Bharucha AE, Burton D, McKinzie S, Rao AS, Zinsmeister AR. Performance characteristics of scintigraphic colon transit measurement in health and irritable bowel syndrome and relationship to bowel functions. Neurogastroenterol Motil. 2010;22:415–23. e95. doi: 10.1111/j.1365-2982.2009.01441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camilleri M. Scintigraphic biomarkers for colonic dysmotility. Clin Pharmacol Ther. 2010;87:748–53. doi: 10.1038/clpt.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burton DD, Camilleri M, Mullan BP, Forstrom LA, Hung JC. Colonic transit scintigraphy labeled activated charcoal compared with ion exchange pellets. J Nucl Med. 1997;38:1807–10. [PubMed] [Google Scholar]
- 28.Cremonini F, Mullan BP, Camilleri M, Burton DD, Rank MR. Performance characteristics of scintigraphic transit measurements for studies of experimental therapies. Aliment Pharmacol Ther. 2002;16:1781–90. doi: 10.1046/j.1365-2036.2002.01344.x. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–91. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Augustin O, Sanchez de Medina F. Intestinal bile acid physiology and pathophysiology. World J Gastroenterol. 2008;14:5630–40. doi: 10.3748/wjg.14.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weir BS. Hardy-Weinberg Disequilibrium. In: Weir BS, editor. Genetic data analysis II : methods for discrete population genetic data. Sinauer Associates; 1996. pp. 98–101. [Google Scholar]
- 32.Wacholder S, Rothman N, Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:513–20. [PubMed] [Google Scholar]
- 33.Wang Y, Localio R, Rebbeck TR. Evaluating bias due to population stratification in epidemiologic studies of gene-gene or gene-environment interactions. Cancer Epidemiol Biomarkers Prev. 2006;15:124–32. doi: 10.1158/1055-9965.EPI-05-0304. [DOI] [PubMed] [Google Scholar]
- 34.Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, Mohammadi M, Rosenblatt KP, Kliewer SA, Kuro-o M. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem. 2007;282:26687–95. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bampton PA, Dinning PG, Kennedy ML, Lubowski DZ, Cook IJ. The proximal colonic motor response to rectal mechanical and chemical stimulation. Am J Physiol Gastrointest Liver Physiol. 2002;282:G443–9. doi: 10.1152/ajpgi.00194.2001. [DOI] [PubMed] [Google Scholar]
- 37.Mekjian HS, Phillips SF, Hofmann AF. Colonic secretion of water and electrolytes induced by bile acids: perfusion studies in man. J Clin Invest. 1971;50:1569–77. doi: 10.1172/JCI106644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triantis V, Saeland E, Bijl N, Oude-Elferink RP, Jansen PL. Glycosylation of fibroblast growth factor receptor 4 is a key regulator of fibroblast growth factor 19-mediated down-regulation of cytochrome P450 7A1. Hepatology. 2010;52:656–66. doi: 10.1002/hep.23708. [DOI] [PubMed] [Google Scholar]
- 39.Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y, Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J Clin Invest. 2005;115:2202–8. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require Klotho beta. J Biol Chem. 2007;282:27277–84. doi: 10.1074/jbc.M704244200. [DOI] [PubMed] [Google Scholar]
- 41.Camilleri M, McKinzie S, Busciglio I, Low PA, Sweetser S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Prospective study of motor, sensory, psychologic, and autonomic functions in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2008;6:772–81. doi: 10.1016/j.cgh.2008.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]