Abstract
The characterization of naturally occurring variations in the human genome has evoked an immense interest during recent years. Variations known as biallelic Single-Nucleotide Polymorphisms (SNPs) have become increasingly popular markers in molecular genetics because of their wide application both in evolutionary relationship studies and in the identification of susceptibility to common diseases. We have addressed the issue of SNP genotype determination by investigating variations within the Renin–Angiotensin–Aldosterone System (RAAS) using pyrosequencing, a real-time pyrophosphate detection technology. The method is based on indirect luminometric quantification of the pyrophosphate that is released as a result of nucleotide incorporation onto an amplified template. The technical platform employed comprises a highly automated sequencing instrument that allows the analysis of 96 samples within 10 to 20 minutes. In addition to each studied polymorphic position, 5–10 downstream bases were sequenced for acquisition of reference signals. Evaluation of pyrogram data was accomplished by comparison of peak heights, which are proportional to the number of incorporated nucleotides. Analysis of the pyrograms that resulted from alternate allelic configurations for each addressed SNP revealed a highly discriminating pattern. Homozygous samples produced clear-cut single base peaks in the expected position, whereas heterozygous counterparts were characterized by distinct half-height peaks representing both allelic positions. Whenever any of the allelic bases of an SNP formed a homopolymer with adjacent bases, the nonallelic signal was added to those of the SNP. This feature did not, however, influence SNP readability. Furthermore, the multibase reading capacity of the described system provides extensive flexibility in regard to the positioning of sequencing primers and allows the determination of several closely located SNPs in a single run.
Since the first report on molecular polymorphism in humans describing the ABO system (Landsteiner 1900) a very large number of genetic variations have been characterized. One of the most common types of genetic diversity is the Single-Nucleotide Polymorphism (SNP), featuring a biallelic situation in which the alternative bases occur at a frequency exceeding 1% (Schafer and Hawkins 1998). The overall occurrence of SNPs in the human genome is not known, but a rough estimate of 0.1–1% of all bases has been reported (Cooper et al. 1985; Collins et al. 1997; Schafer and Hawkins 1998). Recent findings, based on screening of many individuals, reveal occurrences of 1 SNP per ∼ 220–350 base pairs in the human genome (Cargill et al. 1999; Halushka et al. 1999). This diversity provides an excellent tool for a wide panel of genetic analyses, including forensics, studies on population migration, diagnosis of disorders with a significant genetic influence, and various pharmacogenetic/pharmacogenomic applications. Especially in the latter areas, the rapid accumulation of available polymorphisms has elicited a renewed interest in association studies (Lander and Schork 1994; Collins et al. 1997).
Even though various simplified techniques are presented as applicable for primary definition of SNPs, semiautomated sequencing of DNA remains to date the most reliable method for this purpose (Eng and Vijg 1997). Emerging technologies for assessing DNA sequence differences between individuals, however, offer considerable promise for increasing the rate at which such polymorphisms can be defined (Collins et al. 1997). In the case of investigating characterized SNPs, several methods have been extensively evaluated. Except for those directly focused on polymerase chain reaction (PCR) and ligation methods (Landegren et al. 1988; Bottema et al. 1993; Livak 1995), most such approaches can be roughly divided into techniques based on primer extension or on the recognition of heteroduplex DNA. The former category includes several assay types, such as Solid Phase Minisequencing (SPM) and detection of dissimilarly sized extension fragments by MALDI-TOF mass spectroscopy (Little et al. 1997; Syvänen 1999), whereas the latter involves a wide number of applications, including mismatch cleavage detection, oligoarray hybridization, molecular beacon signaling, fluorescence monitoring of PCR, and electronic dot blot assay (Pease et al. 1994; Mashal et al. 1995; Southern 1996; Tyagi and Kramer 1996; Gilles et al. 1999; Nauck et al. 1999; for review, see Graber et al. 1998).
In our study design, we have approached the issue of SNP identification by using the recently described real-time pyrophosphate (PPi) detection method known as pyrosequencing (Nyrén and Lundin 1985; Ronaghi et al. 1996, 1998). This technique is based on an indirect bioluminometric assay of the pyrophosphate (PPi) that is released from each dNTP upon DNA chain elongation. Following Klenow polymerase-mediated base incorporation, PPi is released and used as a substrate, together with adenosine 5′-phosphosulfate (APS), for ATP sulfurylase, which results in the formation of ATP. Subsequently, the ATP accomplishes the conversion of luciferin to its oxi-derivative by the action of luciferase. The ensuing light output becomes proportional to the number of added bases, up to about four bases. To allow processivity of the method dNTP excess is degraded by apyrase, which is also present in the starting reaction mixture, so that only dNTPs are added to the template during the sequencing procedure (Nyrén and Lundin 1985; Ronaghi et al. 1996). The process has been fully automated and adapted to a 96-well format, which allows rapid screening of large SNP panels.
To assess the applicability of pyrosequencing for SNP identification, we selected 10 such variations, distributed throughout the 5′ regulatory and coding sequences of angiotensinogen, angiotensin I-converting enzyme, and angiotensin II type I receptor. These genes represent essential components of the Renin–Angiotensin–Aldosterone System (RAAS), which plays a crucial role in hormonal mechanisms that regulate blood pressure and electrolyte–blood-volume homeostasis (Goodfriend et al. 1995; Sealy and Laragh 1995). Several biallelic variations in this gene cluster have been analyzed in various combinations for correlation to clinical drug response data. Using large sample panels, combinations of three to six polymorphisms were ultimately compiled into genetic signatures that are indicative of patient susceptibility to a class of antihypertensive substances (Sanders 1999). The panel of nucleotide variations selected for this study encompasses several different biallelic bases and a wide variety of sequence contexts. By taking advantage of the assay system's flexibility regarding positioning of the identification primer and the number of bases read for each SNP, several parameters were employed for reinforced assessment of the analyzed genotypes. Using this approach, the entire assembly of tested SNPs proved amenable to unequivocal genotype determination in all configurations tested. The described assay therefore enables a highly reliable SNP identification on an overall basis, because of its ability to relate allelic bases quantitatively to closely situated nonallelic counterparts and because of the low level of background signal. Moreover, the ability to prepare and determine the sequence of 96 samples in parallel enables expedient analysis of large sample numbers.
RESULTS
PCR Optimization and Preparation of Templates for Pyrosequencing
The target sequences employed for PCR-amplification and ensuing SNP genotyping by pyrosequencing were derived from reports on angiotensinogen (AGT) (Fukamizu et al. 1990; Jeunemaitre et al. 1992), angiotensin I-converting enzyme (ACE) (Soubrier et al. 1988; Hubert et al. 1991), and angiotensin II type I receptor (AT1R) (Guo et al. 1994). Among reported polymorphisms, two in AGT (A-20C and G-6A)(Jeunemaitre et al. 1992), five in ACE (A-240T, T1237C, G2215A, G2350A, and T3409C) (Villard et al. 1996; Low et al. 1998) and one in AT1R (A1166C) (Bonnardeaux et al. 1994) were selected for analysis by pyrosequencing. Furthermore, two variations located in the AT1R upstream region (A-227C and C-226G) were identified by Sanger DNA sequencing and were included in the panel of polymorphic positions studied here. Accordingly, selected motifs were separately optimized for PCR product amount and specificity, using a panel of test temperature cycling protocols. As a result of such titrations, a single setting that enabled high-quality outputs was adopted for all 10 fragments.
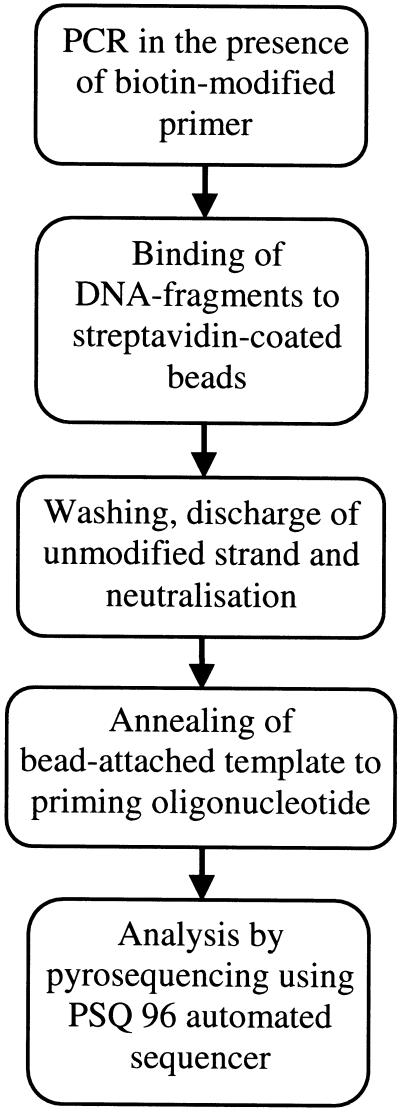
To obtain a sequencing template from PCR one of each PCR primer pair was covalently coupled to biotin. Single-stranded DNA was generated by exposing amplified DNA fragments, immobilized onto streptavidin-coupled beads, to alkali, followed by release of the complementary strand and neutralization. In this experimental setting, bead-attached DNA was consistently employed as a template. The entire sample preparation procedure, including amplification, strand separation, and annealing to the respective primer, was conducted in 96-well microtiter plates. Sample transfer from various solutions in this chain of events was expedited by a magnetic 96-pin manifold. Subsequent to these steps, samples were loaded in special microplates that fitted into a socket of the PSQ 96 instrument. A schematic outline of the procedure is shown in Figure 1.
Figure 1.
Schematic outline of the procedure.
Automated Real-Time Pyrosequencing
The DNA sequencer employed can automatically add small volumes of each of the four nucleotides to DNA templates, according to a prespecified dispensation scheme, and measure signal output from light upon nucleotide incorporation, emitted by the conversion of luciferin to oxiluciferin. The ultimate reaction in this chain of events is triggered by the key substance pyrophosphate (PPi) through several intermediate enzymatic interactions, and the released light quantum is proportional to the amount of released PPi. In practice, the signal intensity maintains a good correlation to the number of incorporated bases up to homopolymers of about 4 bases (Ronaghi et al. 1996, 1998). On average, one base is read every 65 seconds, and 96 samples can be processed in parallel.
Accurate and Expedient Genotype Assessment through SNP/Context-Adapted Nucleotide Addition
It is possible to instruct the PSQ 96 sequencer to add the four bases according to a specified iterative cycle (Ronaghi et al. 1996, 1998). This approach was tested (not shown) for several of the SNPs addressed here, and the data obtained produced results equivalent to those described below. Whenever detailed knowledge of the context target sequences is available, which is typical in a regular SNP scanning study, sequence-specific dispensing protocols can be applied. This approach was consistently preferred here because it requires fewer nucleotide additions and, consequently, leads to a faster sequencing procedure with less deterioration of the reaction mixtures per base read, which may ultimately promote the resolution of pyrogram readouts (Figs. 2–4).
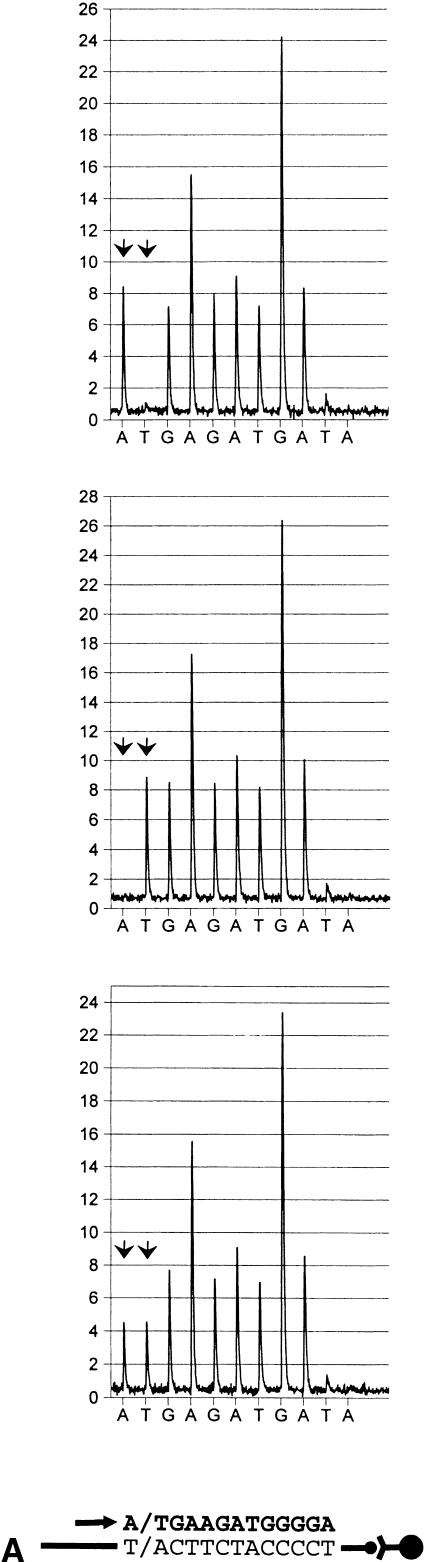
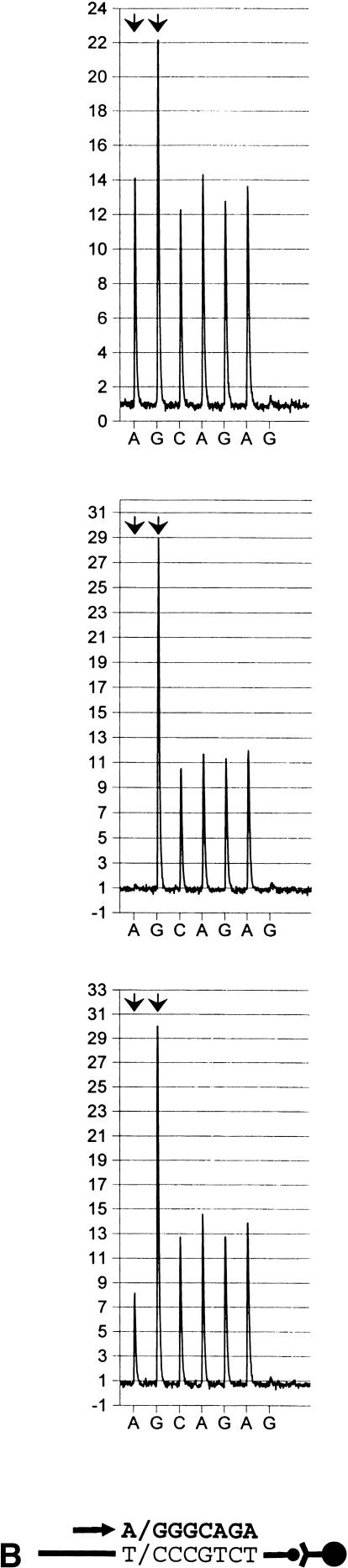
Figure 2.
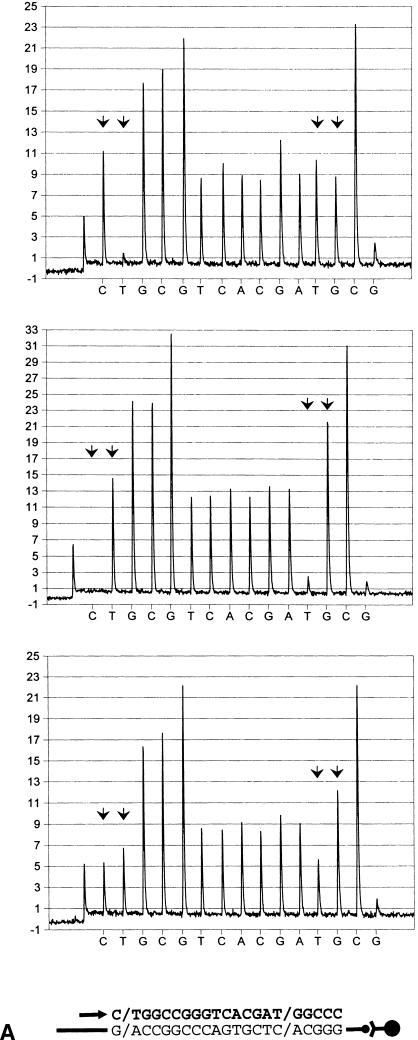
Identification of single nucleotide polymorphisms (SNPs) in biotin-labeled templates bound to streptavidin-coated paramagnetic beads (Dynabeads Streptavidin), by pyrosequencing. Primers were extended in the antisense direction. Sequencing primers and incorporated nucleotides are shown below each pyrogram series as an arrow and bold letters, respectively. Arbitrary luminescense units are shown on the ordinate axis. Each SNP is indicated by an arrowhead doublet. The two top diagrams show the respective homozygous test samples, whereas those below represent heterozygous counterparts. (A) A/T polymorphism, located in the human angiotensin I-converting enzyme promoter (A-240T). (B) C/T variation in the human angiotensin I-converting enzyme gene in exon 8 (T1237C). (C) Identical with (b), except that an allele-separating nucleotide dispensing scheme (within the biallelic variation) was applied.
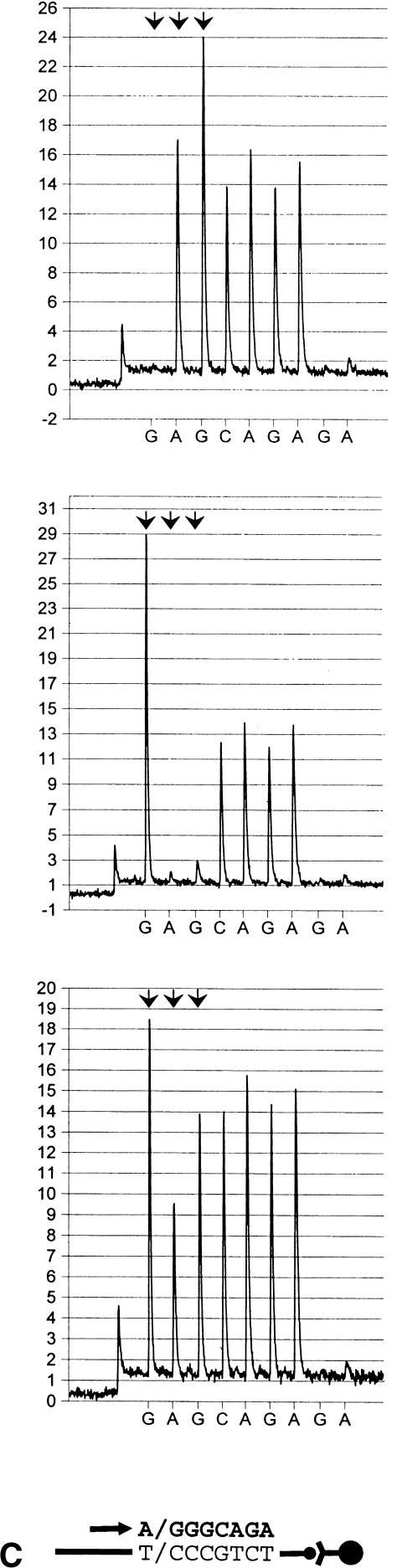
Figure 4.
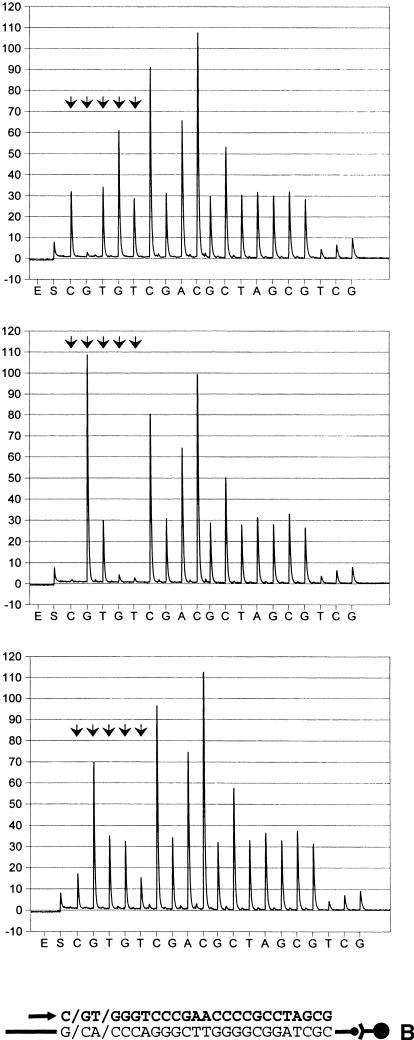
Assessment of distinct SNPs on single fragments using real-time pyrophosphate detection. Arrowhead doublets indicate the location of SNPs. The schematic illustration below each pyrogram series shows the template, the primer location (arrow), and incorporated nucleotides (bold letters). (a) Variations in the human angiotensinogen promoter. Genotypes shown (at bottom) are: G/G, A/A and G/A (G-6A) and A/A, C/C and A/C (A-20C). (b) Polymorphisms, located in the promoter of human angiotensin II type I receptor gene. SNP configurations shown (top to bottom) correspond to G/G, C/C and G/C in position C-226G and A/A, C/C and A/C, in position A-227C.
Discrimination of Alleles with SNP-Proximal Primers
Each of the 10 SNPs tested was evaluated for genotype discrimination, using primers with their 3′ ends annealed adjacent to the respective variable position. For this purpose test samples of each SNP in this panel were employed that harbored the two alternative homozygous and the heterozygous configurations, respectively, as determined either by the semiautomated solid-phase Sanger DNA technique (Ståhl et al. 1988; Lagerkvist et al. 1994) or by solid-phase single-base extension reactions (Syvänen et al. 1990, 1993). To obtain reference luminescence peaks for each of the variable bases, several SNP-succeeding bases were read. Consequently, the sequencing analyses were allowed to proceed to include nonallelic bases equivalent to those of the biallelic position. Moreover, one or two nonproductive nucleotide dispensations, corresponding to bases of the respective SNP, were conducted (Figs. 2–4). The latter approach was added to obtain an estimate of primer elongation asynchronization, which although generally negligible can attain measurable levels, especially when homopolymers of four or more bases become incorporated during synthesis. As evident from Figure 2, showing two typical examples based on sequencing primers located at an adjacent 5′-proximal position of the respective biallelic variation, the different genotypes were easily mapped by visual inspection of pyrograms. Several SNPs described here were followed by at least two bases distinct from those of the SNP position (Fig. 2A). In those cases, allelic peaks from heterozygous DNA were generally evenly distributed and attained about 60% of the height (and area) of the nonvariable counterparts. Whenever one of the alternative bases of an SNP comprised part of a homopolymer, a skewed ratio was obtained because of inherent properties of the pyrosequencing principle (Fig. 2B). Because peak heights were largely proportional to the number of incorporated bases, such genotype configurations were nonetheless readily amenable to unequivocal determination, similar to the aforementioned type of SNP.
To increase the detailed resolution of such SNPs, the base dispensation program can be set to instruct incorporation of each allelic (SNP-succeeding) homopolymer separately. For example, an SNP in exon 8 of ACE (T1237C) is characterized by a T/CCC motif (Villard et al. 1996) and can be resolved either by (antisense orientation) AG or GAG base addition during pyrosequencing. (Figs. 2B,C). When analyzing heterozygous samples, the latter setting enabled a split of the homopolymer into two lower peaks (Fig. 2C). No appreciable difference in T1237C SNP interpretability, however, was seen between the two dispensation protocols (Table 1; see also section on “Evaluation of SNP Identification Accuracy” below). Moreover, it appears that the limited extension of homopolymers addressed here neither impeded genotype interpretation nor suppressed data readout reproducibility when compared with archetypal SNPs (Table 1 and below). Nonetheless, the allele-splitting dispensation approach may prove benefical for optimal determination of polymorphisms in longer homopolymer stretches.
Table 1.
Accuracy and Precision of SNP Genotyping in a Panel of Test Series
| Gene | Position | Heterozygotes | Homozygotes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B1a/B1 | B2a/B2 | B1s/B1 | B2s/B2 | |||||||||||||
| N | ER | AM | RSD | ER | AM | RSD | N | ER | AM | RSD | N | ER | AM | RSD | ||
| ACE | A-240T | 13 | 0.5 | 0.569 | 0.273 | 0.5 | 0.611 | 0.125 | 6 | 1 | 0.958 | 0.08 | 11 | 1 | 0.975 | 0.064 |
| ″ | T1237C a | 12 | 0.5 | 0.573 | 0.046 | 2.5 | 2.232 | 0.023 | 3 | 1 | 1.061 | n.d. | 7 | 3 | 2.605 | 0.029 |
| ″ | T1237C b | 12 | 0.5 | 0.558 | 0.039 | 1 | 0.954 | 0.03 | 3 | 1 | 1.055 | n.d. | 7 | 3 | 2.636 | 0.017 |
| ″ | G2215A | 8 | 0.5 | 0.546 | 0.04 | 0.5 | 0.558 | 0.056 | 4 | 1 | 1.067 | 0.039 | 3 | 1 | 0.896 | n.d. |
| ″ | G2350A | 8 | 0.5 | 0.613 | 0.065 | 0.5 | 0.615 | 0.04 | 4 | 1 | 1.106 | 0.017 | 3 | 1 | 1.032 | n.d. |
| ″ | T3409C c | 7 | 0.5 | 0.569 | 0.068 | 0.5 | 0.578 | 0.11 | 3 | 1 | 0.957 | n.d. | 5 | 1 | 1.053 | 0.056 |
| ″ | T3409C d | 16 | 2.5 | 2.041 | 0.05 | 0.5 | 0.564 | 0.093 | 11 | 3 | 2.379 | 0.089 | 3 | 1 | 0.971 | n.d. |
| AGT | G-6A | 12 | 0.5 | 0.573 | 0.141 | 0.5 | 0.722 | 0.069 | 7 | 1 | 1.071 | 0.082 | 3 | 1 | 1.204 | n.d. |
| ″ | A-20C | 7 | 1.5 | 1.242 | 0.035 | 0.5 | 0.695 | 0.048 | 1 | 2 | 1.606 | n.d. | 14 | 1 | 0.76 | 0.08 |
| AT1R | A-227C | 11 | 0.5 | 0.494 | 0.104 | 2 | 2.07 | 0.115 | 33 | 1 | 0.857 | 0.04 | 3 | 4 | 3.984 | n.d. |
| ″ | C-226G | 11 | 0.5 | 0.473 | 0.101 | 2 | 2.001 | 0.114 | 33 | 1 | 0.939 | 0.038 | 3 | 4 | 3.866 | n.d. |
| ″ | A1166C e | 7 | 0.5 | 0.653 | 0.087 | 0.5 | 0.611 | 0.079 | 1 | 1 | 0.89 | n.d. | 7 | 1 | 1.176 | 0.061 |
| ″ | A1166C f | 4 | 0.5 | 0.609 | 0.054 | 1.5 | 1.196 | 0.059 | 3 | 2 | 1.456 | n.d. | 8 | 1 | 1.128 | 0.044 |
Acronyms used: (ACE) human angiotensinogen-converting enzyme gene; (AGT) human angiotensinogen gene; (AT1R) human angiotensin II type I receptor gene. Designations of the polymorphic positions are as in Hubert et al. 1991, Jeunemaitre et al. 1992, and Bonnardeaux et al. 1994. A-226C and C-227G have been entitled according to standard principles for SNP nomenclature. Abbreviations: (ER) expected ratio, (AM) arithmetic mean; (RSD) relative standard deviation, the number limit for this operation was set to four; (n.d.) not determined; (B1a/B1) ratio between the initially appearing base of the respective SNP (in heterozygous samples) and a non-allelic counterpart; (B2a/B2) definition analogous with that of B1a/B1; (B1s/B1) ratio between the initial base of a homozygous SNP and a reference counterpart; (B2s/B2) definition analogous with that of B1s/B1. Index letters: (A) the dispensing scheme was as specified in Fig. 2B; (B) the nucleotide dispensing protocol was according to specification in Fig. 2C; (C) the 3′ end of the sequencing primer was annealed in 5′ juxtaposition, relative to the SNP; (D) the sequencing primer was located three bases in the upstream direction, in relation to the SNP; (E) the sequencing approach adhered to that of c; (F) the sequencing primer (3′ end) was positioned four bases 5′ of the SNP.
Genotype Determination Using SNP-Distal Primers
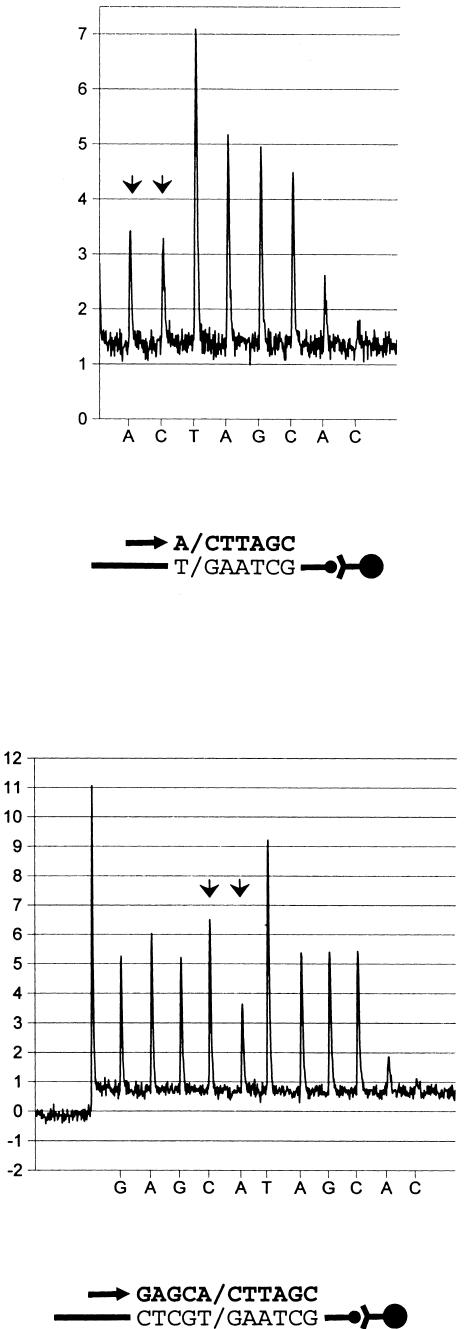
Additional oligonucleotides were employed as primers for strand extension using 3 of the 10 tested fragment types in the real-time pyrophosphate detection procedure. They were designed to direct synthesis at three and four bases upstream of each SNP respectively. Results show an equivalent resolution of SNP genotypes, as compared with aforementioned counterparts, and exemplifies the technique's wide options for design of primers for the elongation reaction (Table 1). A typical example is depicted in Figure 3.
Figure 3.
Genotype assessment of a single SNP by pyrosequencing using two different primer oligonucleotides and heterozygous test samples. Below each pyrogram series, the respective sequencing primer (indicated by arrow) and incorporated bases (bold letters) are shown. Primers were extended in the sense direction. The ordinate represents luminescence (arbitrary units). Arrowhead doublets indicate the location of SNPs. The polymorphic position, addressed in both panels, corresponds to position A1166C in exon 5 of human angiotensin II type I receptor gene. (Upper panel) Extension of a sequencing primer, juxtaposed to the biallelic position. (Lower panel) Synthesis from a primer, located four bases upstream of the SNP.
Identification of Multiple SNPs on a Single Template
To evaluate the feasibility of PSQ 96 for addressing several polymorphic positions in a single run, two separate fragments that harbor SNP pairs were investigated, using an uninterrupted sequencing procedure. One of those comprised a section of the AGT promoter and covered two variations designated G-6A and A-20C (Jeunemaitre et al. 1992) that are located within a 15-base segment. Here, the primer elongation reaction was allowed to proceed over 18 bases in total. Albeit with a slightly reduced conformity between expected and observed peak ratios at the distal end of this sequence context, both variations were accurately determined with high clarity (Fig. 4A and Table 1). As indicated by RSD calculation, the resolving power of the primer-distal SNP compared favorably with that of the proximal counterpart (Table 1). The second DNA fragment harbored two juxtaposed SNPs within the AT1R promoter (A-227C and C-226G). Whenever heterozygosity occurred, peaks from five consecutive nucleotide dispensations became indicative of the genotype configurations of the two polymorphic positions. This is caused by nonsynchronous allelic extensions within the variable region of such samples. At the end of the second SNP, however, the allelic elongations attained correct (synchronous) relative positions which secured readability of the succeeding DNA sequence (Fig. 4Bb). Despite the relatively large numbers tested (Table 1), only three haplotypes were identified: G/G; A/A, C/C; C/C and C/G; A/C (C-227G and A-226C, respectively). Unequivocal identifications of the respective biallelic configurations are shown in Figure 4. Moreover, good readout consistency within hetero- and homozygous SNP configurations was evidenced by low RSD valves (Table 1).
Evaluation of SNP Identification Accuracy
An expanded test sample panel, encompassing human genomic DNA from 15 to 47 individuals, served as a template for additional genotyping experiments by pyrosequencing. The allelic configurations were consistently very clear, with the exception of the rare event of signal levels falling beyond ∼ 1.5–2.0 fluorescence units. To estimate the precision of SNP genotyping, arithmetic mean values and the relative standard deviation (RSD) of ratios between peaks of the variable position as well as between these and selected nonallelic counterparts, were calculated. In most instances, neither of the biallelic bases comprised homopolymers, and ratios close to the expected figure 0.5 were recorded (Table 1). Appreciable deviations from theoretical values occurred, however, when homopolymers were involved. Moreover, RSD values of homozygous and heterozygous SNPs commonly attained figures below 0.1; only a few scattered figures occurred above this level. From Table 1 it is also evident that the tested homopolymer SNPs did not perform less well than other biallelic variations in the range of ratio values. Results in compliance with these findings were obtained from a similar test series, encompassing 44 samples of each of 7 SNPs, distinct from those addressed here (not shown).
DISCUSSION
The accumulated number of identified biallelic variations in the human genome has increased substantially in recent years, largely through the application of novel scanning technologies such as chip microarray hybridization (Pease et al. 1994; Southern 1996), temperature-modulated heteroduplex analysis (Underhill et al. 1997), and other approaches. Reliable determination of allelic configuration of identified or candidate SNPs is, however, a quite different issue, since high-throughput screening assays are prone to reveal both false positive and false negative results. In this study, we present an expedient approach for accurate assessment of biallelic genetic variations which is applicable also to single-base mutations and micro-insertions/deletions. It is essentially based on synthesis of a complementary DNA strand onto a template and real-time detection of incorporated bases.
An inherent capacity of the automated pyrosequencing method to allow the reading of up to 20–25 consecutive bases imparts several advantages to this methodology for SNP identification. First, it provides a considerable flexibility in primer positioning, which enhances the possibility to design feasible primers for the synthesis reaction as compared to techniques based on single-nucleotide extension. Second, it enables the identification of several closely located polymorphisms in a single extension reaction.
Primer extension using the pyrosequencing approach can be conducted either on liquid-phase or solid-phase templates. Although some signal quenching by bead particles can be assumed to occur, the latter approach—based on anchorage of single strands to paramagnetic beads by the well-established biotin–streptavidin interaction—was consistently employed. This is because data readouts from bead-attached DNA strands were more robust when compared with those obtained from unbound counterparts, presumably because neutralization of the liquid phase results in a higher ionic strength in the final reaction mixture. Moreover, elongation of solid phase-attached templates is readily amenable to an uninterrupted manifold preparation procedure that precedes the sequencing analysis. Separation of DNA duplexes can also be accomplished by heat, which would, in principle, eliminate at least one of the aforementioned difficulties. In our work, however, this type of intervention commonly resulted in spurious peaks, presumably because of the release of a significant fraction of the complementary bead-attached DNA strand, even when moderate conditions were employed. In the protocol employed here, several pre- and post-neutralization washing steps were included prior to the sequencing reaction. Lately, however, we have learned that a markedly simplified procedure based on alkalization (with no preceding washes) of bound DNA fragments and subsequent neutralization in annealing buffer provides sequencing readouts of exactly the same quality.
A striking feature of pyrogram readouts from this study is their clear distinction between the various genotypes of each SNP studied. Moreover, RSD values for the ratio between key peaks of the respective SNPs and reference counterparts were almost consistently about 0.1 or lower. This is indicative of low variance from typical values. A similar study, conducted in our laboratory involving different but much larger sample panels for each SNP had an overall outcome that resembled findings presented here. Some notable precautions, however, can be taken from that investigation. Primer concentrations exceeding 10–20 pmoles of each PCR primer reduced luminescence signal output considerably, and this proved detrimental to the accuracy of genotype determination. This particular effect can be attributed to a steeply increasing unfavorable proportion of full-length DNA products versus low-molecular-weight components in the Dynabead binding reaction, with escalating oligonucleotide amounts. Furthermore, it was found that arbitrary signal levels from nonallelic base peaks should preferably not be below 4 luminescence units to allow for high-precision assessment of genetic variations.
Heat-sensitive enzyme components (especially luciferase) in the pyrosequencing reaction mixture require that the extension temperature be set to 28° C. Such conditions indisputably favor interactions within and between primer molecules. Oligonucleotides employed as sequencing primers must accordingly be carefully designed to avoid illegitimate extensions. In contrast to single-base extension technologies, however, pyroseqencing offers a considerable flexibility in primer positioning, which notably alleviates problems confined to the temperature limitation.
In principle, reading of the variable bases only should be sufficient for satisfactory identification of biallelic polymorphism and, in fact, the analyzed SNPs lend themselves to unequivocal assessment simply by reading this section of the pyrogram readouts. Nonetheless, recordings of several nonallelic bases beyond the variable positions in the same reaction are likely to confer additional precision to the analysis and were therefore included on a regular basis. Using this approach, numerical values for each SNP determination were calculated semiautomatically and employed as estimates for accuracy. In this study nonallelic counterparts to variable bases were selected as references, but non-equivalent bases can also serve this purpose. An SNP analysis software package, based on an algorithm that compares SNP-specific bases with several feasible nonvariable reference bases, has been developed. It is equipped with genotype accuracy assessment properties and editing functions. In large test sample series, full conformity between manual and automatic genotype calling was seen (not shown). This improvement substantially enhances the throughput and convenience of SNP identification by pyrosequencing.
METHODS
Solid-Phase Sanger Sequencing
Manifold sequencing reactions of PCR-amplified and biotin-labeled DNA-fragments, employing streptavidin-coupled Sepharose HP attached to teeth of plastic combs (Amersham Pharmacia Biotech AB, Uppsala, Sweden), were conducted essentially as outlined (Ståhl et al. 1988; Lagerkvist et al. 1994). Resulting products were loaded on a sequencing gel and detected using an ALFexpress automated laser fluorescence sequencer. Data interpretation was accomplished with the aid of ALFwin dedicated software (Amersham Pharmacia Biotech AB).
Solid-Phase Minisequencing
Single-nucleotide determinations were conducted largely as described by Syvänenen et al. (Syvänen et al. 1990, 1993). Deviations from the technical details in these reports were confined to fluorescein-12-dNTPs (NEN Life Science Products, USA) and DynaZyme F-500L DNA polymerase (Finnzymes Oy, Helsinki, Finland), which replaced their respective equivalents. In addition, signals indicating the respective genotypes were obtained from p-nitrophenol, which was generated from the substrate p-nitrophenyl phosphate by anti-fluorescein Fab-fragment-conjugated alkaline phosphatase (Boehringer-Mannheim). Yellow p-nitrophenol was detected in a plate-reader at 450 nm (Multiskan EX, Labsystems Oy, Helsinki, Finland).
PCR and Sequencing Primers (for Pyrosequencing Reactions)
Primers for amplification by PCR and for sequencing of amplicons were purchased from Scandinavian Gene Synthesis AB, Köping, Sweden. Selected regions in the human angiotensinogen (AGT), angiotensin I-converting enzyme (ACE) and angiotensin II type I-receptor (AT1R) were amplified from genomic DNA. The following PCR primer pair was employed for AGT targets (b indicates biotin): 5′-b-TGATGTAACCCTCCTCTCCA-3′, 5′-CGGCTTACCTTCTGCTGTAGTA-3′ (142 bp; A-6G and A-20C); ACE targets were amplified using the following primers: 5′-b-GGTCGGGCTGGGAAGAT-3′, 5′-GCTCCCGCAGAGGAAGC-3′ (158 bp; A-240T); 5′-b-GGACCAGCTCTCCACAGTGC-3′, 5′-GCCAGCACGTCCCCAAT-3′ (129 bp; T1237C); 5′-GCCAGGAAGTTTGATGTGAAC-3′, 5′-b-GATTCCCCTCTCCCTGTACCT-3′ (145 bp; G2215A); 5′-b-CTCCCCTTACAAGCAGAGGT-3′, 5′-CCCTCCCATGCCCATAA-3′ (122 bp; G2350A); 5′-CTCGCTCTGCTCCAGGTAC-3′, 5′-b-GCCTCCTTGGACTGGTAGAT-3′ (123 bp; T3409C). The AT1R counterparts were: 5′-b-ACCCCCAGCTCGCTCTC-3′, 5′-GACGGCAGCAAGGATGAT-3′ (111 bp; A-227C and C-226G); 5′-GCCCCTCAGATAATGTAAGC-3′, 5′-b-TTTCTCCTTCAATTCTGAAAAGTA-3′ (177 bp; A1166C). The corresponding sequencing primers were as follows: 5′-ACGGCAGCTTCTTCCCC-3′, 5′-AGAAAGGGCCTCCTCTCTTT-3′, 5′-CCCCGACGCAGGGAGAC-3′, 5′-GACCTAGAACGGGCAGC-3′, 5′-AGGTCTTCATATTTCCGGGA-3′, 5′-TCGCTCTGCTCCAGGTACTT-3′, 5′-TCAACTATCAGATCAGCGTTCAC-3′, and 5′-AGCACTTCACTACCAAATGAGC-3′, respectively.
DNA Amplification and Fragment Analysis
DNA samples were prepared either from blood or from buccal cells of Caucasian individuals. All amplification reactions were performed in a 96-well microtiter plate thermal cycler (PE 9600, Perkin Elmer, USA). The reactions were carried out in 50 μl of 1× PCR buffer II (Perkin Elmer), 1.5 mm MgCl2, 25 μm of each of the four dideoxynucleotides, 10–20 pmoles of primers and Taq polymerase (1.5 units; AmpliTaq Gold, Perkin Elmer). A single fragment type (ACE, 260 bp) was amplified in a slightly modified reaction (0.875 mm MgCl2, 2.5% v/v of DMSO, and 2.5 units of AmpliTaq Gold). Thermal cycling was performed with an initial denaturation for 5 minutes at 95° C, succeeded by 50 cycles of denaturation for 15 seconds at 95°C, primer annealing for 30 seconds, and synthesis for 45 seconds. A final primer extension was conducted for 5 minutes at 72°C.
Each amplicon was inspected with respect to PCR yield and specificity by UV-transillumination of EtBr-stained gels from subsequent electrophoretic separation in 1.5% agarose.
Preparation of Single-Stranded DNA and Annealing to Identification Oligonucleotide
Biotinylated DNA fragments (about 5 pmoles each) were attached to streptavidin-modified paramagnetic beads (Dynabeads™ M-280, Dynal A/S, Skøyen, Norway) by slow revolution of the suspensions in a thermostated (43° C) chamber (HB-1, Techne Ltd, Duxford, England) in a high salt (BW) buffer (0.05% Tween 20, 0.5 mm EDTA, 1 m NaCl, 5 mm Tris-HCl, pH 7.6). Following washing steps in BW buffer and 10 mm Tris-HAc, pH 7.6, respectively, the material was resuspended in 0.15 m NaOH. After a 10-minute incubation period, the separated DNA strands were saved for subsequent processing as follows: Beads were washed twice in 10 mm Tris-HAc, pH 7.6 and transferred to a solution [20 mm Mg(CH3COO)2, 10 mm Tris-HAc pH 7.6], containing 20–25 pmoles of the respective sequencing primer. Subsequently, the annealing procedure was conducted. This step comprised heating for 45 seconds at 95° C followed by cooling at room temperature. Throughout most of the preparation steps, the bead-coupled fragments were processed in parallel using a manifold device equipped with 96 magnetic ejectable microcylinders (PSQ 96 Sample Prep Tool, Pyrosequencing AB), which were protected by a disposable plastic pocketed cover.
Solid-Phase Pyrosequencing
DNA sequence reading was conducted in a P3 PSQ 96 instrument (Pyrosequencing AB), which is a third-generation prototype of an instrument that recently has become commercially available. Its principal components are instrumentation for accurate dispensation of small reagent volumes (0.2 μl), a plate rotating mechanism, a Charged Coupled Device (CCD) camera, data acquisition modules, and an interface for PC connection. Prior to analysis, the enzymes and each of the four dNTPs (PSQ 96 SNP Reagent Kit, Pyrosequencing AB) were loaded in a special cartridge that was mounted in the PSQ 96 instrument.
Samples, annealed with their respective sequencing primers, were transferred to a translucent 96-well microplate, designed to fit onto a socket in the PSQ 96 system. A unique dispensing order, complementary to the template and including both alleles, was specified for each tested SNP. Subsequent to sequencing, readout data were further processed for data evaluation.
Statistical Evaluation
Pyrogram readouts were converted to numerical values for peak heights, using a software module designed for this purpose (Pyrosequencing AB). Each base of the various SNPs was compared with a reference counterpart located elsewhere in the sequence context, and a ratio was calculated. The values thus obtained were subsequently processed for arithmetic means and relative standard deviation (RSD), the latter being a measure of data readout consistency. Hetero- and homozygous samples were tabulated separately.
Acknowledgments
We thank Dr. Rhiannon Sanders at Eurona Medical AB and Dr. Björn Ingemarsson at Pyrosequencing AB for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Present address: National Food Administration, SE-751 26, Uppsala, Sweden.
E-MAIL: ulfh@slv.se; FAX 46 (18) 171433.
REFERENCES
- Bonnardeaux A, Davies E, Jeunemaitre X, Féry I, Charru A, Clauser E, Tiret L, Cambien F, Corvol P, Soubrier F. Angiotensin II type I receptor gene polymorphisms in human essential hypertension. Hypertension. 1994;24:63–69. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- Bottema CDK, Sarkar G, Cassay JD, Ii S, Dutton CM, Sommer SS. PCR-amplification of specific alleles: A general method of rapidly detecting mutations, polymorphisms, and haplotypes. Meth Enzymol. 1993;218:388–402. doi: 10.1016/0076-6879(93)18031-7. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Chakravarti A. Variations on a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Smith BA, Cooke HJ, Niemann S, Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genome. Hum Genet. 1985;69:201–205. doi: 10.1007/BF00293024. [DOI] [PubMed] [Google Scholar]
- Eng C, Vijg J. Genetic testing: The problems and the promise. Nature Biotechnol. 1997;15:422–426. doi: 10.1038/nbt0597-422. [DOI] [PubMed] [Google Scholar]
- Fukamizu A, Takahashi S, Seo MS, Tada M, Tanimoto K, Uehara S, Murakami K. Structure and expression of the human angiotensinogen gene. J Biol Chem. 1990;265:7576–7582. [PubMed] [Google Scholar]
- Gilles PN, Wu DJ, Foster CB, Dillon PJ, Chanock SJ. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nature Biotechnol. 1999;17:365–370. doi: 10.1038/7921. [DOI] [PubMed] [Google Scholar]
- Goodfriend TL, Elliott ME, Catt KJ. Angiotensin receptors and their antagonists. N Engl J Med. 1995;334:1649–1654. doi: 10.1056/NEJM199606203342507. [DOI] [PubMed] [Google Scholar]
- Graber JH, O'Donnell MJ, Smith CL, Cantor CR. Advances in DNA diagnostics. Curr Opin Biotechnol. 1998;9:14–18. doi: 10.1016/s0958-1669(98)80078-5. [DOI] [PubMed] [Google Scholar]
- Guo D-F, Furuta H, Mizukoshi M, Inagami T. The genomic organisation of human angiotensin II type I receptor. Biochem Biophys Res Commun. 1994;200:313–319. doi: 10.1006/bbrc.1994.1450. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan J-B, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Hubert C, Houot A-M, Corvol P, Soubrier F. Structure of the angiotensin I-converting enzyme gene. J Biol Chem. 1991;266:15377–15383. [PubMed] [Google Scholar]
- Jeunemaitre X, Soubrier F, Kotelevtsev YV, Lifton RP, Williams CS, Charru A, Hunt SC, Hopkins PN, Williams RR, Lalouel J-M, et al. Molecular basis of human hypertension: Role of angiotensinogen. Cell. 1992;71:169–180. doi: 10.1016/0092-8674(92)90275-h. [DOI] [PubMed] [Google Scholar]
- Lagerkvist A, Stewart J, Lagerström-Fermér M, Landegren U. Manifold sequencing: Efficient processing of large sets of sequencing reactions. Proc Natl Acad Sci. 1994;91:2245–2249. doi: 10.1073/pnas.91.6.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Landsteiner K. Zur Kenntnis der antifermentativen, lytischen und agglutinierended Wirkungen des Blutserums und der Lymphe. Zbl Bakt. 1900;27:357–362. [Google Scholar]
- Little DP, Braun A, O'Donnell MJ, Köster H. Mass spectrometry from miniaturized arrays for full comparative DNA analysis. Nature Med. 1997;3:1413–1416. doi: 10.1038/nm1297-1413. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridisation. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- Low C-M, Yap EP-H, Lee EJ-D. Characterization of polymorphisms IVS25AS-6A to G, C1237T and T3409C in the human angiotensin converting enzyme gene. Pharmacogenetics. 1998;8:361–364. doi: 10.1097/00008571-199808000-00011. [DOI] [PubMed] [Google Scholar]
- Mashal RD, Koonts J, Sklar J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nature Genet. 1995;9:177–183. doi: 10.1038/ng0295-177. [DOI] [PubMed] [Google Scholar]
- Nauck MS, Gierens H, Nauck MA, März W, Wieland H. Rapid genotyping of human platelet antigen 1 (HPA-1) with fluorophore-labelled hybridisation probes on the LightCycler™. Brit J Haematol. 1999;105:803–810. doi: 10.1046/j.1365-2141.1999.01427.x. [DOI] [PubMed] [Google Scholar]
- Nyrén P, Lundin A. Enzymatic method for continuous monitoring of inorganic pyrophosphatase synthesis. Anal Biochem. 1985;151:504–509. doi: 10.1016/0003-2697(85)90211-8. [DOI] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SPA. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlén M, Nyrén P. Real-time pyrophosphate detection for DNA sequencing. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Sanders R. Mining the Swedish clinical archives to develop pharmacogenomic tests. Mol Diagnosis. 1999;4:319–325. doi: 10.1016/s1084-8592(99)80008-3. [DOI] [PubMed] [Google Scholar]
- Schafer AJ, Hawkins JR. DNA variation and the future of human genetics. Nature Biotechnol. 1998;16:33–39. doi: 10.1038/nbt0198-33. [DOI] [PubMed] [Google Scholar]
- Sealy JE, Laragh JH. The renin-angiotensin-aldosterone system for normal regulation of blood pressure and sodium and potassium homeostasis. In: Laragh JL, Brenner BM, editors. Hypertension: Pathophysiology, Diagnosis, and Management. New York: Raven Press; 1995. pp. 1287–1317. [Google Scholar]
- Soubrier F, Alhenc-Gelas F, Hubert C, Allegrini J, John M, Tregear G, Corvol P. Two putative active centers in human angiotensin I-converting enzyme revealed by molecular cloning. Proc Natl Acad Sci USA. 1988;85:9386–9390. doi: 10.1073/pnas.85.24.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. DNA chips: Analysing sequence by hybridisation to oligonucleotides on a large scale. Trends Genet. 1996;12:110–115. doi: 10.1016/0168-9525(96)81422-3. [DOI] [PubMed] [Google Scholar]
- Ståhl S, Hultman T, Olsson A, Moks T, Uhlén M. Solid phase DNA sequencing using the biotin-avidin system. Nucl Acids Res. 1988;16:3025–3038. doi: 10.1093/nar/16.7.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvänen A-C. From gels to chips: “Minisequencing” primer extension for analysis of point mutations and single nucleotide polymorphisms. Human Mut. 1999;13:1–10. doi: 10.1002/(SICI)1098-1004(1999)13:1<1::AID-HUMU1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Syvänen A-C, Aalto-Setälä K, Harju L, Kontula K, Söderlund H. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics. 1990;8:684–692. doi: 10.1016/0888-7543(90)90255-s. [DOI] [PubMed] [Google Scholar]
- Syvänen A-C, Sajantila A, Lukka M. Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am J Genet. 1993;52:46–59. [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridisation. Nature Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villard E, Tiret L, Visvikis S, Rakotovao R, Cambien F, Soubrier F. Identification of new polymorphisms of the angiotensin I-converting enzyme (ACE) gene, and study of their relationship to plasma ACE levels by two-QTL segregation-linkage analysis. Am J Hum Genet. 1996;58:1268–1278. [PMC free article] [PubMed] [Google Scholar]