1. Introduction
It is established that chronic alcohol ingestion results in liver injury and is a significant cause of morbidity and mortality. This is especially apparent in the United States where a significant number of annual mortalities associated with end stage liver disease are attributable to excess alcohol ingestion [1, 2]. Alcoholic liver disease (ALD) is considered to be progressive and associated with duration and quantity of alcohol consumed [3]. Upwards to 90% of individuals consuming alcohol on a daily basis develop fatty liver (steatosis) which can resolve upon cessation of alcohol consumption. Individuals who continue a daily consumption of 20–80gms of alcohol daily for extended durations of time will progress to steatohepatitis with approximately 40% of the more severely affected patients diagnosed with this inflammatory syndrome dying within 6 months. The remainder of patients who continue excessive alcohol intake on a daily basis will progress to alcoholic cirrhosis.
The onset and progression of ALD is multifactorial. It is now widely accepted that chronic alcohol ingestion initiates a pro-oxidant environment in the liver. One early report suggesting this phenomenon appeared nearly 3 decades ago describing significant elevations of the volatile product of lipid peroxidation, pentane, expired by rats treated with 18% v/v ethanol in their drinking water for 14 weeks and challenged with an acute dose of ethanol [4]. This report noted the quantity of expired pentane was inversely correlated with dietary vitamin-E and positively correlated with hepatic triglyceride content, both of which are important biological variables involved in lipid peroxidation. Since that early report, a significant body of literature has accumulated documenting the occurrence of hepatic lipid peroxidation in animal models and human subjects with ALD. Numerous attempts to quantify concentrations of lipid peroxidation products, such as malondialdehyde (MDA) or 4-hydroxynonenal (4-HNE), in blood or tissues of animals or humans consuming alcohol, have proven problematic due to the spontaneous generation of the products during sample preparation or requirement for use of sophisticated analytical instrumentation. An additional problem arises due to the chemical nature of these electrophiles, which react with cellular nucleophiles in both stable and transient-type mechanisms that can be rapidly biotransformed. The propensity of 4-HNE and other electrophilic products of lipid peroxidation to react with cellular protein nucleophiles is the premise that detection of aldehyde-modified proteins could serve as a biomarker for alcohol-induced oxidative stress. In fact, a number of reports have appeared using immunohistochemcial approaches to detect hepatic proteins modified by 4-HNE or MDA [5–11]. Taken together, these reports demonstrate that despite the highly effective antioxidant systems and redundant biotransformation capacity of liver, the chronic alcohol ingestion generates a pro-oxidative environment in the liver resulting in lipid peroxidation and the covalent modification of hepatic proteins. However, as with many diseases of oxidative stress, the central question remains concerning the association of these molecular lesions with impairment of hepatocellular function.
This report presents a review of the hepatic proteins we have identified to date that are targets for 4-HNE modification in animal models of alcohol-induced oxidative stress and how this covalent modification impacts protein function mechanistically linked to the pathobiology of ALD. In addition, new data are presented describing the modification of liver fatty acid binding protein (L-FABP) in rats chronically ingesting alcohol and the potential role of this modification in the development of steatosis.
2. Materials and Methods
2.1 Reagents
Unless otherwise specified, all chemicals were obtained from Sigma-Aldrich Chemical Co. (St Louis, MO). The synthesis of 4-HNE was as previously described with a purity documented to be greater than 99% by TLC, and UV/Vis spectrophotometry [12]. Antibodies used to detect 4-HNE modified proteins for western blotting or immunohistochemistry were produced using rabbit hosts immunized with 4-HNE-modified keyhole limpet hemocyanin [13].
2.2 Animals
All procedures involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Colorado Denver and performed in accordance with published National Institutes of Health guidelines. Animal models for ALD used for the studies described here consisted of either male Sprague-Dawley rats or male C57BL/6J mice fed the Lieber-DeCarli ethanol or control diet as described elsewhere [14, 15]. Each ethanol-fed animal was paired with a nutritional control animal that received the diet containing equivalent calories derived from maltose-dextran rather than ethanol. At the conclusion of the 60–112 day feeding period for rats and 6-week treatment for mice, the animals were anesthetized by intraperitoneal administration of pentobarbital and euthanized by exsanguination. Blood plasma was prepared for measurement of ALT activity as an index of liver injury and liver tissue was processed for preparation of paraffin-embedded sections and subcellular fractions as described in previous reports [16, 17]. All subcellular fractions were supplemented with a cocktail of protease inhibitors and stored at −80 °C until analysis.
2.3 Two-Dimensional Electrophoresis, In-gel Digest and Mass Spectral Analysis
Electrophoretic separation of rat or mouse hepatic proteins by two-dimensional electrophoresis and spot harvesting were performed as described previously [14–16, 18]. Hepatic proteins resolved by electrophoresis and modified by 4-HNE were detected using antibodies generated against 4-HNE adducts. Harvested proteins were routinely digested overnight at 25 °C in the presence of 100mM ammonium bicarbonate and 0.65 µg sequencing grade trypsin (Promega Corporation). The resulting peptides were subjected to liquid chromatography and tandem mass spectrometry (LC-MS/MS) using an Agilent 1100 Series LC-ESI-MSD Trap as outlined previously [14–16]. The MS-MS ion searches were performed on deconvoluted spectra using MASCOT. In some cases samples were subjected to analysis using an ABI 4800 MALDI-TOF/TOF instrument.
2.4 4-Hydroxynonenal Modification of Recombinant Proteins
The identification of 4-HNE modified proteins in subcellular fractions necessitated in vitro experiments to determine the effects of 4-HNE on protein function. This was accomplished by exposing the respective recombinant proteins to physiologically relevant concentrations of 4-HNE, which have been found to range from 0.1 to 100 µM in soluble fractions of liver and have also been estimated to be 3–10 mM in membrane fractions of hepatocytes [19–22]. At the conclusion of the incubation with 4-HNE, residual 4-HNE was removed and the effect on protein activity was determined. By subjecting each recombinant protein to a range of 4-HNE concentrations it was possible to identify the amino acid targets of modification as well as determine which residues are the most susceptible to modification.
3.0 Results and Discussion
3.1 Profiles of Ethanol Induced Liver Injury In Rats and Mice
The ethanol administration paradigms used in our studies for rats and mice [14–16] provide adequate nutrition for maintenance of body weight with isocaloric control animals gaining weight during the treatment period. Animals receiving ethanol do not display significant losses in body weight. Administration of the ethanol-containing diets reproducibly result in statistically significant, 1.5- to 2-fold increases in plasma ALT activities over those observed in isocaloric control animals indicating mild ethanol-induced hepatocellular injury. Likewise, rats and mice receiving ethanol display significantly increased hepatic triglyceride contents upwards to 30% and greater than those of isocaloric control animals, with the increased hepatic lipid accumulation in ethanol-treated animals resulting in significant increases in liver to body weight ratios. The ethanol-induced lipid deposits are characteristically present as a mixture of macro-and microsteatosis. Hepatic cytochrome P-4502E1 activities are consistently increased 2- to 3-fold in ethanol-treated animals as compared to isocaloric controls suggesting increased reactive oxygen species (ROS) production. The appearance of hepatic lesions indicative of inflammation is seldom observed in the ethanol-treated animals. Likewise, no signs of ethanol-induced hepatic fibrosis have been observed. Taken together, these parameters are consistent with those displayed by individuals during the early stages of ethanol-induced liver injury characterized by sustained increases in plasma ALT activities and hepatomegaly attributable to hepatic triglyceride accumulation.
3.2 Detection and Characterization of Hepatic Proteins Modified by 4-HNE
As summarized in Table 1, our previous proteome-wide scans for hepatic proteins modified by 4-HNE in rats and mice ingesting ethanol have identified protein targets that have potential mechanistic links with the initiation and/or progression of ALD. For example, 4-HNE modifications on Hsp70, Hsp90 and PDI were found to modify enzymatic activity; providing a mechanism for altered protein processing consistent with alcohol induced oxidative stress [14, 16, 18]. It is becoming more evident that Hsp70 and Hsp90 play important and interactive roles as cellular protein chaperones [23, 24] demonstrated by our in vitro assays [14, 16] that indicate significant impairment of protein folding and refolding, respectively. Similarly, our observation that 4-HNE modification of the endoplasmic reticulum protein, PDI, results in a decreased ability of this protein to undergo critical disulfide exchange with target proteins – suggesting a mechanism that would further impair hepatocellular protein processing. As detailed in recent comprehensive reviews [25–27], it is now recognized that sustained oxidative stress and accumulation of unfolded proteins resulting from chronic alcohol ingestion or other metabolic disorders such as nonalcoholic liver disease and insulin resistance, is linked to the phenomenon of endoplasmic reticulum stress. Stressing of the ER initiates complex signaling cascades proposed to be adaptive and pro-survival through coordination of catabolic and metabolic pathways of glucose and lipid metabolism [25, 27]. Interestingly, a prominent response of hepatocytes to ER stress is increased lipid synthesis and storage. This observed pathology is similar to that of chronic ethanol consumption or metabolic disorders such as nonalcoholic liver disease and insulin resistance associated with obesity. Thus, our previous observations that covalent modification of protein chaperones, such as Hsp90, Hsp70 and PDI by 4-HNE, in conjunction with corresponding decreases in their enzymatic activities, provide important information concerning the mechanisms of alcohol-induced liver injury and lipid accumulation. As noted in Table 1, 4-HNE modification of hepatic Erk1/2 was previously observed in rats chronically ingesting alcohol [28, 29]. Interestingly, identification of Erk1/2 as a target for 4-HNE modification originated from the observation of the immunohistochemcial co-localization of hepatic total Erk1/2 protein and 4-HNE-protein adducts in rats chronically ingesting alcohol [29]. Additional experiments using hepatocytes isolated from ethanol-treated rats revealed increased 4-HNE-Erk1/2 adduct formation was associated with the loss of constitutive Erk1/2 phosphorylation. In vitro experiments using hepatocytes exposed to sub-cytotoxic concentrations of 4-HNE confirmed the inhibition of Erk1/2 phosphorylation mechanistically linked with impaired Elk-AP-1 signaling [28]. Using recombinant Erk, the results of this latter study identified His 178 within the phosphorylation loop as the site for 4-HNE modification suggesting a potential mechanism for impaired Erk phosphorylation. The observation of impaired Erk1/2 signaling through AP-1 transcriptional activity provides mechanistic information implicating another mechanism whereby the pro-oxidant effects of chronic ethanol ingestion impact liver function.
Table 1.
Summary of Proteins Modified by 4-HNE in Rat and Mouse Models of Alcoholic Liver Disease
| Protein | Animal Model |
Target Sequence of Modificationa |
Effect of Modification on Protein Function |
Reference |
|---|---|---|---|---|
| Hsp70 | Rat | Impairment of ATPase activity and protein folding | 16 | |
| Hsp90 | Rat | Impairment of refolding of damaged proteins | 14 | |
| PDI | Rat | Dysfunctional restoration of mismatched intraprotein disulfide bonds | 18 | |
| Erk 1/2 | Rat | Inhibition of Erk1/2 phosphorylation and impaired hepatocellular regeneration | 20, 21 | |
| Prx6 | Mouse | Modification of solvent accessible thiol potentially restricting access to active site Cys 47 | 15, 19 |
Numbers below bolded amino acid signify sequence number of modified target residue
Another hepatic protein target of 4-HNE modification in a mouse model of ALD previously identified in our laboratory is peroxiredoxin 6 (Prx6) [15, 30] (Table 1). Although the specific substrates of this protein remain elusive, Prx6 has been proposed to reduce a variety of peroxides and is thus classified as an antioxidant enzyme. Studies from our laboratory indicate that Prx6 is among the more resistant enzymes to modification by 4-HNE in that concentrations of 4-HNE in excess of 200µM are required to inhibit activity by 50% [30]. Our observation that mice deficient in Prx6 are not predisposed to liver injury resulting from chronic alcohol administration indicates that this antioxidant enzyme does not play an important protective role during the early stages of ALD [31].
Taken together, the proteins listed in Table 1 provide important information indicating that a number of hepatocellular proteins are targets for 4-HNE modification resulting from chronic ethanol ingestion. The information in Table 1 indicates that during the early stages of ALD, hepatocytes are challenged with impaired protein folding and processing through dysregulation of Hsp70, Hsp90 and PDI. Given the pathologic consequences of sustained ER stress [25, 27], decreased chaperone-dependent processes may provide a mechanism for the observed increase in lipid synthesis, triglyceride accumulation, and enhanced oxidative stress – all of which are documented to occur during this disease. Likewise, our previous studies suggest impaired hepatocellular proliferation through impairment of Erk1/2 signaling. Of the limited number of proteins we have identified as targets for 4-HNE modification, it is becoming more apparent that, like many diseases of oxidative stress, early ALD is a multifactorial disorder involving the dysregulation of numerous hepatocellular systems that are likely to become more compromised and numerous as the disease progress.
3.3 Identification of 4-HNE Modified Liver Fatty Acid Binding Protein in a Rat Model of ALD
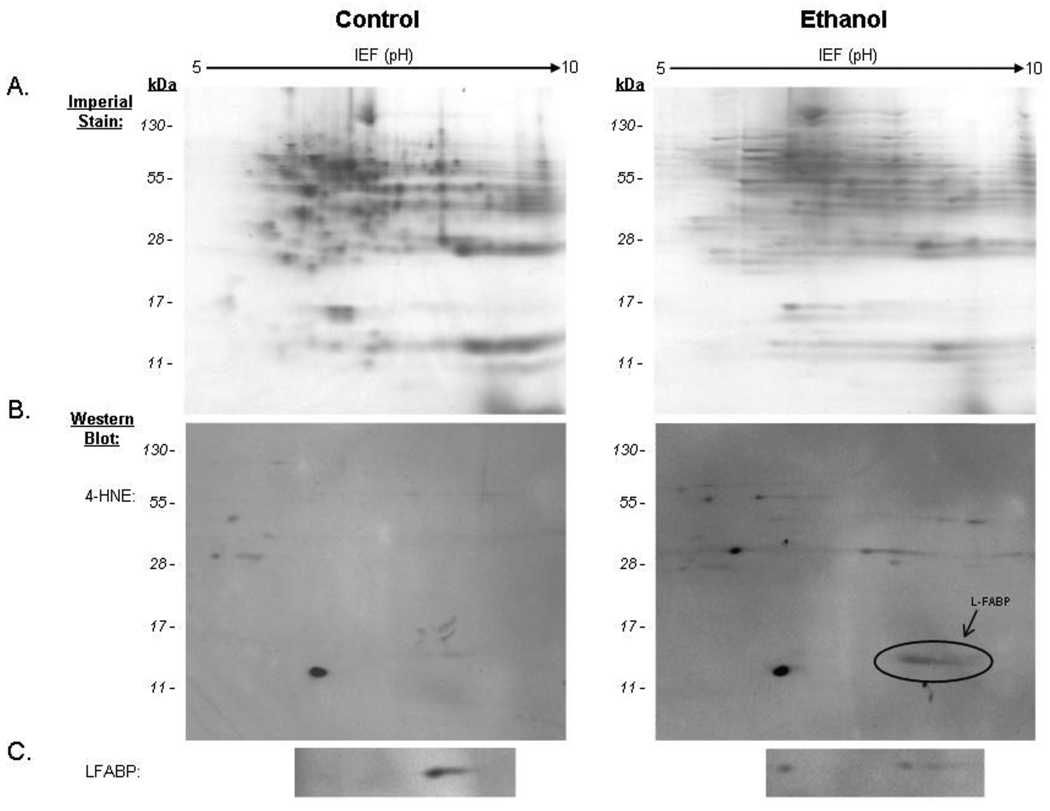
More recently, the experimental approaches described in the previous paragraphs have revealed an additional hepatic protein that is a target for 4-HNE in rats chronically ingesting alcohol. The 2D Imperial-stained gel presented in Figure 1A represents a proteome-wide scan of hepatic 4-HNE modified proteins in cytosolic fractions prepared from rats treated for 16 weeks with either the isocaloric control or alcohol-containing diet. The immunoblot presented in the left panel of Figure 1B displays the presence of a limited number of 4-HNE immunopositive proteins in control rats whereas the representative blot of an ethanol-treated animal in right panel contains a circle insert identifying a unique population of proteins with an approximate molecular weight of 15,000. The in-gel digest of this immunopositive spot from the ethanol-treated animal followed by LC-MS/MS analysis resulted in identification of 6 peptides listed in Table 2 indicating a significant MOWSE score (p<0.05) matching these of these peptides to liver fatty acid binding protein (L-FABP). It is noteworthy that one peptide representing residues 47–90 was not detected in the tryptic digests of the immunopositive spots obtained from the 2D gels prepared from ethanol treated rats. Therefore, additional conformation of protein identify was obtained by probing paired blots with an antibody against L-FABP (Figure 1C).
Figure 1.
Two-dimensional electrophoresis and immunoblot detection of 4-HNE-modified cytosolic proteins from isocaloric-control and ethanol-fed rat liver. L-FABP, indicated by the arrow, was identified following in-gel digest and LC-MS/MS analysis interfaced with the MASCOT search engine. Probes against L-FABP are shown for the control and ethanol-fed rats.
Table 2.
Summary of tryptic digest from rat L-FABP identified by LC-MS/MS
| Protein ID* | Mass | pI | Amino Acid Sequence |
Peptides Matched |
|---|---|---|---|---|
| Rat LFABP | 14,192 | 6.67 | 7–20 | YQVQSQENFEPFMK |
| 21–31 | AMGLPEDLIQK | |||
| 37–46 | LTTTYGSK | |||
| 91–96 | MVTTFK | |||
| 100–121 | SVTEFNGDTITNTMLGDIVYK |
Significant protein match based on -10* LogP MOWSE score of 84 (p< 0.05). Minimal score of significance is 54
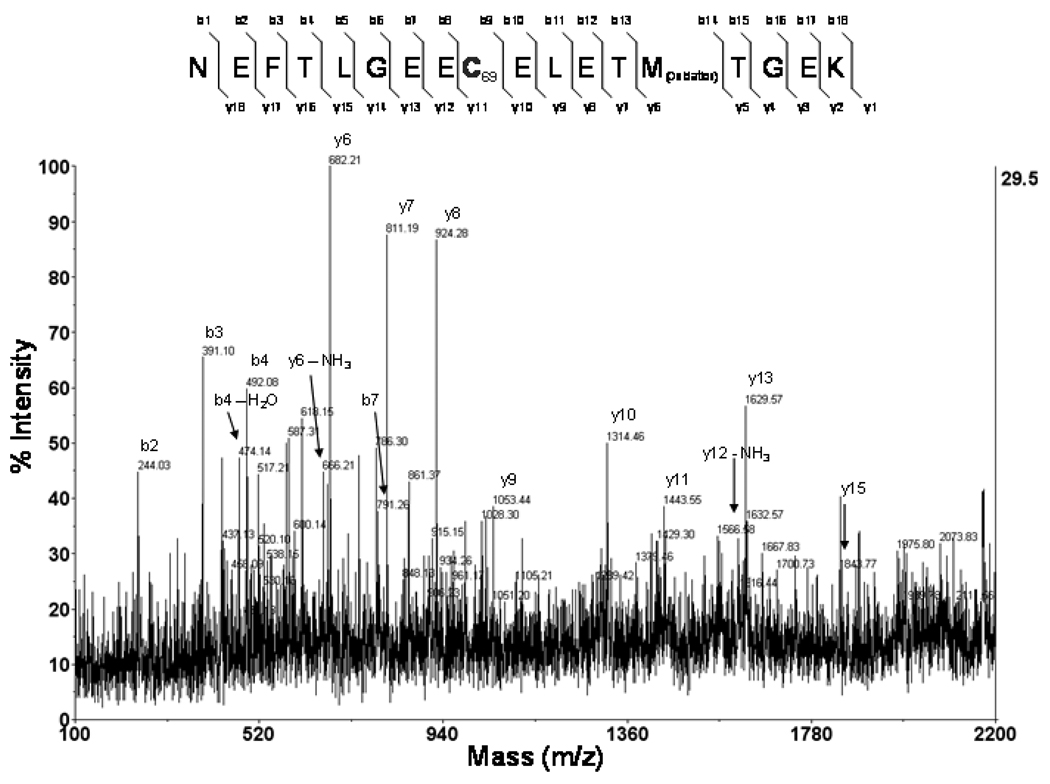
Additional experiments were performed using recombinant mouse L-FABP protein to begin identifying nucelophilic amino acids that are potential targets for 4-HNE modification. This was accomplished by incubating of recombinant mouse L-FABP with 117.03µM 4-HNE, representing a 10-fold excess of this aldehyde. Modified protein was run on SDS-PAGE, stained with Coomassie blue, digested with trypsin and analyzed using MALDI-TOF/TOF Mass Spectrometry. The spectra presented in Figure 2 and data presented in Table 3 confirmed modification of a peptide corresponding to amino acids 61–68. The mass of this peptide (m/z 2231.91) represents a shift equivalent to the mass of 4-HNE (156 Da) as compared to the parent peptide. It is noteworthy that when 4-HNE-modified L-FABP was reduced with 10mM NaBH4, the mass of the peptide was increased, as predicted, to m/z 2233.96 (total mass shift of 158 Da). The spectra of y and b fragment ions presented in Figure 2 confirm modification of Cys 69 as a stable adduct of the primary residues of L-FABP modified by 4-HNE. Interestingly, molecular modeling simulations of L-FABP [32] highlight a possible portal at the bottom of the beta-barrel structure localized near Cys 69 that may regulate the exit of bound hydropholic ligands – suggesting that modification of this residue may alter release and exit of lipid ligands. At present, the functional consequences of L-FABP modification by 4-HNE are currently under investigation in our laboratory. Previous reports have described decreased affinity of adipocyte FABP (A-FABP) for the fluorescent ligand 1-Anilinonaphthalene-8-Sulfonic Acid following 4-HNE modification [33] whereas 4-HNE modification of epithelial FABP (E-FABP) reportedly stabilizes the protein when subjected to chemical denaturation with guanidine hydrochloride [34]. Therefore, the overall impact of 4-HNE modification on the functional properties of L-FABP are difficult to predict and complicated by structural characteristics and complexities of L-FABP as compared to adipocyte and epithelial fatty acid binding proteins.
Figure 2.
MS/MS Spectra of 4-HNE modified Cys69 of L-FABP. MALDI-TOF/TOF Mass Spectrometry was used to identify sites of 4-HNE modification on digested peptides of recombinant mouse L-FABP after incubation with a 10-fold molar excess of the aldehyde. A mass shift was detected on peptide NEFTLGEECELETMTGEK with a total mass of 2233.8 Da. Relative y and b ions show the presence of the 4-HNE adduct on the nuclophillic residue Cys69 and oxidation of Met74.
Table 3.
Identification of 4-HNE adducted peptide and specific residue modified in recombinant mouse L-FABP.
| Treatment* | Peptide Identified |
Mass | Sequence** |
|---|---|---|---|
| 10X 4-HNE | 61–78 | 2231.91 | |
| 10X 4-HNE and 10 mM NHBH4 | 61–78 | 2233.96 |
Recombinant mouse L-FABP was modified with 10X molar excess of 4-HNE (117.03µM) or 10X 4-HNE followed by reduction with 10 mM NHBH4
Numbers below bolded amino acid signify sequence number of modified target residue
The characteristics of the L-FABP protein are presented in reviews elsewhere [35, 36]. Briefly, L-FABP represents 2–5% of the cytosolic protein present in liver and belongs to a larger family of intracellular lipid binding proteins (iLBP). Cellular localization of L-FABP also includes the nucleus. Relevant to the data presented here, L-FABP is localized in the organ that is highly active in fatty acid metabolism. This observation makes L-FABP extremely relevant to the observed lipid accumulation in early stages of ALD. This protein binds a number of chemically diverse hydrophobic ligands including saturated and unsaturated long-chain fatty acids as well as their CoA thioesters. Additional ligands include lysophosphatidic acid, bile salts and at least two forms of peroxisome proliferators. A unique characteristic of L-FABP is its ability to simultaneously bind 2 ligands via 2 different binding sites within the cavity of the protein, different from the remaining members of the iLBP family that are capable of only binding a single ligand. L-FABP binds fatty acids as well as certain peroxisome proliferators and translocates to the nucleus, indicating that the protein is important in transporting not only lipids but also in activating nuclear receptors. Our observation that L-FABP is modified by 4-HNE in an animal model of ALD is novel and consistent with one other report of a carbonylated iLBP present in adipose tissue of mice ingesting a high-fat diet [33]. Interestingly, these investigators reported that in vitro modification of the adipose fatty acid binding protein (A-FABP) by 4-HNE significantly impaired fatty acid binding kinetics; suggesting that sustained cellular oxidative stress may result in impaired fatty acid processing. Studies in our laboratory are presently underway to determine additional sites of 4-HNE modification on L-FABP and the corresponding effects on ligand binding parameters.
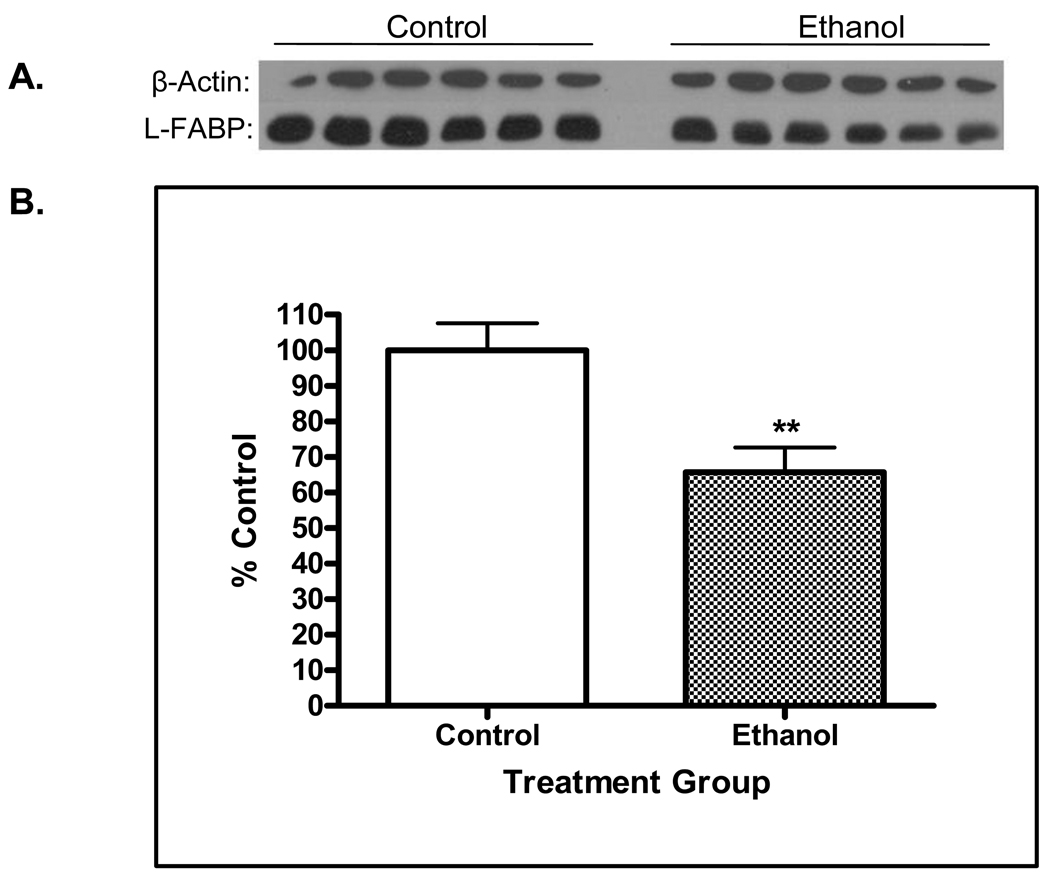
The data presented in Figure 3 suggest additional implications associated with 4-HNE modification on L-FABP. Specifically, it was noted in the 2D blots for L-FABP that the immunostaining was much more dense in the control animals (Figure 1C). Therefore, additional experiments were conducted using western blots for L-FABP on standard 1-dimensional electrophoresis. It is evident from Figure 3A and B that chronic alcohol ingestion resulted in an approximated 30% decrease (p<0.01) of L-FABP protein. These results indicate that 4-HNE modification may result in enhanced cellular clearance of the protein. An additional possibility is chronic ethanol ingestion results in translocation of L-FABP to other cellular compartments such as the nucleus. Both of these possibilities are currently under investigation in our laboratory. These results suggest that the modification of L-FABP by 4-HNE may have significant mechanistic implications for the development and progression of steatosis during the early stages of the disease.
Figure 3.
A. Western blot analysis for L-FABP in cytosolic fractions prepared from control and ethanol-treated rats. Protein load was normalized using β-actin. B. Densitometetric quantification detected an approximate 30% decrease (p<0.01, n = 6) decrease of L-FABP in rats chronically ingesting ethanol.
Concluding remarks
We have used proteomic approaches to uncover molecular mechanisms of hepatic oxidative stress induced by chronic ethanol ingestion. Using LC-MS/MS and MALDI-TOF/TOF spectroscopy, we have identified hepatic proteins modified with 4-HNE in rat and mouse models of ALD. Identification of Hsp70, Hsp90 and PDI as in vivo targets for modification by 4-HNE suggest that impairment of their activity could contribute to the initiating events of ER stress and subsequent hepatic lipid accumulation. Likewise, identification of Erk1/2 as an in vivo target for 4-HNE modification led to identification of the specific site of adduction explaining the observed dysregulation of phosphorylation events potentially associated with impaired liver regeneration. The recent identification of 4-HNE modified L-FABP in a rat model of ALD is providing important information concerning additional mechanisms contributing to dysregulation of lipid homeostasis associated with the early stages of ALD.
Acknowledgments
This work was supported in part by Grants NIH/NIAAA R37AA09300, NIH/NIAAA R01 DK074487-01 and NIH/NIAAA NRSA (RLS) 5 F31 AA018898-02.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Arteel G, Marsano L, Mendez C, Bentley F, McClain CJ. Advances in alcoholic liver disease. Best Pract Res Clin Gastroenterol. 2003;17(4):625–647. doi: 10.1016/s1521-6918(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 2.Bergheim I, McClain CJ, Arteel GE. Treatment of alcoholic liver disease. Dig Dis. 2005;23(3–4):275–284. doi: 10.1159/000090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucey MR, Mathurin P, Morgan TR. Alcoholic hepatitis. N Engl J Med. 2009;360(26):2758–2769. doi: 10.1056/NEJMra0805786. [DOI] [PubMed] [Google Scholar]
- 4.Litov RE, Gee DL, Downey JE, Tappel AL. The role of peroxidation during chronic and acute exposure to ethanol as determined by pentane expiration in the rat. Lipids. 1981;16(1):52–63. doi: 10.1007/BF02534921. [DOI] [PubMed] [Google Scholar]
- 5.Kono H, Arteel GE, Rusyn I, Sies H, Thurman RG. Ebselen prevents early alcohol-induced liver injury in rats. Free Radic Biol Med. 2001;30(4):403–411. doi: 10.1016/s0891-5849(00)00490-1. [DOI] [PubMed] [Google Scholar]
- 6.Niemela O. Aldehyde-protein adducts in the liver as a result of ethanol-induced oxidative stress. Front Biosci. 1999;4:D506–D513. doi: 10.2741/niemela. [DOI] [PubMed] [Google Scholar]
- 7.Niemela O, Parkkila S, Yla-Herttuala S, Halsted C, Witztum JL, Lanca A, Israel Y. Covalent protein adducts in the liver as a result of ethanol metabolism and lipid peroxidation. Lab Invest. 1994;70(4):537–546. [PubMed] [Google Scholar]
- 8.Niemela O, Parkkila S, Yla-Herttuala S, Villanueva J, Ruebner B, Halsted CH. Sequential acetaldehyde production, lipid peroxidation, and fibrogenesis in micropig model of alcohol-induced liver disease. Hepatology. 1995;22(4 Pt 1):1208–1214. [PubMed] [Google Scholar]
- 9.Paradis V, Kollinger M, Fabre M, Holstege A, Poynard T, Bedossa P. In situ detection of lipid peroxidation by-products in chronic liver diseases. Hepatology. 1997;26(1):135–142. doi: 10.1053/jhep.1997.v26.pm0009214462. [DOI] [PubMed] [Google Scholar]
- 10.Sampey BP, Korourian S, Ronis MJ, Badger TM, Petersen DR. Immunohistochemical characterization of hepatic malondialdehyde and 4-hydroxynonenal modified proteins during early stages of ethanol-induced liver injury. Alcohol Clin Exp Res. 2003;27(6):1015–1022. doi: 10.1097/01.ALC.0000071928.16732.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukamoto H, Horne W, Kamimura S, Niemela O, Parkkila S, Yla-Herttuala S, Brittenham GM. Experimental liver cirrhosis induced by alcohol and iron. J Clin Invest. 1995;96(1):620–630. doi: 10.1172/JCI118077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15(11):1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 13.Hartley DP, Kroll DJ, Petersen DR. Prooxidant-initiated lipid peroxidation in isolated rat hepatocytes: detection of 4-hydroxynonenal- and malondialdehyde-protein adducts. Chem Res Toxicol. 1997;10(8):895–905. doi: 10.1021/tx960181b. [DOI] [PubMed] [Google Scholar]
- 14.Carbone DL, Doorn JA, Kiebler Z, Ickes BR, Petersen DR. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J Pharmacol Exp Ther. 2005;315(1):8–15. doi: 10.1124/jpet.105.088088. [DOI] [PubMed] [Google Scholar]
- 15.Roede JR, Stewart BJ, Petersen DR. Decreased expression of peroxiredoxin 6 in a mouse model of ethanol consumption. Free Radic Biol Med. 2008;45(11):1551–1558. doi: 10.1016/j.freeradbiomed.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 16.Carbone DL, Doorn JA, Kiebler Z, Sampey BP, Petersen DR. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem Res Toxicol. 2004;17(11):1459–1467. doi: 10.1021/tx049838g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little RG, 2nd, Petersen DR. Subcellular distribution and kinetic parameters of HS mouse liver aldehyde dehydrogenase. Comp Biochem Physiol C. 1983;74(2):271–279. doi: 10.1016/0742-8413(83)90101-9. [DOI] [PubMed] [Google Scholar]
- 18.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18(8):1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 19.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. 4-hydroxynonenal: a membrane lipid oxidation product of medicinal interest. Med Res Rev. 2008;28(4):569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 20.Benedetti A, Comporti M, Fulceri R, Esterbauer H. Cytotoxic aldehydes originating from the peroxidation of liver microsomal lipids. Identification of 4,5-dihydroxydecenal. Biochim Biophys Acta. 1984;792(2):172–181. doi: 10.1016/0005-2760(84)90219-4. [DOI] [PubMed] [Google Scholar]
- 21.Benedetti A, Esterbauer H, Ferrali M, Fulceri R, Comporti M. Evidence for aldehydes bound to liver microsomal protein following CCl4 or BrCCl3 poisoning. Biochim Biophys Acta. 1982;711(2):345–356. doi: 10.1016/0005-2760(82)90044-3. [DOI] [PubMed] [Google Scholar]
- 22.Benedetti A, Fulceri R, Ferrali M, Ciccoli L, Esterbauer H, Comporti M. Detection of carbonyl functions in phospholipids of liver microsomes in CCl4- and BrCCl3-poisoned rats. Biochim Biophys Acta. 1982;712(3):628–638. doi: 10.1016/0005-2760(82)90292-2. [DOI] [PubMed] [Google Scholar]
- 23.Pratt WB, Morishima Y, Peng HM, Osawa Y. Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Exp Biol Med (Maywood) 235(3):278–289. doi: 10.1258/ebm.2009.009250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 25.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 140(6):900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplowitz N, Than TA, Shinohara M, Ji C. Endoplasmic reticulum stress and liver injury. Semin Liver Dis. 2007;27(4):367–377. doi: 10.1055/s-2007-991513. [DOI] [PubMed] [Google Scholar]
- 27.Tsang KY, Chan D, Bateman JF, Cheah KS. In vivo cellular adaptation to ER stress: survival strategies with double-edged consequences. J Cell Sci. 123(Pt 13):2145–2154. doi: 10.1242/jcs.068833. [DOI] [PubMed] [Google Scholar]
- 28.Sampey BP, Carbone DL, Doorn JA, Drechsel DA, Petersen DR. 4-Hydroxy-2-nonenal adduction of extracellular signal-regulated kinase (Erk) and the inhibition of hepatocyte Erk-Est-like protein-1-activating protein-1 signal transduction. Mol Pharmacol. 2007;71(3):871–883. doi: 10.1124/mol.106.029686. [DOI] [PubMed] [Google Scholar]
- 29.Sampey BP, Stewart BJ, Petersen DR. Ethanol-induced modulation of hepatocellular extracellular signal-regulated kinase-1/2 activity via 4-hydroxynonenal. J Biol Chem. 2007;282(3):1925–1937. doi: 10.1074/jbc.M610602200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roede JR, Carbone DL, Doorn JA, Kirichenko OV, Reigan P, Petersen DR. In vitro and in silico characterization of peroxiredoxin 6 modified by 4-hydroxynonenal and 4-oxononenal. Chem Res Toxicol. 2008;21(12):2289–2299. doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- 31.Roede JR, Orlicky DJ, Fisher AB, Petersen DR. Overexpression of peroxiredoxin 6 does not prevent ethanol-mediated oxidative stress and may play a role in hepatic lipid accumulation. J Pharmacol Exp Ther. 2009;330(1):79–88. doi: 10.1124/jpet.109.152983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long D, Mu Y, Yang D. Molecular dynamics simulation of ligand dissociation from liver fatty acid binding protein. PLoS One. 2009;4(6):e6081. doi: 10.1371/journal.pone.0006081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimsrud PA, Picklo MJ, Sr., Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6(4):624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Bennaars-Eiden A, Higgins L, Hertzel AV, Kapphahn RJ, Ferrington DA, Bernlohr DA. Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J Biol Chem. 2002;277(52):50693–50702. doi: 10.1074/jbc.M209493200. [DOI] [PubMed] [Google Scholar]
- 35.Thompson J, Reese-Wagoner A, Banaszak L. Liver fatty acid binding protein: species variation and the accommodation of different ligands. Biochim Biophys Acta. 1999;1441(2–3):117–130. doi: 10.1016/s1388-1981(99)00146-8. [DOI] [PubMed] [Google Scholar]
- 36.Zimmerman AW, van Moerkerk HT, Veerkamp JH. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int J Biochem Cell Biol. 2001;33(9):865–876. doi: 10.1016/s1357-2725(01)00070-x. [DOI] [PubMed] [Google Scholar]