Abstract
Hsc82 and Hsp82, the Hsp90 family proteins of yeast, are both required for fermentative growth at 37°C. Inactivation of either of the mitochondrial AAA proteases, Yme1 or Yta10/12, allows fermentative growth of hsc82Δ or hsp82Δ strains at 37°C. Genetic evidence indicates interaction of Hsc82/Hsp82 with the Yme1 and Yta10/Yta12 complexes in promoting F1Fo-ATPase activity, with Hsc82 specifically required for F1-ATPase assembly. A previously reported mutation in Rpt3, one of the six ATPases of the proteasome, suppresses yme1Δ phenotypes and increases transcription of HSC82 but not HSP82. These genetic interactions describe a functional role for Hsp90 proteins in mitochondrial biogenesis.
Keywords: Hsp90, F1Fo-ATPase, Assembly, Yeast, AAA protease, Yme1, Yta10, Yta12
1. Introduction
Chaperones in mitochondria are associated with numerous activities, including uptake of nuclear-encoded proteins, FeS cluster synthesis, protein folding, and prevention of protein aggregation (Voos and Rottgers, 2002). Hsp90 proteins have an established function in late-stage protein folding and early-stage unfolding, but an emerging theme is that they also function in assembly and disassembly of oligomeric protein complexes (Mayer et al., 2009). In mammalian cells one Hsp90 isoform, Trap1, is localized to the mitochondria (Leskovar et al., 2008). Hsp90 is also present in mitochondria of Tetrahymena thermophila (Smith et al., 2007). Hsc82 (constitutively expressed) and Hsp82 (heat inducible) are the Hsp90 equivalent in Saccharomyces cerevisiae, and they are localized primarily in the yeast cytoplasm and nucleus (Borkovich et al., 1989). Neither has been reported to enter yeast mitochondria, nor do they have typical mitochondrial targeting sequences. Hsc82 and Hsp82 are 97% identical in their amino acid sequences and have many overlapping functions. Deletion of just one of them yields a viable cell, but deletion of both does not (Borkovich et al., 1989). However, different responses of hsp82Δ/hsp82Δ and hsc82Δ/hsc82Δ to the inhibitor macbecin II and improved efficiency of cell cycle progression during thermal stress in the presence of Hsp82 compared to Hsc82 suggest that they are not functionally equivalent in vivo (McClellan et al., 2007; Morano et al., 1999). Genetic interactions between Atp1 and Hsc82 or Hsp82 (McClellan et al., 2007) and physical interactions between Atp2 and Hsc82 or Hsp82, and between Atp3 and Hsc82 (Krogan et al., 2006) suggest a possible role for these heat shock proteins in F1Fo-ATPase assembly. In human cancer cells, F1Fo-ATPase functions as a co-chaperone of Hsp90-substrate protein complexes (Papathanassiu et al., 2006).
Genetic and physical interactions of Hsp90 proteins with mitochondrial ATP synthase subunits support a role for these chaperones in ATP synthase assembly, however their precise functions in this complex process are unknown. ATP synthase, also known as F1Fo-ATPase, is assembled from three subcomplexes: F1 containing the central stalk proteins (Atp3, Atp15, and Atp16) surrounded by a trimer of Atp1-Atp2 dimers (Atp1Atp2)3, the peripheral or stator stalk of Fo (Atp4, Atp5, Atp7 and Atp14) and the rotor component of Fo containing oligomeric Atp9 (for recent review, see (Rak et al., 2009). F1Fo-ATPase assembly occurs in a stepwise process. First the Atp9 oligomer and F1 associate to form F1-Atp9, which then associates with the peripheral stalk. Incorporation of Atp6, the proton channel component of Fo, is believed to be the last step in assembly of F1Fo-ATPase (Goyon et al., 2008). Assembly of Fo involves complex regulation and integration of mitochondrial- and nuclear-encoded subunits. Atp9 oligomerization is assisted by Atp25 (Zeng et al., 2008). Specific chaperones, Atp11, Atp12, and Fmc1, are required for formation of (Atp1Atp2)3 in the assembly of F1 (Ackerman and Tzagoloff, 2005), and two other chaperones, Atp10 and Atp23, are involved in incorporation of Atp6 into the complex (Osman et al., 2007; Tzagoloff et al., 2004). Strains lacking Fmc1 do not respire at elevated temperature due to compromised in ATP synthase assembly. This defect is rescued by increased substrate level ATP synthesis by the citric acid cycle (Schwimmer et al., 2005).
The AAA (ATPases associated with various cellular activities) proteases of mitochondria, Yme1 and Yta10/Yta12, have also been implicated in determining the activity of ATP synthase. Yme1 and Yta10/Yta12 complexes have chaperone and proteolytic functions and behave as quality control agents that ensure correct formation of multienzyme complexes (Koppen and Langer, 2007; Langer et al., 2001; Leidhold and Voos, 2007). Both are localized to the mitochondrial inner membrane. Yme1 probably acts as a hexameric complex similar to bacterial FtsH, with ATPase and protease domains in the intermembrane space (IMS) and N-terminal domains in the matrix. The closely related subunits of the Yta10/Yta12 complex have ATPase and protease domains in the matrix and extended regions between their two transmembrane domains in the IMS. Deletion of either Yta10 or Yta12 inactivates the complex and leads to respiratory defects that are attributed to decreased assembly of electron transport and oxidative phosphorylation complexes (Guélin et al., 1994; Paul and Tzagoloff, 1995; Tauer et al., 1994; Tzagoloff et al., 1994). Yta10/Yta12 functions as both a chaperone and protease in the assembly of the oligomeric Atp9 complex (Arlt et al., 1996) and has a role in F1-ATPase assembly that does not overlap that of Atp11 or Atp12 (Paul and Tzagoloff, 1995). The functions of Yme1 are still poorly understood. Only a few endogenous substrates for proteolysis have been clearly identified (Phb1, Yme2, and Cox2) (Kambacheld et al., 2005; Leonhard et al., 2000; Nakai et al., 1995; Pearce and Sherman, 1995; Weber et al., 1996). Atp4 may be another substrate for Yme1 because its level increases in yme1Δ strains (Jia et al., 2007; Kominsky et al., 2002; Lemaire et al., 2000). In contrast, Atp4 and Atp7 are at near normal concentrations in the absence of Yta10/Yta12 (Paul and Tzagoloff, 1995). Yme1 also directs nuclear-encoded proteins to the intermembrane space (Rainey et al., 2006).
Deletion of Yme1 leads to several phenotypes: slow growth on YPEG at 37°C, slow growth on YPD at 16°C slow growth in the absence of mitochondrial DNA (mtDNA), increased escape of mtDNA to the nucleus (Thorsness and Fox, 1993; Thorsness et al., 1993), altered mitochondrial morphology (Campbell and Thorsness, 1998), decreased chronological life span and decreased long term spore viability (Francis et al., 2007). Loss of Yme1 also increases the level of phosphatidylethanolamine by reducing turnover of phosphatidylserine decarboxylase, Psd1 (Nebauer et al., 2007). A previously isolated suppressor of the growth defects of yme1Δ carries a mutation in the proteasomal ATPase subunit, Rpt3 (also known as Ynt1) (Campbell et al., 1994). Disruption of the Yta10/Yta12 complex is sometimes more detrimental to growth than disruption of the Yme1 complex. For example, cells lacking either Yta10 or Yta12 lose respiratory competence whereas respiration in the absence of Yme1 is only impaired at 37°C (Tauer et al., 1994; Thorsness and Fox, 1993; Tzagoloff et al., 1994), but growth by fermentation at 16° and in the absence of mtDNA is more detrimental in the absence of Yme1 (Thorsness and Fox, 1993).
Because Yme1 genetically interacts with Hsp82 (Zhao et al., 2005) we hypothesized that Yme1 may interact with Hsc82 and Hsp82 in mitochondria, perhaps during assembly and disassembly of protein complexes, particularly under stressful conditions, and that this interaction may be a primary function of Yme1. In support of this proposition, Yme1 has recently been shown to be synthetically lethal with the chaperone Atp12 that is involved in F1-ATPase assembly (Costanzo et al., 2010). In this study we show that hsc82Δ and hsp82Δ yeast containing intact mtDNA are inviable or very slow-growing when grown by fermentation at 37°C, and that yme1Δ or yta10Δ suppress this phenotype. Suppression of hsc82Δ or hsp82Δ by yme1Δ is eliminated by introduction of atp1Δ, atp2Δ, atp4Δ, or by growth in the absence of mtDNA, indicating a role for Yme1 and these heat shock proteins in assembly of F1Fo-ATPase. The observations reported here, combined with previous studies, support a model for assembly of F1Fo-ATPase at 37°C that involves participation of Hsc82, Hsp82, Yme1, and Yta10/12.
2. Materials and methods
2.1. Strains, media and DNA techniques
Yeast strains used in this study are listed in Table 1. Standard genetic techniques were used to construct and analyze the various strains (Sherman et al., 1986). Yeast were grown in complete glucose medium (YPD) containing 2% glucose, 2% bacto peptone, 1% yeast extract, 40 mg/l tryptophan, 40 mg/l adenine; complete ethanol-glycerol medium (YPEG) containing 3% glycerol, 3% ethanol, 2% bacto peptone, 1% yeast extract, 40 mg/l tryptophan, 40 mg/l adenine; minimal glucose medium (SD) containing 2% glucose, 6.7 g/l yeast nitrogen base without amino acids (Difco), supplemented with the appropriate nutrients. The complete set of nutrients included: uracil at 40 mg/l, adenine at 40 mg/l, tryptophan at 40 mg/l, lysine at 60 mg/l, leucine at 100 mg/l, histidine at 20 mg/l, isoleucine at 30 mg/l, and valine at 150 mg/l. For plates, bacteriological agar (US Biological) was added at 16 g/l. Where indicated, geneticin was added at 300 μg/ml (US Biological), and ethidium bromide (EtBr) at 25 ug/ml. Null alleles for particular genes were generated by polymerase chain reaction (PCR) amplification of sequences from a deletion library (Open Biosystems) in which individual genes have been replaced by the kanomycin resistance gene, followed by transformation of strains in the D273-10 genetic background with the PCR products and selection of geneticin resistance, and subsequent isolation of transformants by streaking three times to single colonies on geneticin plates. Knockout strains were verified by PCR. The PCR primers used in this study are shown in Table 2.
Table 1.
Strains
| Strain | Genotypea | Source |
|---|---|---|
| BFY138 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1-Δ1::URA3 ATP1-111[ρ+ , TRP1] | (Francis et al., 2007) |
| BFY145 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1Δ1::URA3 atp1-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY178 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1Δ1::URA3 atp2-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY190 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1Δ1::URA3 atp5-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY222 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ cox4-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY234 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1Δ1::URA3 por1-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY236 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ por1-Δ1::KanMX6 rpt3-215 [ρ+, TRP1] | This study |
| BFY238 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ rpt3-215 [ρ+, TRP1] | This study |
| BFY240 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1-Δ1::URA3 por1-Δ1::KanMX6 rpt3-215 [ρ+, TRP1] | This study |
| BFY261 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY263 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1-Δ1::URA3 hsc82-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY265 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY267 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1-Δ1::URA3 hsp82-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY291 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 atp1-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY293 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 atp2-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY296 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 atp1-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY298 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 atp2-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY300 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 yme1-Δ::URA3 atp1-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY303 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 yme1-Δ::URA3 atp2-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY306 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 yme1-Δ::URA3 atp1-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY308 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 yme1-Δ::URA3 atp2-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY310 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 rpt3-215 [ρ+, TRP1] | This study |
| BFY312 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 yme1-Δ1::URA3 rpt3-215 [ρ+, TRP1] | This study |
| BFY314 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 rpt3-215 [ρ+, TRP1] | This study |
| BFY316 | MATα ura3-52 ade2 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 yme1-Δ1::URA3 rpt3-215 [ρ+, TRP1] | This study |
| BFY322 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 atp3-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY324 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 atp3-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY327 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 atp4-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY328 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 yme1-Δ1::URA3 atp4-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY330 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 atp4-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY332 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 yme1-Δ1::URA3 atp4-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY334 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 yta10-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY340 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 cox4-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY343 | MATα ura3-52 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 cox4-Δ1::KanMX6 yme1-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY344 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 cox4-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY347 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 cox4-Δ1::KanMX6 yme1-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY350 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 yta10-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY351 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 atp5-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY353 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 atp5-Δ1::KanMX6 [ρ+, TRP1] | This study |
| BFY357 | MATα ura3-52 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 atp5-Δ1::KanMX6 yme1-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY358 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 atp5-Δ1::KanMX6 yme1-Δ1::URA3 [ρ+, TRP1] | This study |
| BFY361 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsc82-Δ1::KanMX6 atp1-Δ1::KanMX6 rpt3-215 [ρ+, TRP1] | This study |
| BFY365 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 hsp82-Δ1::KanMX6 atp1-Δ1::KanMX6 rpt3-215 [ρ+, TRP1] | This study |
| DKY40 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp4-Δ1::URA3 [ρ+, TRP1] | This study |
| DKY48 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1-Δ1::URA3 atp4-Δ1::URA3 [ρ+, TRP1] | This study |
| JTY3 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp3-Δ1::KanMX6 [ρ+, TRP1] | (Smith and Thorsness, 2005) |
| KWY44 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 por1Δ::KanMX6 [ρ+, TRP1] | This study |
| NTY1 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1-Δ1::URA3 rpt3-215 [ρ+, TRP1] | (Campbell et al., 1994) |
| PTY33 (Wild type) | MATa ura3-52 ade2 leu2-3,112 trp1-Δ1 [ρ+, TRP1] | (Thorsness and Fox, 1993) |
| PTY44 (Wild type) | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 [ρ+, TRP1] | (Thorsness and Fox, 1993) |
| PTY52 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yme1-Δ1::URA3 [ρ+, TRP1] | (Thorsness et al., 1993) |
| PTY206 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 yme1-6 [ρ+, TRP1] | This study |
| TCY47 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp1-Δ1::KanMX4 [ρ+, TRP1] | This study |
| TCY49 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp2-Δ1::KanMX4 [ρ+, TRP1] | This study |
| TCY71 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp5-Δ1::KanMX4 [ρ+, TRP1] | This study |
| TCY86 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 yta10-Δ1::KanMX6 [ρ+, TRP1] | This study |
The mitochondrial genotype is bracketed.
Table 2.
| Primers used for PCR amplification of genes in the Open Biosystems Yeast deletion library: | |
| Hsc82F | 5′-AAGCGTTGGGTAATGAGGGA |
| Hsc82R | 5′-GGACAGCTGGTAGGACAATTT |
| Hsp82F | 5′-TAATACCAACCAGGTCCTTCC |
| Hsp82R | 5′-CGATTTCAGATTCTTCGCGT |
| Cox4F | 5′-CAGATCGAACGAAACGGAAA |
| Cox4R | 5′-AACTTCGCAACAACCTCTAGC |
| Primers used for PCR amplification and sequencing of RPT3: | |
| Rpt3F | 5′-TGGACAAACCAATTCTGCCT |
| Rpt3R | 5′-CGTGATAAAAAAGTGGCGTC |
| Rpt3seq1 | 5′-GGCAATCGGATAAAATTC |
| Rpt3seq2 | 5′-GAACCTATTGATCAGAAC |
| Rpt3seq3 | 5′-CCGGTAAGACGATGCTTG |
| Rpt3seq4 | 5′-GATTAGATAGAAAGATTGAG |
| Primers used for RT-PCR amplification of ~150 base segments of HSC82, HSP82, and ACT1: | |
| Hsc82F | CTGCCATCAGAACTGGTCAA |
| Hsc82R | TATCTTGTGCACCACCCTCA |
| Hsp82F | ACTCCAAAGCCAGAGCAAAA |
| Hsp82R | ACCTGAACTCTGTCGGCAAC |
| Act1F | GCCTTCTACGTTTCCATCCA |
| Act1R | GGCCAAATCGATTCTCAAAA |
Escherichia coli strain XL-1 Blue (Stratagene) was used for preparation and manipulation of recombinant DNA. Plasmid-containing E. coli were grown in LB broth (10g bactotryptone, 10g NaCl, 5g yeast extract per liter) supplemented with 125 μg/ml of ampicillin (US Biological). All manipulations of DNA were performed using standard techniques (Sambrook et al., 1989). Plasmid DNA was prepared by the boiling lysis method (Maniatis et al., 1982). Column-purified (Qiagen) plasmids or PCR products were sequenced by the Davis Sequencing Facility (University of California, Davis). Primers used for sequencing Rpt3 are shown in Table 2.
2.2. Dilution analysis for growth and measurement of the percentage of cells containing mtDNA
Dilution assays were performed on strains grown overnight in YPD. Using the approximation that OD600 = 107 cells/ml, 5 μl aliquots containing 5×104, 5×103, 5×102 or 50 cells of each strain assayed were plated on YPD, YPEG, or SD-EtBr. Approximately 50 cells of each strain were spread on two YPD plates. After 2 days at 30°C colonies were counted and transferred to YPD plates spread with PYT33 lacking mtDNA. After one day at 30°C, cells were transferred to YPEG plates and incubated at 30°C for one additional day and the number of growing colonies recorded. The percentage of yeast cultured on the original media that contained mtDNA was calculated by dividing the number of colonies growing on YPEG by the number of colonies that grew on the YPD plates.
2.3. Isolation of mitochondria and immuno-detection of mitochondrial proteins.
Cells were grown in 1 liter of YPD or YPEG media at 30°C to an OD600 of approximately 1.5. Mitochondrial isolation was performed essentially as described (Yaffe, 1991). Western blots of mitochondrial proteins were performed as previously described (Kominsky et al., 2002). Antibodies against Por1 (a gift from C. Koehler), mtHsp70 (a gift from T. Lithgow), and Tom40 (a gift from C. Koehler) were used for immuno-blotting. Levels of protein in the lanes of immuno- blots were normalized using Ponceau Red staining, and relative levels of protein in individual bands in the blots were calculated after exposure of blots treated with a chemiluminescent detection reagent (GE Healthcare) to film. Standard error of the mean for each protein was derived from at least three independent densitometric measurements.
2.4. Real time PCR
For semi-quantitative RT-PCR analysis of HSC82 and HSP82 gene expression, individual colonies were picked from YPD plates and grown overnight at 30°C in 5 ml YPD and then for 6 hr at 37°C. Cultures were centrifuged and cell pellets frozen at −80°C. To isolate RNA, cells were resuspended in five volumes of Trizol (Invitrogen, Carlsbad, California) per volume of cell pellet. The suspension was thawed and frozen 10 times using a 40°C water bath and liquid nitrogen, then vortexed 5 times for 30 sec separated by 30 sec incubations at room temperature, extracted with 0.2 ml chloroform per ml Trizol, incubated at room temperature for 3 min and centrifuged at 4°C for 15 minutes. 0.5 ml isopropanol per ml Trizol was added to the recovered aqueous phase, incubated for 10 minutes at room temperature, and centrifuged for 15 min at 4°C. The nucleic acid pellets were washed with 75% ethanol, air dried, and dissolved in 50 μl RNase-free water. RNA was purified using the RNeasy protocol (Qiagen Inc, Sanata Clara, California) with on-column DNase I removal of contaminating genomic DNA. RNA was eluted in 30μl RNase-free water and quantitated using a NanoDrop spectrophotometer. To generate cDNA, 2 μg RNA was mixed with 4 μL of reverse transcription buffer and 1 μL of IScript reverse transcriptase (Bio-Rad Laboratories) in a total volume of 20 μL. The mixture was incubated in a thermocycler for 5 min at 25°C, 30 min at 42°C, 5 min at 85°C, then held at 4°C. The resulting cDNA was diluted with 100 μL of nuclease-free water and stored at −20°C.
Diluted cDNA (10 μL) was used as a template for RT-PCR amplification using SYBR Green Supermix (Bio-Rad). Primers for PCR amplification of internal fragments of Hsc82, Hsp82, and Act1 mRNAs were designed using Primer 3 software (http://frodo.wi.mit.edu) to generate amplicons of approximately 150 bases (Table 2). RT-PCR was performed by incubating at 95°C for 3 min, followed by 95°C for 10 s, and 60°C for 30 s for 40 cycles. Following RT-PCR amplification, melting curve analysis was performed to ensure the quality of amplification by incubating for 10s at each step with an increase in temperature of 0.5°C in each cycle over a range of 55°C to 95°C. The resulting Hsc82 and Hsp82 mRNA expression levels were reported relative to Act1 as described by (Livak and Schmittgen, 2001).
3. Results
3.1. hsc82Δ and hsp82Δ impair growth by fermentation at 37°C.
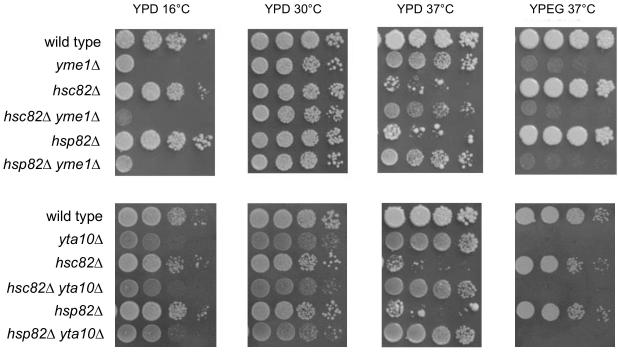
We created hsp82Δ and hsc82Δ in wild type and yme1Δ yeast and determined how these deletions affected yeast cell growth. Growth was assessed on both fermentation media (complete glucose; YPD) and respiration media (complete ethanol/glycerol; YPEG) at three temperatures (16°C, 30°C, 37°C). As reported previously (Borkovich et al., 1989), respiratory growth of hsc82Δ and hsp82Δ yeast was similar to wild type at all temperatures, as was fermentative growth at 16°C and 30°C (Fig. 1 and data not shown). However, hsc82Δ and hsp82Δ mutants did not grow by fermentation at 37°C (Fig. 1). In addition, of about 600 hsp82Δ cells plated on YPD for 2 days at 37°C, 2% produced colonies. Of a similar number of hsc82Δ cells, 0 or 1 colony grew. When these plates were transferred to 30°C for 2 days, 75% of the hsp82Δ colonies and 5% of the hsc82Δ colonies grew. Therefore, hsc82Δ was more deleterious to growth at 37°C than hsp82Δ. Suppressors arise quickly in hsc82Δ and hsp82Δ strains grown at 37°C on YPD and the growth of these strains observed in Figure 1 was due to colonies bearing suppressors.
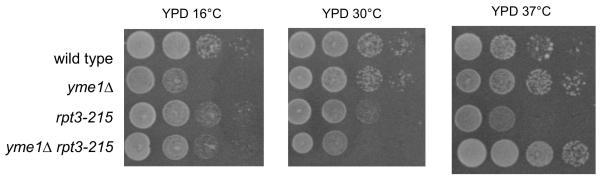
Fig. 1.
Growth of strains bearing hsc82Δ, hsp82Δ, yme1Δ, and yta10Δ on YPD at 16°C, 30°C, and 37°C, and YPEG at 37°C. Strains compared: wild type (PTY44), yme1Δ (PTY52), hsc82Δ (BFY261), hsp82Δ (BFY265), hsc82Δ yme1Δ (BFY263), hsp82Δ yme1Δ (BFY267), yta10Δ (TCY86), hsc82Δ yta10Δ (BFY350), hsp82Δ yta10Δ (BFY334).
3.2. Inactivation of Yme1 or Yta10 suppresses slow growth of hsc82Δ and hsp82Δ by fermentation at 37°C.
The extremely poor growth of hsc82Δ and hsp82Δ by fermentation at 37°C was suppressed by yme1Δ (Fig 1). The yme1Δ proved to be epistatic to either hsc82Δ or hsp82Δ as hsc82Δ yme1Δ and hsp82Δ yme1Δ grew like yme1Δ under all conditions (Fig. 1 and data not shown). yme1Δ also suppressed the reduced viability of hsc82Δ and hsp82Δ yeast. When hsc82Δ yme1Δ and hsp82Δ yme1Δ cells were spread on YPD plates and incubated for 2 days at 37°C, slow growing colonies were visible, and after transfer to 30°C 95% of the cells produced colonies. The inviability of haploid spores bearing both hsc82Δ and hsp82Δ (i.e. no Hsp90 proteins) was not suppressed by inactivation of yme1Δ.
Like yme1Δ, yta10Δ suppressed slow growth of hsc82Δ or hsp82Δ by fermentation at 37°C (Fig. 1). yta10Δ grew progressively slower by fermentation compared to wild type as the temperature was reduced from 37°C to 16°C as did the hsc82Δ yta10Δ and hsp82Δ yta10Δ double mutants (Fig. 1). All yta10Δ strains were unable to grow by respiration (Fig. 1 and data not shown). When hsc82Δ yta10Δ or hsp82Δ yta10Δ cells were plated to YPD and incubated for 2 days at 37°C, no colonies were visible. After transfer to 30°C for 2 days, more than 95% of both double mutants produced colonies. yta10Δ thus improved viability of hsc82Δ and hsp82Δ yeast grown by fermentation at 37°C.
Simultaneous inactivation of both Yme1 and Yta10 is highly deleterious to yeast cell growth (Dunn et al., 2006). The yme1Δ yta10Δ double mutant as well as the hsc82Δ yme1Δ yta10Δ and hsp82Δ yme1Δ yta10Δ triple mutants grew only as microcolonies from isolated germinating spores (data not shown).
3.3. yme1Δ-directed suppression of slow growth of hsc82Δ and hsp82Δ by fermentation at 37°C requires a functional F1Fo-ATPase.
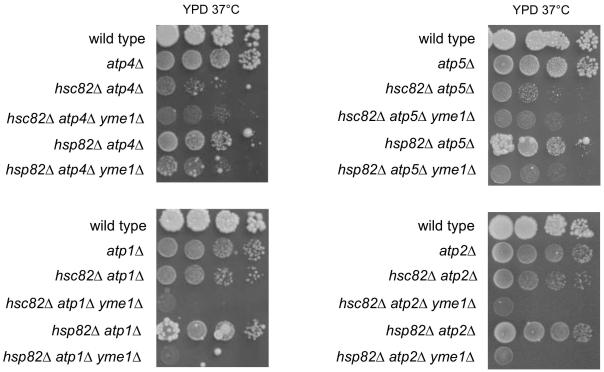
There is significant interplay between Yme1 and the F1Fo-ATPase, with mutations in one affecting phenotypes or activities of the other (Francis et al., 2007; Kominsky et al., 2002; Weber et al., 1995). This observation together with the established function of Hsp90 proteins in assembly of multisubunit complexes (Mayer et al., 2009), suggested that the growth defect of hsc82Δ and hsp82Δ could reflect a role for these proteins in assembly of the F1Fo-ATPase during fermentation at 37°C. In yme1Δ yeast, this growth defect might be mitigated by alterations in assembly or activity of the F1Fo-ATPase. In this model, suppression of hsc82Δ or hsp82Δ by yme1Δ would require functional F1Fo-ATPase. As the only membrane-embedded component of the peripheral stalk, Atp4 appears to play a central role in stalk assembly and the stalk’s interaction with F1-ATPase and other F0 subunits (Rak et al., 2009). Atp5 is a component of the peripheral stalk that binds to an Atp1 subunit of F1-ATPase (Carbajo et al., 2007; Walker and Dickson, 2006). Atp1 and Atp2 are components of F1-ATPase. To test whether yme1Δ suppression required a functional F1Fo-ATPase, triple mutants combining hsp82Δ yme1Δ and hsp82Δ yme1Δ with either atp4Δ, atp5Δ, atp1Δ, or atp2Δ were isolated and their phenotypes determined. Growth of each triple mutant was clearly slower than the corresponding hsc82Δ yme1Δ and hsp82Δ yme1Δ strains (Figs. 1 and 2). Therefore, suppression of hsc82Δ and hsp82Δ by yme1Δ was reduced in the absence of peripheral stalk or F1-ATPase subunits.
Fig. 2.
Growth at 37°C on YPD of hsc82Δ yme1Δ and hsp82Δ yme1Δ in the presence of atp4Δ, atp5Δ, atp1Δ, and atp2Δ.
Strains compared: wild type (PTY44), atp4Δ (DKY40), hsc82Δ atp4Δ (BFY327), hsc82Δ atp4Δ yme1Δ (BFY328), hsp82Δ atp4Δ (BFY330), hsp82Δ atp4Δ yme1Δ (BFY332), atp5Δ (TCY71), hsc82Δ atp5Δ (BFY351), hsc82Δ atp5Δ yme1Δ (BFY357), hsp82Δ atp5Δ (BFY353), hsp82Δ atp5Δ yme1Δ (BFY358), atp1Δ (TCY47), hsc82Δ atp1Δ (BFY291), hsc82Δ atp1Δ yme1Δ (BFY300), hsp82Δ atp1Δ (BFY296), hsp82Δ atp1Δ yme1Δ (BFY306), atp2Δ (TCY49), hsc82Δ atp2Δ (BFY293), hsc82Δ atp2Δ yme1Δ (BFY303), hsp82Δ atp2Δ (BFY298), hsp82Δ atp2Δ yme1Δ (BFY308).
Interestingly, deletion of some F1Fo-ATPase subunits acted to suppress hsc82Δ or hsp82Δ, allowing for growth of these mutants by fermentation at 37°C. Specifically, atp4Δ, atp5Δ, atp1Δ and atp2Δ suppressed hsp82Δ slow growth, while atp1Δ and atp2Δ also suppressed hsc82Δ (Fig. 1 and 2). Suppression of hsc82Δ or hsp82Δ by deletion of F1-ATPase subunits was dependent on Yme1 because suppression by atp1Δ, and atp2Δ was reduced in triple mutants with yme1Δ. Suppression of hsp82Δ by atp4Δ and atp5Δ was also reduced in the presence of yme1Δ but hsc82Δ atp4Δ and hsc82Δ atp5Δ were largely unaffected by yme1Δ. (Fig. 2). Thus, there was an interdependence of Yme1 and F1Fo-ATPase subunits in their suppression of hsc82Δ and hsp82Δ growth defects.
One possible explanation for the poor growth of hsc82Δ and hsp82Δ strains on YPD at 37° was because mtDNA became unstable, and the suppressive effects brought by the loss of F1Fo-ATPase subunits or Yme1 was due to increasing the stability of mtDNA. However, no significant destabilization of mtDNA was observed for hsc82Δ and hsp82Δ strains (Table 3), and suppressors could either have no effect on mtDNA stability (compare hsc82Δ to hsc82Δ yme1Δ) or actually destabilized mtDNA (compare hsc82Δ to hsc82Δ atp4Δ). Consequently, we concluded that changes in mtDNA stability were not the basis for suppression of growth defects of hsc82Δ and hsp82Δ strains. Strains that contained atp4Δ or atp5Δ did have unstable mtDNA, but when either of these mutations were combined with yme1Δ or yme1Δ plus hsc82Δ or hsp82Δ, the percentage of cells retaining their mtDNA increased to greater than 90%, showing that the tendency of atp4Δ or atp5Δ strains to lose mtDNA was dependent on the presence of Yme1.
Table 3. Proportion of cells containing mtDNA.
| Strain | % ρ+ cells |
|---|---|
| wild type | 99 |
| yme1 Δ | 99 |
| hsc82 Δ | 99 |
| hsc82Δ yme1Δ | 100 |
| hsp82 Δ | 99 |
| hsp82Δ yme1Δ | 100 |
| atp4 Δ | 0 |
| atp4Δ yme1Δ | 98 |
| hsc82Δ atp4Δ | 33 |
| hsc82Δ atp4Δ yme1Δ | 98 |
| hsp82Δ atp4Δ | 30 |
| hsp82Δ atp4Δ yme1Δ | 92 |
| atp5 Δ | 31 |
| atp5Δ yme1Δ | 100 |
| hsc82Δ atp5Δ | 30 |
| hsc82Δ atp5Δ yme1Δ | 99 |
| hsp82Δ atp5Δ | 10 |
| hsp82Δ atp5Δ yme1Δ | 100 |
| atp1 Δ | 97 |
| atp1Δ yme1Δ | 96 |
| hsc82Δ atp1Δ | 97 |
| hsc82Δ atp1Δ yme1Δ | 98 |
| hsp82Δ atp1Δ | 100 |
| hsp82Δ atp1Δ yme1Δ | 98 |
| atp2 Δ | 97 |
| atp2Δ yme1Δ | 97 |
| hsc82Δ atp2Δ | 99 |
| hsc82Δ atp2Δ yme1Δ | 98 |
| hsp82Δ atp2Δ | 98 |
| hsp82Δ atp2Δ yme1Δ | 100 |
Strains used: wild type, PTY44; yme1Δ, PTY52; hsc82Δ, BFY261; hsc82Δ yme1Δ, BFY263; hsp82Δ, BFY265; hsp82Δ yme1Δ, BFY267; atp4Δ, DKY40; atp4Δ yme1Δ, DKY48; hsc82Δ atp4Δ, BFY327; hsc82Δ atp4Δ yme1Δ, BFY328; hsp82Δ atp4Δ, BFY330; hsp82Δ atp4Δ yme1Δ, BFY332; atp5Δ, TCY71; atp5Δ yme1Δ, BFY190; hsc82Δ atp5Δ, BFY351; hsc82 atp5Δ yme1Δ, BFY357; hsp82Δ atp5Δ, BFY353; hsp82Δ atp5Δ yme1Δ, BFY358; atp1Δ, TCY47; atp1Δ yme1Δ, BFY145; hsc82Δ atp1Δ, BFY291; hsc82Δ atp1Δ yme1Δ, BFY300; hsp82Δ atp1Δ, BFY296; hsp82Δ atp1Δ yme1Δ, BFY306; atp2Δ, TCY49; atp2Δ yme1Δ, BFY178; hsc82Δ atp2Δ, BFY293; hsc82Δ atp2Δ yme1Δ, BFY303; hsp82Δ atp2Δ, BFY298; hsp82Δ atp2Δ yme1Δ, BFY308.
To determine whether removal of a subunit from a different inner membrane complex affected growth of hsc82Δ yme1Δ and hsp82Δ yme1Δ by fermentation at 37°C similar to removing subunits of F1Fo-ATPase, hsc82Δ, hsp82Δ, and the corresponding double mutants with yme1Δ were crossed with a strain lacking subunit 4 of cytochrome c oxidase (cox4Δ) to generate triple mutants. cox4Δ had no effect on hsc82Δ or hsp82Δ growth at 37°C and did not alter suppression of hsc82Δ or hsp82Δ by yme1Δ at 37°C (data not shown).
3.4. Hsc82 is more critical than Hsp82 for growth at 37°C in the absence of mitochondrial DNA.
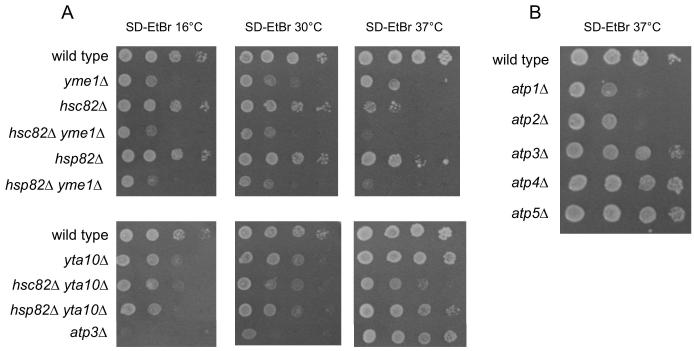
To assess the importance of mtDNA in growth of unsuppressed and suppressed hsc82Δ and hsp82Δ yeast, strains were grown on media containing ethidium bromide to induce loss of the mitochondrial genome (SD-EtBr). hsc82Δ ρ0 and hsp82Δ ρ0 grew similar to wild type ρ0 at 30°C and 16°C, but at 37°C their growth was slowed relative to wild type ρ0 (Fig. 3A). The 37°C growth defect was more severe for hsc82Δ ρ0 than hsp82Δ ρ0. Survival at elevated temperature was also more dependent on Hsc82 than Hsp82. When plated on SD-EtBr at 37°C for 4 days, no hsc82Δ ρ0 cells grew into colonies, whereas 65% of hsp82Δ ρ0 cells produced colonies. After transfer to 30°C for 2 days, 3% of hsc82Δ ρ0 and 75% of hsp82Δ ρ0 cells produced colonies. Interestingly, while the effects of hsc82Δ were similar in ρ+ and ρ0, hsp82Δ growth and viability were less impaired in ρ0 than in ρ+.
Fig. 3.
Growth of strains lacking mtDNA at 16°C, 30°C, and 37°C.
Strains compared: A. wild type (PTY44), yme1Δ (PTY52), hsc82Δ (BFY261), hsp82Δ (BFY265), hsc82Δ yme1Δ (BFY263), hsp82Δ yme1Δ (BFY267), yta10Δ (TCY86), hsc82Δ yta10Δ (BFY350), hsp82Δ yta10Δ (BFY334), atp2-227 (TCY55), atp3Δ (JTY3), atp3Δ atp2-227 (JTY6). B. wild type (PTY44), atp1Δ (TCY47), atp2Δ (TCY49), atp3Δ (JTY3), atp4Δ (DKY40), atp5Δ (TCY71).
3.5. yta10Δ but not yme1Δ suppresses the inviability of hsc82Δ ρ0 at 37°C.
As shown in previous studies (Thorsness et al., 1993), yme1Δ grew very slowly as a ρ0 strain at 30°C and 37°C, and in a ρ0 background yme1Δ hsc82Δ or yme1Δ hsp82Δ did not grow (Fig. 3A). No colonies of yme1Δ ρ0, hsc82Δ yme1Δ ρ0, and hsp82Δ yme1Δ ρ0 grew on SD-EtBr after 4 days at 37°C, or when these plates were subsequently transferred to 30°C, indicating that the absence of Yme1 is lethal for ρ0 cells at 37°C, which is in contrast to yeast with the same genotypes but containing mtDNA.
Like yta10Δ ρ+, growth of yta10Δ ρ0 was temperature dependent, with faster growth relative to wild type ρ0 at 37°C than at 30°C or 16°C (Fig. 3A). Unlike their ρ+ counterparts, hsc82Δ ρ0 and hsp82Δ ρ0 strains bearing yta10Δ grew more slowly than yta10Δ ρ0 at 37°C with hsc82Δ being clearly more deleterious to growth of yta10Δ ρ0 than hsp82Δ. Nonetheless, yta10Δ suppressed the slow growth of hsc82Δ ρ0 yeast and hsp82Δ ρ0 yeast (Fig. 3A). Of approximately 600 plated cells on SD-EtBr, less than 0.1% of hsc82Δ yta10Δ ρ0 cells and greater than 95% of hsp82Δ yta10Δ ρ0 cells produced visible colonies after 4 days at 37°C. Following transfer to 30°C for 2 days, greater than 95% of hsc82Δ yta10Δ ρ0 cells produced colonies. hsc82Δ yta10Δ ρ0 cells did not grow but did not die at 37°C. yta10Δ prevented death of both ρ+ and ρ0 hsc82Δ cells.
3.6. Loss of central or peripheral stalk proteins does not affect ρ0 growth at 37°C.
ATP3 encodes a subunit of the F1F0-ATPase central stalk. atp3Δ yeast grow only in the absence of mtDNA, and grow more slowly than wild type ρ0 at 30°C. As was found for yta10Δ strains, growth of atp3Δ yeast was temperature-dependent, with growth increasing relative to wild type as the temperature increased from 16°C and 37°C (Fig. 3A). Loss of the peripheral stalk proteins, Atp4 or Atp5, also showed little difference from wild type in ρ0 growth at 37°C, whereas loss of Atp1 or Atp2 substantially, but not completely, reduced growth (Fig. 3B).
3.7. Hsc82 is required for suppression of yme1Δ by rpt3-215.
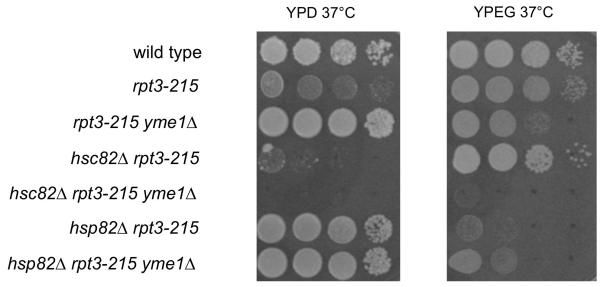
In addition to the well-characterized function of the six ATPases in the proteasome, the proteasomal ATPase subcomplex by itself has a separate function in transcription, including transcription of heat shock proteins (Gonzalez et al., 2002; Kodadek, 2010; Sulahian et al., 2006). Interestingly, a mutant proteasomal ATPase subunit, Rpt3-215, was previously isolated as a suppressor of yme1Δ phenotypes (Campbell et al., 1994). Therefore, we examined whether the growth-altering effects of rpt3-215 were influenced by the presence of hsc82Δ or hsp82Δ. hsc82Δ yme1Δ rpt3-215 grew more slowly than hsp82Δ yme1Δ rpt3-215 on YPD and YPEG at 37°C (Fig. 4). Therefore, Hsc82 was required for suppression of yme1Δ by rpt3-215. The genetic interactions of rpt3-215 and HSP82/HSC82 could potentially be explained either through alteration of transcription of HSP82/HSC82 or a direct effect on the levels of outer mitochondrial membrane proteins via proteasome activity.
Fig. 4.
Growth of strains bearing hsc82Δ, hsp82Δ, yme1Δ, and rpt3-215 on YPD at 37°C and YPEG at 37°C. Strains compared: wild type (PTY44), rpt3-215 (BFY238), yme1Δ rpt3-215 (NTY1), hsc82Δ rpt3-215 (BFY310), hsc82Δ yme1Δ rpt3-215 (BFY312), hsp82Δ rpt3-215 (BFY314), hsp82Δ yme1Δ rpt3-215 (BFY316).
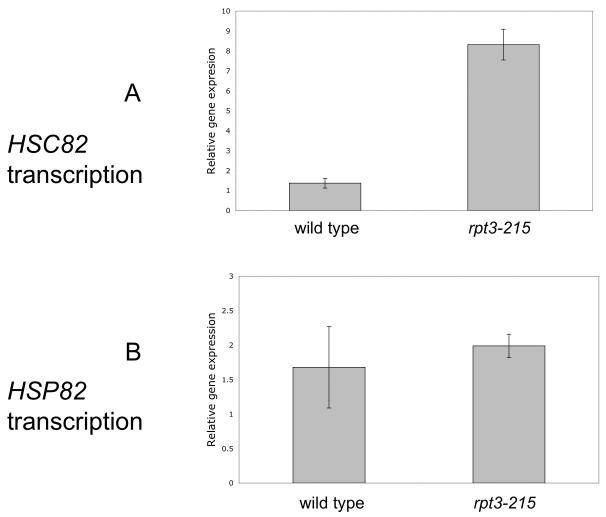
3.8. rpt3-215 increases the level of HSC82 mRNA in strains grown at 37°C
The requirement for Hsc82 to elicit suppression of yme1Δ by Rpt3-215 suggested that Rpt3-215 might be altering transcription of HSC82 but not HSP82. mRNA was extracted from wild type and rpt3-215 yeast and probed by real-time PCR with primers that amplified sequences within the coding regions of HSC82, HSP82, and ACT1. The relative level of HSC82 mRNA increased 5-fold in rpt3-215 compared to wild type, but the level of HSP82 mRNA did not (Fig. 5A and B). Strains bearing hsc82Δ or hsp82Δ did not produce the corresponding mRNAs, demonstrating that the primers used for amplification of mRNA gene fragments were specific for HSC82 and HSP82 mRNAs.
Fig. 5.
Relative gene expression of HSC82 and HSP82. Analysis of HSC82 (A) and HSP82 (B) mRNAs in wild type (PTY44) and rpt3-215 (BFY238).
3.9. The presence of rpt3-215 correlates with increases in Por1 and Tom40 during fermentation.
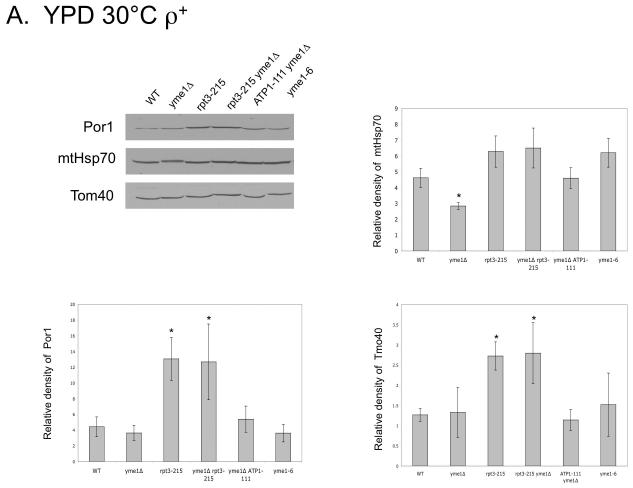
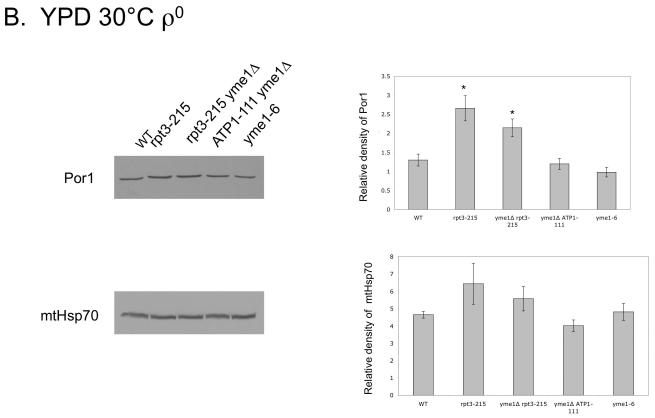
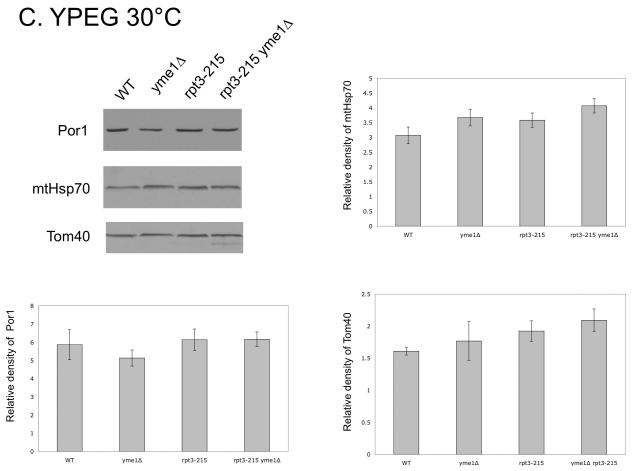
We postulated that the rpt3-215 mutation might change levels of mitochondrial outer membrane proteins through altered turnover by the proteasome (Neutzner et al., 2007). To test this hypothesis, we used immuno-blots to measure levels of Por1, a major protein involved in metabolite exchange (Crompton et al., 1999; Halestrap and Brennerb, 2003; Vyssokikh and Brdiczka, 2003), and of Tom40, a protein primarily involved in translocation of nuclear-encoded proteins into mitochondria (reviewed in Ryan et al., 2000) from ρ+ and ρ0 mitochondria of wild type and mutant yeast. Protein levels were normalized using Ponceau Red staining, and mtHsp70, a matrix protein involved in protein translocation into mitochondria, served as a control. The extremely slow growth of yme1Δ ρ0 at 30°C by fermentation precluded its inclusion in the ρ0 comparison, however two strains were included as partial substitutes for yme1Δ ρ0. Yeast carrying the yme1-6 point mutation have some phenotypes in common with yme1Δ (Francis and Thorsness, unpublished results), while yme1Δ ATP1-111 yeast have a suppressor mutation in Atp1 that allows faster growth of yme1Δ ρ0 (Francis et al., 2007). In ρ+ strains, a statistically significant increase in Por1 was observed in rpt3-215 and yme1Δ rpt3-215 yeast grown by fermentation at 30°C as compared to wild type, yme1Δ, yme1Δ ATP1-111 and yme1-6 (Fig. 6A). The level of Tom40 in strains containing rpt3-215 also increased (Fig. 6A). In contrast, the level of mtHsp70 was constant in all stains except yme1Δ strain, which had a reduced amount of Hsp70 (Fig. 6A). Comparison of the corresponding ρ0 strains showed that Por1 increased in strains bearing rpt3-215 and yme1Δ rpt3-215 whereas mtHsp70 did not (Fig. 6B). In contrast, strains containing rpt3-215 grown in YPEG at 30°C did not have increased levels of Por1, Tom40 or mtHsp70 (Fig. 6C).
Fig. 6.
Western blots using antibodies against Por1, mtHsp70, and Tom40 of mitochondrial proteins from ρ+ strains (A) and ρ0 strains (B) grown in YPD at 30°C, and strains grown in YPEG at 30°C (C). * indicates significant difference. Strains compared: wild type (PTY44), yme1Δ (PTY52), rpt3-215 (BFY238), yme1Δ rpt3-215 (NTY1), yme1Δ ATP1-111 (BFY138), yme1-6 (PTY206).
Because por1Δ yeast are viable (Michejda et al., 1990), we were able to examine the suppressive effects of rpt3-215 in the absence of Por1. rpt3-215 suppressed slow growth of yme1Δ por1Δ and yme1Δ por1Δ ρ0 (data not shown), indicating that changes in Por1 concentration is not the basis for suppression of yme1Δ phenotypes.
3.10. The suppressor mutation in RPT3 is in the Walker A ATP binding loop.
A mutation in RPT3, an essential subunit of the 26S proteasome, suppresses specific yme1Δ phenotypes (Campbell et al., 1994). Sequencing this suppressing allele, designated rpt3-215, identified a T for C mutation at position 644 that produced a leucine for proline substitution at position 215 in Rpt3. This mutation was in the conserved Walker A (or P-loop) ATP-binding sequence motif of Rtp3 (208-GVLLYGPLGTGKTML-222). Such a mutation might be expected to interfere with the ATPase cycle of Rpt3, thereby decreasing the activity of the proteasome and affecting growth. In accord with this expectation, rpt3-215 and yme1Δ rpt3-215 exhibited slow growth by fermentation at 30°C compared to wild type and yme1Δ yeast. yme1Δ and yme1Δ rpt3-215 increased growth of yeast relative to wild type as the temperature increased from 16 to 37°C but growth of rpt3-215 yeast decreased. In fact yme1Δ rpt3-215 yeast grew faster than wild-type yeast. (Fig. 7).
Fig. 7.
Growth of strains bearing yme1Δ and rpt3-215 at 16°C, 30°C, and 37°C. Strains compared: wild type (PTY44), yme1Δ (PTY52), rpt3-215 (BFY238), yme1Δ rpt3-215 (NTY1).
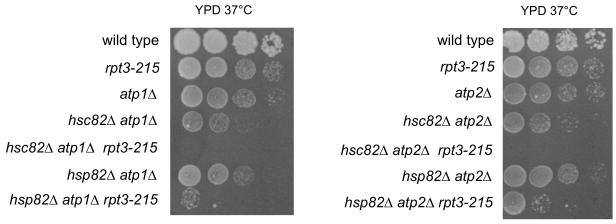
3.11. rpt3-215 prevents growth by fermentation at 37°C of hsc82Δ atp1Δ and hsc82Δ atp2Δ, but not hsp82Δ atp1Δ and hsp82Δ atp2Δ.
After showing that rpt3-215 affected growth of hsc82Δ yme1Δ and hsp82Δ yme1Δ strains differently, we attempted to determine whether rpt3-215 produced a similar outcome when F1-ATPase was disrupted by deletion of ATP1, ATP2, or ATP3. hsc82Δ atp3Δ rpt3-215 and hsp82Δ atp3Δ rpt3-215 strains, grew as microcolonies from germinating spores at 30°C, and were not further studied. hsc82Δ atp1Δ rpt3-215 and hsc82Δ atp2Δ rpt3-215 strains were inviable when grown by fermentation at 37°C. Slow growth of hsp82Δ atp1Δ and hsp82Δ atp2Δ strains was reduced further by the combination with rpt3-215 (Fig. 8). Consequently, HSC82 was also required for growth of atp1Δ or atp2Δ yeast in the presence of rpt3-215.
Fig. 8.
Growth on YPD at 37°C of strains bearing hsc82Δ, hsp82Δ, rpt3-215, atp1Δ, and atp2Δ. Strains compared: wild type (PTY44), rpt3-215 (BFY238), atp1Δ (TCY47), hsc82Δ atp1Δ (BFY291), hsc82Δ atp1Δ rpt3-215 (BFY361), hsp82Δ atp1Δ (BFY296), hsp82Δ atp1Δ rpt3-215 (BFY365), atp2Δ (TCY49), hsc82Δ atp2Δ (BFY293), hsc82Δ atp2Δ rpt3-215 (BFY366), hsp82Δ atp2Δ (BFY298), hsp82Δ atp2Δ rpt3-215 (BFY368).
4. Discussion
4.1. Growth of strains lacking Hsc82 or Hsp82.
Although Hsp82 expression is heat inducible and Hsc82 is constitutively expressed, both hsp82Δ and hsc82Δ grow like wild type by respiration at 16, 30, and 37 °C and by fermentation at 16 and 30°C. For growth by fermentation of ρ+ strains at 37°C there is a requirement for both of these proteins (Fig. 1A). Loss of Hsc82 is more harmful to growth at elevated temperatures than loss of Hsp82. These results support previous studies with diploid strains showing that loss of three out of four HSC82 and HSP82 genes results in virtually no growth (Borkovich et al., 1989). Those hsc82Δ and hsp82Δ colonies that grew at 37°C (Fig. 1A) likely bear suppressor mutations. The decreased growth of hsc82Δ and hsp82Δ strains on YPD plates at 37°C differs from the moderate growth inhibition reported for comparable homozygous diploid strains in liquid fermentation medium at 37°C after inoculation with cells grown at 25°C (Borkovich et al., 1989). This difference may be explained by the generation of spontaneous suppressors in liquid media. The fact that respiration is unimpeded at 37°C in these strains (Fig. 1B) suggests that the detrimental effects on fermentation are the result of different responses to heat shock during fermentation and respiration (Patriarca and Maresca, 1990).
Neither Hsc82 nor Hsp82 has previously been reported in mitochondria, although surveys of genetic and proteomic relationships in yeast suggest roles for Hsc82 and Hsp82 in mitochondrial membrane biogenesis (Gavin et al., 2006; Krogan et al., 2006; McClellan et al., 2007; Zhao et al., 2005). Our genetic data are best explained by the presence of these proteins in yeast mitochondria. After communicating our results with Dr. Susan Lindquist and Dr. Dan Tardiff, they confirmed by protein immunoblot that Hsc(p)82 was present in yeast mitochondrial fractions (Tardiff and Lindquist, personal communication).
4.2. Roles for Hsc82, Hsp82, Yme1, and Yta10/12 in F1Fo-ATPase assembly
Many aspects of F1Fo-ATPase assembly have been discovered and specialized chaperones identified (Rak et al., 2009), but some steps, such as the assembly of F1-ATPase and the assembly of the F1-ATPase with the peripheral stalk, are not fully understood. Figure 9 shows a model for assembly of F1Fo-ATPase that is consistent with earlier studies and the genetic evidence presented here. Although the order of events in Figure 9 has considerable support (Ackerman and Tzagoloff, 2005; Goyon et al., 2008; Kucharczyk et al., 2009; Velours and Arselin, 2000), some events, such as the order of incorporation of Atp8, must still be regarded as provisional (Rak et al., 2009).
Fig. 9.
Model for assembly of F1Fo-ATPase at 37°C. The inner membrane (IM) is shown with the intermembrane space above the membrane and the matrix below. One subunit of the Yme1 and Yta10/12 complexes is shown. A. Assembly of F1. Yme1 interacts genetically with Hsc82 or Hsp82 to which Atp2 is bound with Atp11 as chaperone. Yta10/12 interacts in an unknown way with the central stalk to which only Hsc82 is bound. Atp1 is bound to chaperone Atp12, which in turn is bound to Fmc1. Assembly of Atp1 and Atp2 around Atp3 begins with interaction of Atp3 with Atp1 displacing Atp12 and Fmc1. As assembly proceeds Atp11, Atp12, and Yta10/12 are displaced and Hsc82 is displaced from Atp3 to produce B. The interaction of Yme1 with Atp2 via Hsc82/Hsp82 remains while F1 associates with oligo-Atp9 that has been formed with assistance from Atp25 to form C. F1-Atp9 associates with Atp8 and the peripheral stalk complex (only Atp4 and Atp5 are shown). Yme1 and Hsc82/Hsp82 are displaced to form D. Atp6 is incorporated with assistance from Atp10, Atp23 and Oxa1 to produce F1F0-ATPase, E.
Step A to B of Fig. 9: Formation of F1-ATPase.
F1-ATPase, can be assembled in the presence or absence of mtDNA. Formation of (Atp1Atp2)3 requires specialized assembly factors: Atp12 and Atp11, for Atp1 and Atp2, respectively, and Fmc1 at elevated temperatures which binds to Atp12 (Ackerman, 2002; Lefebvre-Legendre et al., 2001; Smith and Thorsness, 2005). No specialized chaperones/assembly factors are known for Atp3, or the other F1 components of the central stalk, Atp15, or Atp16. A subcomplex containing Atp15 and Atp16 has been purified from pig heart mitochondria (Penin et al., 1990), and the stable subcomplex has been isolated when these proteins are co-expressed in E. coli (Orriss et al., 1996) suggesting that chaperones may not be required for this step in assembly of the central stalk. Assembly of Atp15/Atp16 with Atp3 has not been studied. The ability of (Atp1Atp2)3 to be separated from the central stalk using mild detergent conditions (Wittig et al., 2008) suggests that Atp3 can be inserted into pre-formed (Atp1Atp2)3. More persuasively, incorporation of Atp3 that has been C-terminally fused to GFP into active ATP synthase (Prescott et al., 2003), impaired ATP synthase assembly produced by N- and C-terminal deletions of Atp3 (Dian et al., 2008), and involvement of Atp3 in formation of (Atp1Atp2)3 (Ludlam et al., 2009) argue for assembly of (Atp1Atp2)3 around the central stalk.
yta10Δ and yta12Δ inhibit assembly of F1-ATPase in a manner that does not overlap with Atp11 and Atp12 (Paul and Tzagoloff, 1995). Three lines of evidence from this study and others suggest that loss of Yta10/12 has a similar effect to loss of Atp3. First, loss of these proteins leads to destabilization of mitochondrial DNA. Strains lacking any of the three central stalk proteins are viable only as ρ0 strains (Giraud and Velours, 1994; Lai-Zhang and Mueller, 2000; Smith and Thorsness, 2005). yta10Δ is also unstable with regard to loss of mtDNA (Lemaire et al., 2000; Tauer et al., 1994). Another genetic connection between atp3Δ and yta10Δ is a similar increase in growth of ρ0 strains relative to wild type as the temperature increases from 16 to 37°C. A temperature dependent increase in growth is not observed with atp1Δ ρ0 and atp2Δ ρ0 showing that in these cases another ATPase does not compensate for the loss of the ATPase activity from (Atp1Atp2)3. If Atp3 is required for assembly of (Atp1Atp2)3 (Ludlam et al., 2009), no (Atp1Atp2)3 would be present in atp3Δ and hence there would be no F1-ATPase activity to assist in maintenance of membrane potential. Nonetheless, atp3Δ has a higher growth rate at 37°C than atp1Δ ρ0 and atp2Δ ρ0. Among the genes up-regulated in ρ0 compared to ρ+ cells are Atp11 and Atp12 (Traven et al., 2001). Fmc1 is a chaperone that is required for proper functioning of Atp12 at high temperatures (37°C) in the incorporation of Atp1 and Atp2 into F1-ATPase (Lefebvre-Legendre et al., 2001). A possible explanation for the temperature dependence of atp3Δ and yta10Δ growth rate is that as Fmc1 becomes increasingly expressed (Atp1Atp2)3 can form independently of Atp3 using Atp11 and Atp12. –Taken together, the evidence suggests a function for Yta10/12 in the step by which the central stalk becomes incorporated into F1-ATPase, although the protein(s) that interacts with Yta10/12 has yet to be identified.
In several instances, a difference is observed between the effects of hsc82Δ and hsp82Δ on growth. For example, hsp82Δ ρ0 and hsp82Δ yta10Δ ρ0 yeast grow faster at 37°C than hsc82Δ ρ0 and hsc82Δ yta10Δ ρ0 yeast (Fig. 3A). Loss of Yta10/12 has a subtle effect on ρ+ or ρ0 growth of hsc82Δ at 37°C; it prevents hsc82Δ death but not its slow growth. Suppression of slow growth at 37°C of hsp82Δ yeast but not hsc82Δ yeast could be achieved by removal of peripheral stalk proteins, Atp4 or Atp5 (Fig. 2). In the case of hsp82Δ atp4Δ and hsp82Δ atp5Δ strains, a reduced requirement for Hsc82 during association of the peripheral stalk to F1-ATPase (discussed below) may allow increased cycling of Hsc82 for F1-ATPase assembly thereby allowing suppression. The preference of F1-ATPase assembly for Hsc82 means that suppression is not observed with hsc82Δ atp4Δ or hsc82Δ atp5Δ. A possible explanation for the greater growth defects conferred by hsc82Δ yeast compared to hsp82Δ yeast would involve Hsc82 interacting with Atp3 (Krogan et al., 2006).
Loss of mtDNA from yme1Δ is more detrimental for growth than it is for either yta10Δ or atp3Δ, and yme1Δ strains do not lose mtDNA as readily as atp3Δ or yta10Δ. The double mutations, yta10Δ yme1Δ and atp3Δ yme1Δ, only allow growth of yeast as micro-colonies after spore germination showing that the combination of these nulls is more severe than each of the nulls individually, and suggesting that yme1Δ acts separately from atp3Δ or yta10Δ. One effect of yme1Δ is to allow an increase in the level of Atp4 (Lemaire et al., 2000). In the absence of Atp6, Atp8, and Atp9 encoded by the mtDNA, increased Atp4 might allow greater assembly of F1-ATPase with the peripheral stalk. However, atp4Δ ρ0 and atp5Δ ρ0 strains grow almost as well as wild type ρ0 at 30°C and 37°C (Fig. 3B and data not shown) showing that F1-ATPase binding to the peripheral stalk is not important for ρ0 growth. In addition, yme1Δ atp4Δ ρ0 yeast grow extremely slowly, similar to yme1Δ ρ0 yeast (Kominsky et al., 2002), showing that yme1Δ is controlling ρ0 growth and has an effect that is independent of atp4Δ. The severe effect of yme1Δ on ρ0 growth is counteracted by mutations in F1-ATPase subunits that increase ATPase activity (Francis et al., 2007). This is consistent with a role for Yme1 in F1-ATPase assembly: a lower level of correctly assembled F1-ATPase in yme1Δ ρ0 yeast can be offset by increased F1-ATPase activity. This suggests that Yme1, like Yta10/12, has a role in F1-ATPase assembly that involves incorporation of the central stalk into F1-ATPase. Combination of yme1Δ with hsc82Δ or hsp82Δ does not suppress ρ0 yeast growth at 37°C, and at 16°C, 30°C, and 37°C yme1Δ produces a similar decrease in ρ0 yeast growth of hsc82Δ and hsp82Δ strains (Fig. 3A). The role for Yme1 in F1-ATPase assembly may involve an interaction with either Hsc82 or Hsp82, which in turn binds to Atp2 (Krogan et al., 2006). In summary, the proposed mechanism for this step involves assembly of Atp1 and Atp2 around the central stalk, with release of Hsc82 and Yta10/12 from the central stalk and Atp11, Atp12, and Fmc1 from Atp1 and Atp2. Yme1 remains associated with Atp2 via Hsc82/Hsp82 after assembly of F1-ATPase prior to association with Atp9 and the peripheral stalk.
Step B to C of Fig.9: Interaction of F1-ATPase with oligoAtp9.
Atp25 is the chaperone for assembly of the Atp9 oligomer (Zeng et al., 2008). Newly synthesized oligomeric Atp9 forms an F1-oligoAtp9 subcomplex prior to its association with Atp6 and the peripheral stalk complex (Ackerman and Tzagoloff, 2005).
Step C to D of Fig.9: Interaction of F1-oligoAtp9 with the peripheral stalk.
Association of F1-oligoAtp9 with the peripheral stalk occurs with a preformed peripheral stalk complex (Rak et al., 2009). An interesting parallel exits between the effects of yme1Δ on hsc82Δ/hsp82Δ and oxa1Δ. Addition of yme1Δ to either hsc82Δ or hsp82Δ imparts the corresponding yme1Δ phenotypes and for hsc82Δ and hsp82Δ this means suppression of slow fermentative growth at 37°C (Fig. 1A). The only other reported strain for which yme1Δ improves growth is oxa1Δ (Lemaire et al., 2000). Oxa1 is required for assembly of F1Fo-ATPase (Altamura et al., 1996), and assists in the correct assembly of the Atp9 oligomer with Atp6. In the absence of Oxa1, Atp6 and Atp4 levels in mitochondria are greatly reduced (Jia et al., 2007). yme1Δ restores assembly of F1Fo-ATPase in oxa1Δ but does not restore respiratory growth or increase steady state levels of cytochrome c oxidase subunits (Lemaire et al., 2000). Apparently, yme1Δ suppresses the effects of oxa1Δ by diminishing turnover of Atp4 and Atp6 (Lemaire et al., 2000). The level of Atp4 also increases in yme1Δ ρ0 (Kominsky et al., 2002). A common link for these effects of yme1Δ on oxa1Δ and hsc82Δ/hsp82Δ may be increased levels of Atp6 and peripheral stalk complex made possible by the absence of the proteolytic activity of Yme1, which in turn allows a higher level of active F1Fo-ATPase to be assembled. We have no evidence to suggest a difference between Hsc82 and Hsp82 in this regard. yme1Δ does not suppress slow growth of hsc82Δ or hsp82Δ yeast in the presence of atp1Δ, atp2Δ, atp4Δ, or atp5Δ (Fig. 2), suggesting a role for Yme1 in sensing Atp4 through the ATPase/protease domains in the IMS, and in sensing F1-ATPase via matrix-localized Hsc82 or Hsp82 through its N-terminal domain. Hsc82 and Hsp82 interact physically with Atp2 (Krogan et al., 2006), and have a synthetic growth defect with Atp1 (McClellan et al., 2007). Therefore, Yme1 may assist in assembly of the F1-Atp9 subcomplex with the peripheral stalk through an interaction with Hsc82/Hsp82 that is bound to Atp2. In this step of the model, Hsc82/Hsp82 is displaced from Atp2 during assembly of the peripheral stalk with the F1-oligoATP9 complex. One obvious possibility is that the N-terminal domain of Yme1 is a co-chaperone for Hsc82/Hsp82. Incorporation of Atp8 into the complex may occur before or after assembly of the peripheral stalk.
A role for Yme1 and Hsc82/Hsp82 in assembly of F1-ATPase with the peripheral stalk is supported by the changes in the percentage of ρ+ cells of atp4Δ or atp5Δ yeast cultures in the presence of yme1Δ and hsc82Δ/hsp82Δ. Nearly all of the loss of mtDNA observed for atp4Δ and atp5Δ strains is prevented in the double mutants with yme1Δ or the triple mutants with yme1Δ and hsc82Δ or hsp82Δ (Table 3). Loss of central or peripheral stalk proteins allows a proton leak across the inner mitochondrial membrane via Atp6 that is counteracted by the loss of mtDNA (Mueller, 2000; Uh et al., 1990). In the case of atp4Δ and atp5Δ strains this leak is apparently unable to form in the absence of Yme1, presumably because the F1-ATPase/oligoAtp9 complex is unable to form a complex with Atp6 under these conditions.
Step D to E of Fig. 9: Incorporation and processing of Atp6.
The last step in F1Fo-ATPase assembly is incorporation of Atp6, the subunit that contains the proton channel that drives rotation of the central stalk (Goyon et al., 2008). Assembly of Atp6 involves three other proteins, Oxa1 (Jia et al., 2007), Atp10 (Tzagoloff et al., 2004), and Atp23 (Osman et al., 2007), the last of which encodes a protease that cleaves part of Atp6.
4.3. The Rpt3-215 suppressor increases transcription of Hsc82
The rpt3-215 mutation is in the active site of an essential proteasomal subunit. The Pro215Leu mutation in the highly conserved ATP binding loop of Rpt3 may behave similarly to the oncogenic Gly12Val mutation in the sequence, 7-VVVGAVGVGKS-17, of the GTP binding loop of ras p21 that inhibits GTP hydrolysis and acts as a permanent ‘on’ switch (Tong et al., 1991). If so, activation of HSC82 transcription by Rpt3-215 would require the ATP bound conformation of Rpt3.
rpt3-215 generally decreases growth consistent with an altered ATPase cycle for Rpt3-215 that decreases proteasomal function. Growth is increases when yme1Δ is combined with rpt3-215. The double mutant grows better than either of the single mutants by fermentation and respiration at 37°C. A particularly informative observation is that rpt3-215 suppresses slow growth at 37°C of hsp82Δ yme1Δ but not hsc82Δ yme1Δ (Fig. 4). This led to the discovery that rpt3-215 increases transcription of HSC82 but not HSP82. Increased HSC82 transcription can account for the increased growth of hsp82Δ yme1Δ rpt3-215 compared to hsp82Δ yme1Δ at 37°C. The very slow growth of hsc82Δ yme1Δ rpt3-215 at 37°C compared to hsc82Δ yme1Δ shows that heat induced transcription of HSP82 cannot compensate for the loss of constitutively expressed HSC82. Therefore, Hsc82 supports growth in a way Hsp82 does not, consistent with a specific role for Hsc82 in F1Fo-ATPase assembly involving Atp3. Increased HSC82 transcription by Rpt3-215 is notably similar to induction of two other heat shock proteins, HSP26 and HSP104, by two other proteasomal ATPases, Rpt6 (Sug1) and Rpt4 (Sug2), acting independently of their roles in proteolysis (Kodadek, 2010; Sulahian et al., 2006).
Another possibility is that Rpt3-215 alters proteasomal turnover of proteins in the outer mitochondrial membrane leading perhaps to increased metabolite exchange. Approximately a 2-fold increase in levels of Por1 and Tom40 is found in mitochondria isolated from rpt3-215 and yme1Δ rpt3-215 compared to those from wild type when grown by fermentation, but not when grown by respiration. However, suppression of slow growth of por1Δ strains by rpt3-215 suggests that Rpt3-215 does not act through this mechanism alone.
4.4 Growth in the absence of F1-ATPase.
Yeast lacking Atp1 or Atp2 grow by fermentation, albeit slower than wild type. Previous studies have demonstrated that loss of either one of these subunits results in aggregation of the other subunit into mitochondrial inclusion bodies (Lefebvre-Legendre et al., 2005). Because growth requires maintenance of a membrane potential across the inner membrane, and the membrane potential during fermentation depends primarily upon ATP hydrolysis in the matrix, another mechanism for ATP hydrolysis must compensate for the absence of F1-ATPase. Remarkably, slow growth of both hsc82Δ and hsp82Δ is suppressed by atp1Δ or atp2Δ (Fig 2). It appears that when Hsc82 or Hsp82 is not cycling through F1Fo-ATPase assembly it becomes available to assist an alternate mechanism for ATP hydrolysis. Whatever this mechanism may be, it also requires Yme1 because yme1Δ eliminates the suppression by atp1 or atp2Δ (Fig. 2). rpt3-215 suppresses slow fermentative growth at 37°C of hsp82Δ atp1Δ and hsp82Δ atp2Δ but not hsc82Δ atp1Δ and hsc82Δ atp2Δ, showing that suppression can occur in the absence of F1-ATPase. Loss of ATP synthase assembly at elevated temperature is suppressed by increased substrate level mitochondrial ATP synthesis via the citric acid cycle (Schwimmer et al., 2005), suggesting that growth by fermentation in the absence of F1-ATPase, may be compensated by improved substrate level ATP hydrolysis in mitochondria by succinyl-CoA ligase, an enzyme of the citric acid cycle, operating in the reverse direction. Future studies may discover specific functions for Hsc82/Hsp82 in mitochondria in addition to the proposed role in F1Fo-ATPase assembly.
Acknowledgements
Thanks to William Murdoch for assistance with statistical analyses, and Kathy Austin and Kumaran Mani for assistance with RT-PCR. Drs. Carla Koehler and Trevor Lithgow supplied antisera. We also thank Drs. Susan Lindquist and Dan Tardiff for communicating results prior to publication. This work was supported by grants from the National Institutes of Health (GM068066) and National Center for Research Resources and the Wyoming INBRE (P20RR016474).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackerman SH. Atp11p and Atp12p are chaperones for F(1)-ATPase biogenesis in mitochondria. Biochim Biophys Acta. 2002;1555:101–105. doi: 10.1016/s0005-2728(02)00262-1. [DOI] [PubMed] [Google Scholar]
- Ackerman SH, Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog Nucleic Acid Res Mol Biol. 2005;80:95–133. doi: 10.1016/S0079-6603(05)80003-0. [DOI] [PubMed] [Google Scholar]
- Altamura N, Capitanio N, Bonnefoy N, Papa S, Dujardin G. The Saccharomyces cerevisiae OXA1 gene is required for the correct assembly of cytochrome c oxidase and oligomycin-sensitive ATP synthase. FEBS Lett. 1996;382:111–115. doi: 10.1016/0014-5793(96)00165-2. [DOI] [PubMed] [Google Scholar]
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Tanaka N, White KH, Thorsness PE. Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell. 1994;5:899–905. doi: 10.1091/mbc.5.8.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci. 1998;111(Pt 16):2455–2464. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- Carbajo RJ, Kellas FA, Yang JC, Runswick MJ, Montgomery MG, Walker JE, Neuhaus D. How the N-terminal domain of the OSCP subunit of bovine F1Fo-ATP synthase interacts with the N-terminal region of an alpha subunit. J Mol Biol. 2007;368:310–318. doi: 10.1016/j.jmb.2007.02.059. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton M, Virji S, Doyle V, Johnson N, Ward JM. The mitochondrial permeability transition pore. Biochem Soc Symp. 1999;66:167–179. doi: 10.1042/bss0660167. [DOI] [PubMed] [Google Scholar]
- Dian EA, Papatheodorou P, Emmrich K, Randel O, Geissler A, Kolling R, Rassow J, Motz C. Role of gamma-subunit N- and C-termini in assembly of the mitochondrial ATP synthase in yeast. J Mol Biol. 2008;377:1314–1323. doi: 10.1016/j.jmb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Dunn CD, Lee MS, Spencer FA, Jensen RE. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol Biol Cell. 2006;17:213–226. doi: 10.1091/mbc.E05-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis BR, White KH, Thorsness PE. Mutations in the Atp1p and Atp3p subunits of yeast ATP synthase differentially affect respiration and fermentation in Saccharomyces cerevisiae. J Bioenerg Biomembr. 2007;39:127–144. doi: 10.1007/s10863-007-9071-4. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Giraud MF, Velours J. ATP synthase of yeast mitochondria. Isolation of the F1 delta subunit, sequence and disruption of the structural gene. Eur J Biochem. 1994;222:851–859. doi: 10.1111/j.1432-1033.1994.tb18932.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- Goyon V, Fronzes R, Salin B, di-Rago JP, Velours J, Brethes D. Yeast cells depleted in Atp14p fail to assemble Atp6p within the ATP synthase and exhibit altered mitochondrial cristae morphology. J Biol Chem. 2008;283:9749–9758. doi: 10.1074/jbc.M800204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guélin E, Rep M, Grivell LA. Sequence of the AFG3 gene encoding a new member of the FtsH/Yme1/Tma subfamily of the AAA-protein family. Yeast. 1994;10:1389–1394. doi: 10.1002/yea.320101016. [DOI] [PubMed] [Google Scholar]
- Halestrap AP, Brennerb C. The adenine nucleotide translocase: a central component of the mitochondrial permeability transition pore and key player in cell death. Curr Med Chem. 2003;10:1507–1525. doi: 10.2174/0929867033457278. [DOI] [PubMed] [Google Scholar]
- Jia L, Dienhart MK, Stuart RA. Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex. Mol Biol Cell. 2007;18:1897–1908. doi: 10.1091/mbc.E06-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambacheld M, Augustin S, Tatsuta T, Muller S, Langer T. Role of the novel metallopeptidase Mop112 and saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J Biol Chem. 2005;280:20132–20139. doi: 10.1074/jbc.M500398200. [DOI] [PubMed] [Google Scholar]
- Kodadek T. No Splicing, no dicing: non-proteolytic roles of the ubiquitin-proteasome system in transcription. J Biol Chem. 2010;285:2221–2226. doi: 10.1074/jbc.R109.077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominsky DJ, Brownson MP, Updike DL, Thorsness PE. Genetic and biochemical basis for viability of yeast lacking mitochondrial genomes. Genetics. 2002;162:1595–1604. doi: 10.1093/genetics/162.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42:221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kucharczyk R, Zick M, Bietenhader M, Rak M, Couplan E, Blondel M, Caubet SD, di Rago JP. Mitochondrial ATP synthase disorders: molecular mechanisms and the quest for curative therapeutic approaches. Biochim Biophys Acta. 2009;1793:186–199. doi: 10.1016/j.bbamcr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Lai-Zhang J, Mueller DM. Complementation of deletion mutants in the genes encoding the F1-ATPase by expression of the corresponding bovine subunits in yeast S. cerevisiae. Eur J Biochem. 2000;267:2409–2418. doi: 10.1046/j.1432-1327.2000.01253.x. [DOI] [PubMed] [Google Scholar]
- Langer T, Kaser M, Klanner C, Leonhard K. AAA proteases of mitochondria: quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem Soc Trans. 2001;29:431–436. doi: 10.1042/bst0290431. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Salin B, Schaeffer J, Brethes D, Dautant A, Ackerman SH, di Rago JP. Failure to assemble the alpha 3 beta 3 subcomplex of the ATP synthase leads to accumulation of the alpha and beta subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J Biol Chem. 2005;280:18386–18392. doi: 10.1074/jbc.M410789200. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Vaillier J, Benabdelhak H, Velours J, Slonimski PP, di Rago JP. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J Biol Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- Leidhold C, Voos W. Chaperones and proteases--guardians of protein integrity in eukaryotic organelles. Ann N Y Acad Sci. 2007;1113:72–86. doi: 10.1196/annals.1391.011. [DOI] [PubMed] [Google Scholar]
- Lemaire C, Hamel P, Velours J, Dujardin G. Absence of the mitochondrial AAA protease Yme1p restores F0-ATPase subunit accumulation in an oxa1 deletion mutant of Saccharomyces cerevisiae. J Biol Chem. 2000;275:23471–23475. doi: 10.1074/jbc.M002045200. [DOI] [PubMed] [Google Scholar]
- Leonhard K, Guiard B, Pellecchia G, Tzagoloff A, Neupert W, Langer T. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol Cell. 2000;5:629–638. doi: 10.1016/s1097-2765(00)80242-7. [DOI] [PubMed] [Google Scholar]
- Leskovar A, Wegele H, Werbeck ND, Buchner J, Reinstein J. The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J Biol Chem. 2008;283:11677–11688. doi: 10.1074/jbc.M709516200. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ludlam A, Brunzelle J, Pribyl T, Xu X, Gatti DL, Ackerman SH. Chaperones of F1-ATPase. J Biol Chem. 2009;284:17138–17146. doi: 10.1074/jbc.M109.002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1982. [Google Scholar]
- Mayer MP, Prodromou C, Frydman J. The Hsp90 mosaic: a picture emerges. Nat Struct Mol Biol. 2009;16:2–6. doi: 10.1038/nsmb0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Michejda J, Guo XJ, Lauquin GJ. The respiration of cells and mitochondria of porin deficient yeast mutants is coupled. Biochem Biophys Res Commun. 1990;171:354–361. doi: 10.1016/0006-291x(90)91401-d. [DOI] [PubMed] [Google Scholar]
- Morano KA, Santoro N, Koch KA, Thiele DJ. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol Cell Biol. 1999;19:402–411. doi: 10.1128/mcb.19.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DM. Partial assembly of the yeast mitochondrial ATP synthase. J Bioenerg Biomembr. 2000;32:391–400. doi: 10.1023/a:1005532104617. [DOI] [PubMed] [Google Scholar]
- Nakai T, Yasuhara T, Fujiki Y, Ohashi A. Multiple genes, including a member of the AAA family, are essential for degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol Cell Biol. 1995;15:4441–4452. doi: 10.1128/mcb.15.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebauer R, Schuiki I, Kulterer B, Trajanoski Z, Daum G. The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p. Febs J. 2007;274:6180–6190. doi: 10.1111/j.1742-4658.2007.06138.x. [DOI] [PubMed] [Google Scholar]
- Neutzner A, Youle RJ, Karbowski M. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found Symp. 2007;287:4–14. discussion 14-20. [PubMed] [Google Scholar]
- Orriss GL, Runswick MJ, Collinson IR, Miroux B, Fearnley IM, Skehel JM, Walker JE. The delta- and epsilon-subunits of bovine F1-ATPase interact to form a heterodimeric subcomplex. Biochem J. 1996;314(Pt 2):695–700. doi: 10.1042/bj3140695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Wilmes C, Tatsuta T, Langer T. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol Biol Cell. 2007;18:627–635. doi: 10.1091/mbc.E06-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanassiu AE, MacDonald NJ, Bencsura A, Vu HA. F1F0-ATP synthase functions as a co-chaperone of Hsp90-substrate protein complexes. Biochem Biophys Res Commun. 2006;345:419–429. doi: 10.1016/j.bbrc.2006.04.104. [DOI] [PubMed] [Google Scholar]
- Patriarca EJ, Maresca B. Acquired thermotolerance following heat shock protein synthesis prevents impairment of mitochondrial ATPase activity at elevated temperatures in Saccharomyces cerevisiae. Exp Cell Res. 1990;190:57–64. doi: 10.1016/0014-4827(90)90143-x. [DOI] [PubMed] [Google Scholar]
- Paul MF, Tzagoloff A. Mutations in RCA1 and AFG3 inhibit F1-ATPase assembly in Saccharomyces cerevisiae. FEBS Lett. 1995;373:66–70. doi: 10.1016/0014-5793(95)00979-j. [DOI] [PubMed] [Google Scholar]
- Pearce DA, Sherman F. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of yme1. J Biol Chem. 1995;270:20879–20882. doi: 10.1074/jbc.270.36.20879. [DOI] [PubMed] [Google Scholar]
- Penin F, Deleage G, Gagliardi D, Roux B, Gautheron DC. Interaction between delta and epsilon subunits of F1-ATPase from pig heart mitochondria. Circular dichroism and intrinsic fluorescence of purified and reconstituted delta epsilon complex. Biochemistry. 1990;29:9358–9364. doi: 10.1021/bi00492a008. [DOI] [PubMed] [Google Scholar]
- Prescott M, Nowakowski S, Gavin P, Nagley P, Whisstock JC, Devenish RJ. Subunit gamma-green fluorescent protein fusions are functionally incorporated into mitochondrial F1F0-ATP synthase, arguing against a rigid cap structure at the top of F1. J Biol Chem. 2003;278:251–256. doi: 10.1074/jbc.M204556200. [DOI] [PubMed] [Google Scholar]
- Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Zeng X, Briere JJ, Tzagoloff A. Assembly of F0 in Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1793:108–116. doi: 10.1016/j.bbamcr.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Schwimmer C, Lefebvre-Legendre L, Rak M, Devin A, Slonimski PP, di Rago JP, Rigoulet M. Increasing mitochondrial substrate-level phosphorylation can rescue respiratory growth of an ATP synthase-deficient yeast. J Biol Chem. 2005;280:30751–30759. doi: 10.1074/jbc.M501831200. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1986. [Google Scholar]
- Smith CP, Thorsness PE. Formation of an energized inner membrane in mitochondria with a gamma-deficient F1-ATPase. Eukaryot Cell. 2005;4:2078–2086. doi: 10.1128/EC.4.12.2078-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Gawryluk RM, Spencer DF, Pearlman RE, Siu KW, Gray MW. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J Mol Biol. 2007;374:837–863. doi: 10.1016/j.jmb.2007.09.051. [DOI] [PubMed] [Google Scholar]
- Sulahian R, Sikder D, Johnston SA, Kodadek T. The proteasomal ATPase complex is required for stress-induced transcription in yeast. Nucleic Acids Res. 2006;34:1351–1357. doi: 10.1093/nar/gkl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauer R, Mannhaupt G, Schnall R, Pajic A, Langer T, Feldmann H. Yta10p, a member of a novel ATPase family in yeast, is essential for mitochondrial function. FEBS Lett. 1994;353:197–200. doi: 10.1016/0014-5793(94)01045-5. [DOI] [PubMed] [Google Scholar]
- Thorsness PE, Fox TD. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993;134:21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong LA, de Vos AM, Milburn MV, Kim SH. Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J Mol Biol. 1991;217:503–516. doi: 10.1016/0022-2836(91)90753-s. [DOI] [PubMed] [Google Scholar]
- Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J Biol Chem. 2001;276:4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Barrientos A, Neupert W, Herrmann JM. Atp10p assists assembly of Atp6p into the F0 unit of the yeast mitochondrial ATPase. J Biol Chem. 2004;279:19775–19780. doi: 10.1074/jbc.M401506200. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Yue J, Jang J, Paul MF. A new member of a family of ATPases is essential for assembly of mitochondrial respiratory chain and ATP synthetase complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:26144–26151. [PubMed] [Google Scholar]
- Uh M, Jones D, Mueller DM. The gene coding for the yeast oligomycin sensitivity-conferring protein. J Biol Chem. 1990;265:19047–19052. [PubMed] [Google Scholar]
- Velours J, Arselin G. The Saccharomyces cerevisiae ATP synthase. J Bioenerg Biomembr. 2000;32:383–390. doi: 10.1023/a:1005580020547. [DOI] [PubMed] [Google Scholar]
- Voos W, Rottgers K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta. 2002;1592:51–62. doi: 10.1016/s0167-4889(02)00264-1. [DOI] [PubMed] [Google Scholar]
- Vyssokikh MY, Brdiczka D. The function of complexes between the outer mitochondrial membrane pore (VDAC) and the adenine nucleotide translocase in regulation of energy metabolism and apoptosis. Acta Biochim Pol. 2003;50:389–404. [PubMed] [Google Scholar]
- Walker JE, Dickson VK. The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Weber ER, Hanekamp T, Thorsness PE. Biochemical and functional analysis of the YME1 gene product, an ATP and zinc-dependent mitochondrial protease from S. cerevisiae. Molec. Biol. Cell. 1996;7:307–317. doi: 10.1091/mbc.7.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber ER, Rooks RS, Shafer KS, Chase JW, Thorsness PE. Mutations in the mitochondrial ATP synthase gamma subunit suppress a slow-growth phenotype of yme1 yeast lacking mitochondrial DNA. Genetics. 1995;140:435–442. doi: 10.1093/genetics/140.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Velours J, Stuart R, Schagger H. Characterization of domain interfaces in monomeric and dimeric ATP synthase. Mol Cell Proteomics. 2008;7:995–1004. doi: 10.1074/mcp.M700465-MCP200. [DOI] [PubMed] [Google Scholar]
- Zeng X, Barros MH, Shulman T, Tzagoloff A. ATP25, a new nuclear gene of Saccharomyces cerevisiae required for expression and assembly of the Atp9p subunit of mitochondrial ATPase. Mol Biol Cell. 2008;19:1366–1377. doi: 10.1091/mbc.E07-08-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]