Summary
Coordinated regulation of protection mechanisms against environmental abiotic stress and pathogen attack is essential for plant adaptation and survival. Initial abiotic stress can interfere with disease resistance signaling [1–6]. Conversely, initial plant immune signaling may interrupt subsequent ABA signal transduction [7, 8]. However, the processes involved in cross talk between these signaling networks have not been determined. By screening a 9,600 compound chemical library, we identified a small molecule DFPM that rapidly down-regulates ABA-dependent gene expression and also inhibits ABA-induced stomatal closure. Transcriptome analyses show that DFPM also stimulates expression of plant defense-related genes. Major early regulators of pathogen resistance responses, including EDS1, PAD4, RAR1, and SGT1b, are required for DFPM- and notably also for Pseudomonas-interference with ABA signal transduction, whereas salicylic acid, EDS16 and NPR1 are not necessary. While DFPM does not interfere with early ABA perception by PYR/RCAR receptors or ABA-activation of SnRK2 kinases, it disrupts cytosolic Ca2+ signaling and downstream anion channel activation in a pad4-dependent manner. Our findings provide evidence that activation of EDS1/PAD4-dependent plant immune responses rapidly disrupts ABA signal transduction and this occurs at the level of Ca2+ signaling, illuminating how the initial biotic stress pathway interferes with ABA signaling.
Results
Novel compound DFPM isolated from a randomly synthesized chemical library inhibits ABA signaling
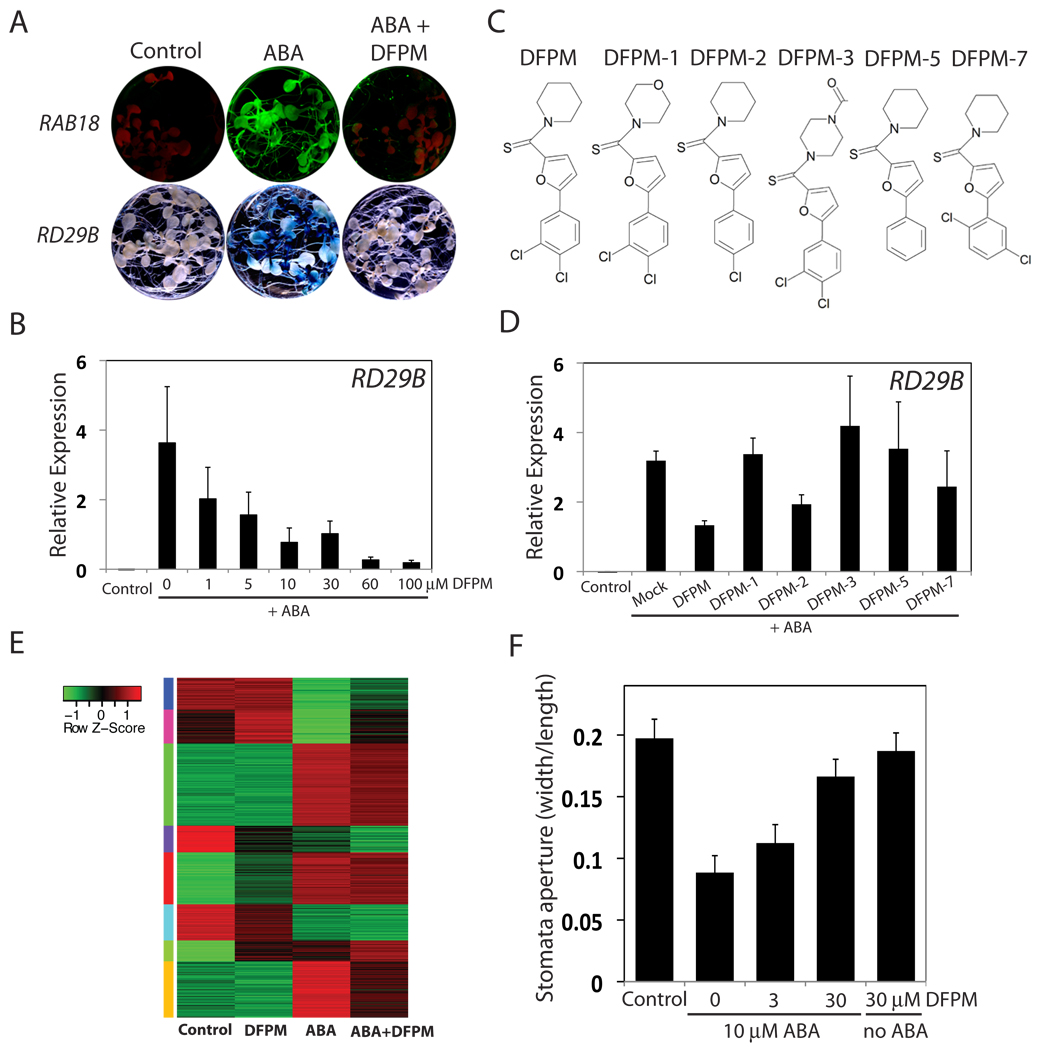
A chemical library of 9,600 randomly synthesized compounds was screened using the WT-RAB18 reporter line grown in 96 well tissue culture plates. Candidate chemicals that antagonized ABA-induced gene expression were selected (Fig. 1A and S1; ID5535396, ID5935873, ID5958440, and ID6015316). Here we report a detailed characterization of the small molecule [5-(3,4-Dichlorophenyl)Furan-2-yl]-Piperidin-1-ylMethanethione (DFPM, ID6015316), which effectively inhibits ABA-induced RAB18 expression (Fig. 1A). In contrast to frequently isolated auxin-related structures in this DIVERSET library, DFPM treatment did not produce auxin-related growth defects or alter auxin-induction of the DR5 promoter expression [9, 10] (Fig. S1C). The inhibitory effect of DFPM on ABA-induced gene expression was confirmed using an alternative GUS reporter line under the control of the RD29B promoter [11] (Fig. 1A). DFPM inhibits ABA-induction of gene expression in a dose-dependent manner (IC50= 3 µM and 1.5 µM for inhibition of ABA-induction of the endogenous RD29B and RAB18 promoters, respectively) (Fig. 1B and S2A). To determine functional relevant residues of the DFPM structure, derivatives of DFPM were analyzed (Fig. 1C). Modification of any ring structure and deleting or changing positions of the chloride groups reduced DFPM activity (Fig. 1D). Thus DFPM was the most effective among the derivatives analyzed. ATH1 GeneChip microarray analyses showed that DFPM down-regulates ABA-induction of more than 40% of ABA-responsive genes, showing that that DFPM affects a subset of the ABA signaling network (Fig. 1E, S3 Table S1).
Figure 1. Small molecule DFPM inhibits ABA-induced gene expression and stomatal closing.
(A) DFPM treatment reduces ABA-induction of GFP and GUS reporter gene expression in RAB18-GFP (green fluorescence protein) and RD29B-GUS (β-glucuronidase) promoter reporter lines. (B) Concentration-dependent effects of DFPM in inhibition of ABA-induced RD29B gene expression measured by quantitative-PCR (q-PCR). (C–D) Structures and test of DFPM derivatives for inhibition of ABA-induced RD29B gene expression as quantified by q-PCR. (E) Transcriptomic analysis shows that groups of ABA-induced genes are down-regulated by DFPM (30 µM) (n=3 microarrays per condition). The heat map contains 470 probesets regulated by ABA (292 up-regulated / 178 down-regulated; 45 probesets are also affected by DFPM shown in Fig. 2A) (F) DFPM exposure 30 min prior to ABA exposure inhibits ABA-induced stomatal closing. Error bars mean ± s.e.m. (n=3 experiments, 30 stomata per each experiment and condition). ABA was applied at 10 µM (A–F).
DFPM also inhibited ABA-mediated physiological responses, including ABA-induced stomatal closure (Fig. 1F) and ABA-inhibition of stomatal opening (Fig. S4C). In contrast, DFPM hardly affected ABA-induced delay in seed germination (Fig. S2C), indicating that DFPM does not control the entire ABA signaling network but rather acts preferably on a subset of ABA responses. In addition, ABA content measurements under non-stress conditions or in response to osmotic stress showed that DFPM does not affect endogenous ABA concentrations (Fig. S2D), suggesting that DFPM disrupts ABA signaling steps rather than ABA metabolism.
DFPM inhibition of ABA responses requires plant immune signaling
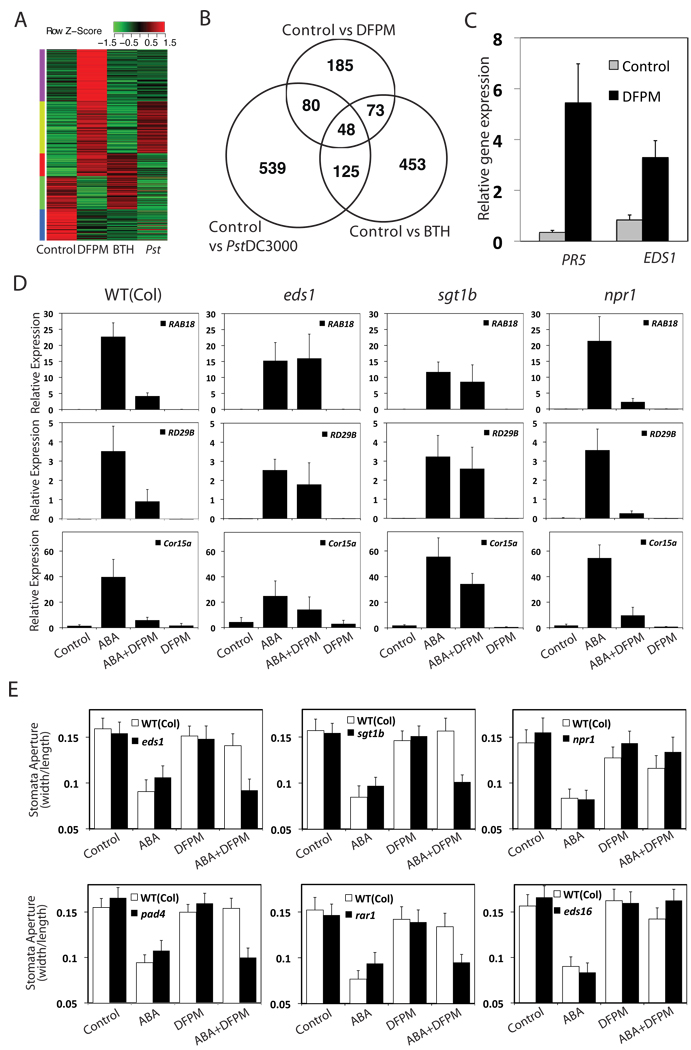
To validate microarray analysis results, expression of several ABA-induced genes was tested by quantitative-PCR, including RAB18, RD29B, Cor15a, and ABI1 (Fig. 2D and Fig. S4B). ABA induction of RAB18, RD29B and Cor15a was reduced by pre-treatment (30 min) with DFPM (Fig. 2D). However DFPM did not affect the ABA-induction of ABI1 in both microarray and q-PCR experiments (Fig. S4B).
Figure 2. DFPM-inhibition of ABA signaling requires early signaling components of effector-triggered immune signal transduction.
(A) Heat map of 386 probesets regulated by DFPM. (B) DFPM-regulated genes overlap with BTH (benzothiadiazole)-regulated and Pseudomonas syringae pv tomato (Pst) DC3000-regulated gene expression. (C) DFPM-induction of PR5 and EDS1 gene expression was quantified by q-PCR. (D) DFPM inhibition of ABA-inducible RAB18, RD29B, and COR15a expression requires functional EDS1 and SGT1b but not NPR1. Error bars show ± s.e.m (n=3). (E) DFPM-inhibition of ABA-induced stomatal closing requires EDS1, PAD4, SGT1b, and RAR1 but not NPR1 or EDS16. Error bars mean ± s.e.m. (n=3 experiments, 30 stomata per experiment and condition). DFPM was applied at 30 µM and ABA was applied at 10 µM (A–E).
In addition to the inhibitory effect of DFPM on ABA-responsive gene induction, transcriptome analyses also revealed that DFPM alone regulates the transcript levels of 386 genes (Fig. 2A). Signaling Pathway Impact Analysis revealed that DFPM induces components in the plant pathogen signaling network (KEGG: ath04626) (Fig. 2B and Table S1). Strong DFPM-induction of typical pathogen responsive genes PR5 and EDS1 [12, 13] were confirmed using q-PCR (Fig. 2C).
To address whether the transcriptional activation of plant defense genes by DFPM is linked to inhibition of ABA signaling, genetic mutations in components of plant disease resistance pathways were analyzed. Notably, DFPM’s inhibitory activity on ABA-induction of RAB18 and RD29B expression was compromised in the eds1-22 [14], pad4-1 [15], sgt1b(eta3) [16, 17] and rar1-21 [18] mutants (Fig. 2D and S4A), indicating that EDS1, PAD4, SGT1b, and RAR1 are required for the inhibitory activity of DFPM on ABA signal transduction. Because EDS1, PAD4, SGT1b, and RAR1 are important early components of plant NB-LRR (nucleotide binding-leucine rich repeat)-triggered immunity [16, 18–20], these data suggest that activation of NB-LRR proteins or early steps of resistance signaling pathways antagonize ABA signal transduction. EDS1 and PAD4 control both salicylic acid (SA)-dependent and SA-independent pathways [21, 22]. A critical SA response regulator, NPR1 [23], was not required for DFPM-disruption of ABA signaling (Fig. 2D), suggesting that SA signaling is not involved in the DFPM inhibition.
Pre-incubation with DFPM for 30 minutes inhibited the rapid response of ABA-induced stomatal closure (Fig. 1F). To test whether DFPM inhibition of this rapid ABA response also requires early pathogen signaling components, ABA-induced stomatal responses of disease resistance mutants were examined (Fig. 2E). DFPM inhibition of ABA-induced stomatal closure required functional EDS1, PAD4, SGT1b, and RAR1, but not NPR1 or the SA biosynthetic gene EDS16/SID2 [24] (Fig. 2E). DFPM also disrupted ABA-inhibition of stomatal opening and the inhibition was impaired in eds1, pad4, rar1, and sgt1b mutants, but not in npr1 (Fig. S4C). These data suggest that the rapid action of DFPM in disrupting stomatal responses to ABA requires EDS1/PAD4-dependent signaling but is independent of salicylic acid.
Constitutively activated NB-LRR receptor SNC1-1 inhibits ABA signaling
The requirement for EDS1, PAD4, SGT1b, and RAR1 during DFPM-inhibition of ABA signaling (Fig. 2D, 2E, S4), and the transcriptional activation of defense-related gene expression by DFPM (Fig. 2A, 2B), led us to hypothesize that DFPM stimulates immune pathways activated by NB-LRR receptors. We therefore tested whether activation of an NB-LRR protein can also inhibit ABA responses. ABA-induction of gene expression and ABA-induced stomatal closure were examined in the snc1-1 (suppressor of npr1-1, constitutive1) mutant [25]. In snc1-1, a point mutation in a TIR (Toll/Interleukin-1 Receptor domain)-NB-LRR protein creates an auto-activated receptor, which triggers constitutive pathogen resistance through EDS1 and PAD4 [25]. ABA-induction of RAB18, RD29B, and Cor15a was reduced in snc1-1 (Fig. S5A)). SNC1 is expressed in guard cells [26, 27] and stomata of snc1-1 were less responsive to ABA during ABA-induced stomatal closing (Fig. S5B; 2-tail T-test p=0.0059 for WT+ABA vs. snc1-1+ABA). These data demonstrate that constitutive activation of a NB-LRR protein antagonizes ABA-induction of gene expression and stomatal closure.
Pseudomonas syringae infection mimics DFPM inhibitory effects on ABA responses
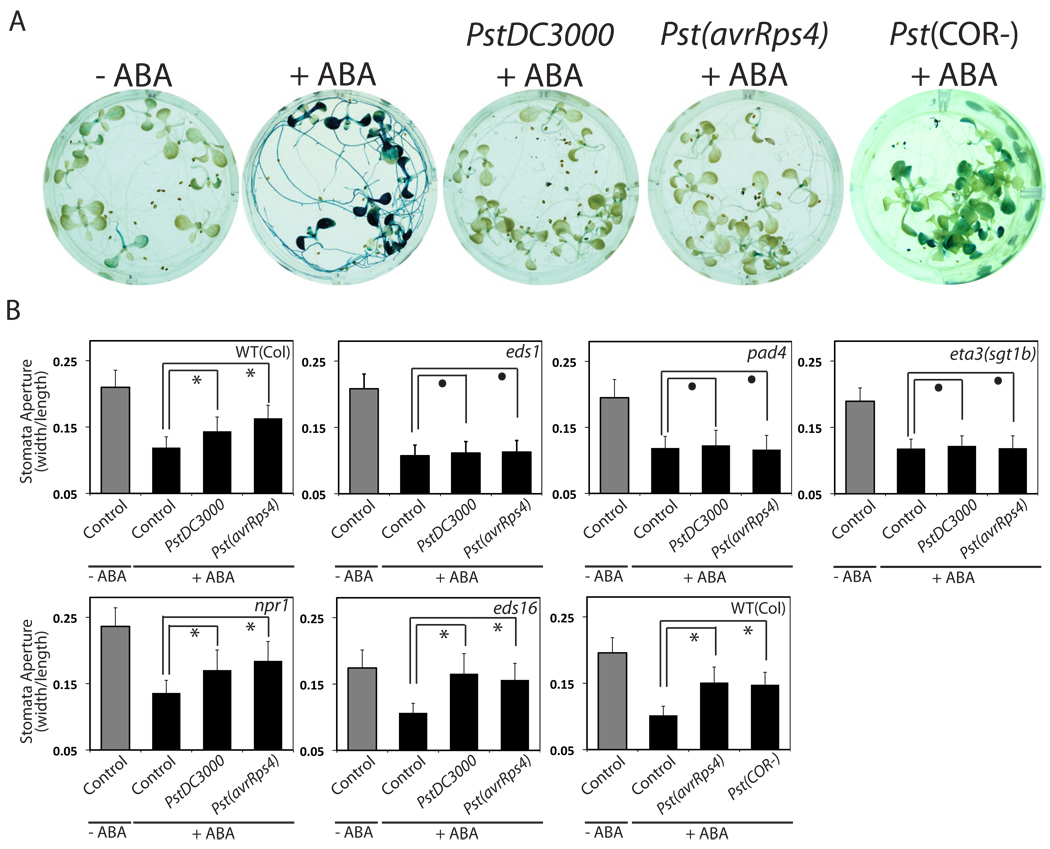
We have shown that chemical activation of EDS1/PAD4-dependent signaling has a negative impact on ABA-induced gene expression and physiological ABA responses. We therefore tested whether EDS1/PAD4 signaling in response to authentic pathogen infection can inhibit ABA signal transduction. ABA-induction of RD29B gene expression was examined after exposure of Arabidopsis seedlings to the virulent Pseudomonas syringae pv tomato (Pst) strain DC3000, which induces EDS1/PAD4-dependent basal (low level) immunity or the avirulent PstDC3000/avrRps4 strain, which induces EDS1/PAD4-dependent effector-triggered immunity after TIR-NB-LRR receptor activation [19, 21]. Infection by either strain led to strong reduction of ABA-induced RD29B gene expression (Fig. 3A).
Figure 3. P. syringae infection inhibits ABA signaling through the EDS1/PAD4 pathway.
(A) Infections by Pseudomonas syringae pv tomato (Pst) DC3000, Pst(avrRps4) and Pst(COR-) inhibit ABA-induced RD29B reporter gene expression. (B) ABA-induced stomatal closing is inhibited by PstDC3000 and Pst(avrRps4) infection in an EDS1/PAD4/SGT1b-dependent manner but independently of NPR1 and EDS16. Infections by Pst(COR-) also inhibit ABA-induced stomatal closing. Symbols * and • represent p< 0.025 and p> 0.2, respectively (n=3 experiments, 30 stomata per experiment and condition, 2-tail T-test). Error bars mean ± s.e.m. (n=3). ABA was applied at 10 µM (A–B).
As reported previously, P. syringae infection causes a transient stomatal closing and reopening [28, 29]. ABA-induced stomatal closing was significantly reduced by infection with PstDC3000 or PstDC3000/avrRps4 (Fig. 3B), indicating that immune signaling triggered by these pathogens can also down-regulate ABA signaling in guard cells. As with the DFPM treatment, Pst-infection inhibited guard cell ABA responses in npr1 and eds16 mutants but failed to do so in eds1, pad4, and sgt1b (Fig. 3B). ABA induction of RD29B gene expression (Fig. 3A) and ABA activation of stomatal closing responses (Fig. 3B) were also inhibited by infection with a PstDC3000(COR-) strain lacking the virulence factor coronatine [30] which mediates stomatal re-opening after pathogen-mediated stomatal closing [28, 31]. This result suggests that the inhibition of ABA signaling by P. syringae infection observed here occurs largely independently of coronatine production.
Analyses of DFPM-inhibition of early ABA signaling mechanisms
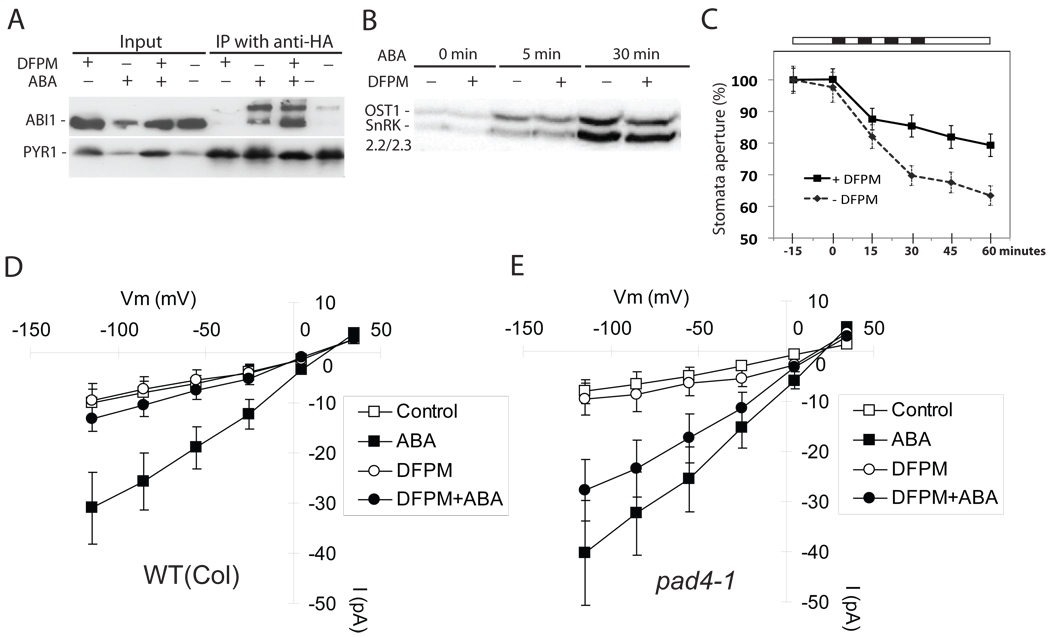
We examined which step in the ABA signal transduction pathway is targeted by DFPM. ABA signal transduction begins with ABA binding to PYR/RCAR receptors and interactions with PP2C protein phosphatases [32, 33]. Co-immunoprecipitation analyses showed that DFPM did not affect ABA-dependent PYR1 interaction with the PP2C ABI1 (Fig. 4A), indicating that ABA perception by PYR/RCAR receptors and PYR1-PP2C complex formation is not directly interrupted by DFPM. ABA perception causes activation of three SnRK2 protein kinases [34–36] by de-activation of the negatively regulating PP2Cs [32, 33, 37–40]. DFPM did not interfere with ABA-activation of these SnRK2 protein kinases (Fig. 4B, S6), indicating that DFPM interferes with downstream processes of SnRK2 kinase activation.
Figure 4. DFPM inhibits guard cell ABA signal transduction at the level of Ca2+ signaling.
(A) ABA-dependent protein-protein interaction between the PYR1 ABA receptor and the ABI1 PP2C-type phosphatase is not disrupted by DFPM pre-treatment. HA-PYR1 and YFP-ABI1 were co-immunoprecipitated in the presence of ABA (100 µM) and DFPM (50 µM). (B) ABA-activation (10 µM) of SnRK2 kinases, OST1, SnRK2.2, and SnRK2.3 [32] was not disrupted by DFPM treatment (50 µM). (C) DFPM (30 µM) inhibits stomatal closing mediated by repetitive imposed Ca2+-transients. Black bars represent periods in which stomata were exposed to buffer containing 1 mM CaCl2 + 1 mM KCl and white bars indicate periods with application of 0 mM CaCl2 + 50 mM KCl [43]. Each black bar corresponds to 5 minute time scale. Stomatal apertures at time = 0 (100%) correspond to average stomatal apertures of 4.02±0.25µm in control treatments and 3.53±0.26µm in DFPM pre-treatments (30 min prior to first Ca2+ pulse). Error bars show ± s.e.m (n=4 experiments). (D) ABA activation of S-type anion channel currents is significantly inhibited by DFPM in Columbia wildtype guard cells (Control: n=6; 10 µM ABA: n=10; 30 µM DFPM: n=4; 30 µM DFPM+10 µM ABA: n=10; p = 0.032, 2-tail T-test). (E) DFPM inhibition of ABA activation of S-type anion channels is not visible in pad4-1 guard cells (Control: n=6; 10 µM ABA: n=10; 30 µM DFPM: n=6; 30 µM DFPM+10 µM ABA: n=10; P = 0.314; 2-tail T-test). Guard cell protoplasts were pretreated with 0.06% DMSO (Control) or DFPM for 30 min before ABA+DMSO or ABA+DFPM treatment. Error bars show ± s.e.m.
Guard cells enable dissection of further steps in early ABA signal transduction [41]. To further investigate which step of ABA signaling can be impaired by DFPM, guard cells were exposed to four repetitive 5 min Ca2+ pulses known to cause Ca2+–induced stomatal closing [42–44]. DFPM partially inhibited imposed repetitive Ca2+ pulse-mediated stomatal closing (Fig. 4C), indicating that DFPM-triggered signaling disrupts stomatal closing at the level or downstream of Ca2+ signaling.
Elevated ABA enhances the cytosolic [Ca2+] sensitivity of S-type anion channel activation in Arabidopsis guard cells [45]. To test whether DFPM impairs ABA-regulation of S-type anion channel activities, ABA-activation of S-type anion channels was analyzed at 2 µM free cytosolic [Ca2+] [43, 45]. DFPM pre-treatment significantly reduced ABA-induced Ca2+-activated S-type anion channel currents (Fig. 4D). DFPM inhibition of ABA-induced Ca2+-activated S-type anion channel activity was significantly impaired in pad4-1 mutant guard cells (Fig. 4E).
Discussion
With the aim to dissect new mechanisms in the ABA signaling network, a small molecule antagonist of ABA signaling, DFPM, was identified by screening a 9,600 compound-containing chemical library (Fig.1, S1). DFPM effectively inhibits ABA-induced gene expression without producing any noticeable growth and developmental defects (Fig. 1, 2). In addition to the long-term inhibitory effect of DFPM on ABA-dependent gene expression, 30 minute pretreatment with DFPM interferes with rapid guard cell ABA responses such as ABA-induced and repetitive Ca2+ pulse-induced stomatal closing (Fig. 1, 4, S4C).
Identification of DFPM as an activator of plant immunity-related gene expression (Fig. 2A, B) provided evidence that DFPM negatively affects ABA signal transduction through activation of plant immune signaling. Many studies have shown that the converse cross-talk occurs from initial ABA/abiotic stimulation which subsequently antagonizes plant pathogen/biotic stress signaling [1–6]. Here we show that initial plant disease resistance signaling by application of the small molecule, DFPM, or P. syringae infection interferes with subsequent ABA signal transduction, indicating that biotic stress responses restrict plant abiotic stress signal transduction.
Our analyses of defense signaling mutants reveal that impairment of ABA signal transduction by DFPM pretreatment requires EDS1 and PAD4, major regulators of effector-triggered and basal immunity in plants (Fig. 2C, D) [19, 21]. Overlap between genes induced by DFPM and the SA analogue BTH (Fig. 2A, 2B) suggests that DFPM activates both SA-dependent and SA-independent defenses. However, the dispensability of SA biosynthesis (eds16/sid2) and downstream signaling (npr1) components for DFPM interference with ABA-responses (Fig. 2D, E, S4C) delineates the DFPM effect to an SA-independent branch of the EDS1/PAD4 pathway which is important for both basal and TIR-NB-LRR receptor-triggered resistance responses [21, 22].
Notably, salicylic acid is necessary for the “reverse cross-talk”, in which initial ABA signal transduction interferes with biotic stress signaling [5, 46], suggesting differences in the underlying mechanisms mediating abiotic to biotic signaling interference [1–6, 46]. The EDS1/PAD4-dependent and SA-independent disruption of ABA responses identified here interferes with early ABA signaling mechanisms since DFPM inhibition of both ABA-triggered stomatal closing and ABA-inhibition of stomatal opening are strongly reduced in the eds1 or pad4 mutants (Fig. 2E) and DFPM inhibition of ABA-activation of anion channel is compromised in pad4 mutant guard cells (Fig. 4E).
A requirement for RAR1 and SGT1b in DFPM-mediated negative regulation of ABA-induced responses (Fig. 2D, E, S4A, S4C) suggests that the antagonism occurs via NB-LRR immune receptors since a major function of RAR1 and SGT1b is to assist the accumulation and activation of plant NB-LRR complexes [47]. This would not, however, explain the effectiveness of virulent PstDC3000 in inhibiting ABA-induced RD29B gene expression (Fig. 3A), which induces ‘basal’ resistance in the absence of obvious NB-LRR recognition. One possibility is that the EDS1/PAD4 basal immunity barrier is triggered by low activity NB-LRR receptors. Alternatively, SGT1 and RAR1 function at an early intersection between NB-LRR activation and EDS1/PAD4 basal resistance signaling. Either scenario is supported by sgt1b and rar1 defects reported for basal resistance to virulent pathogen infection [48–50]. Together, the data favor inhibition of a sector of ABA signaling proceeding through the plant EDS1/PAD4 basal resistance pathway that can be effectively activated by NB-LRR receptors such as RPS4 and SNC1 (Fig. 3, S5).
Investigation of the mechanism mediating DFPM disruption of ABA signal transduction showed that DFPM interferes with events at the level of or downstream of intracellular Ca2+ signaling, whereas upstream ABA perception by PYR/RCAR receptors [32, 33] and subsequent activation of the major ABA signaling kinases, OST1, SnRK2.2, and SnRK2.3 were not affected by DFPM treatment (Fig. 4, S6). It is notable that intracellular Ca2+ has been proposed as an important transducer of plant immunity [51–55]. One hypothesis is that distinct Ca2+ signals generated during biotic stress signaling interfere with those produced during ABA signal transduction. Alternatively, depletion of Ca2+ binding proteins that are shared by pathogen-induced and ABA responses may limit ABA signal transduction. For example, the Ca2+-dependent protein kinases, CPK6, 4, and 11 have been shown to be required for ABA signal transduction [43, 56] and recent research shows that CPK4, 5, 6, and 11 function in flg22-induced resistance to the bacterial pathogen PstDC3000 [54]. However, other associated proteins or mechanisms may also trigger the identified biotic to ABA signaling interference identified here.
In summary, our findings define negative regulation of ABA signal transduction by rapid activation of plant innate immune responses by the small molecule DFPM and by P. syringae infection in part independently of SA signaling. Combined genetic and guard cell signaling analyses show that activation of resistance signaling antagonistically regulates ABA responses downstream of ABA-activated SnRK2 kinase activation, at the level of or downstream of Ca2+ signaling. Further investigation of how the small molecule DFPM modulates Ca2+ signaling during ABA signaling will shed light on regulatory mechanisms that adjust plant adaptive responses against combined biotic and abiotic stress exposures.
Supplementary Material
Acknowledgments
We thank Dr. Chris Somerville (UC Berkeley/Energy Biosciences Institute) for providing access to the chemical library. We also thank Drs. Jane Glazebrook (Univ. of Minnesota), William Gray (Univ. of Minnesota), Jeff Dangl (Univ. of North Carolina), Xin Li (Univ. of British Columbia), Erwin Grill (Technische Universität München), and Taku Demura (RIKEN) for providing mutants and materials and Drs. Yunde Zhao (UCSD) and Aurelien Boisson-Dernier for helpful discussions. This research was supported by NIH (R01GM060396) and NSF (MCB0918220) and in part by Chemical Sciences, Geosciences, and Biosciences Division of the Office of Basic Energy Sciences of the DOE (DE-FG02-03ER15449) grants (J.I.S.) and a DFG ‘SFB 670’ grant (J.E.P.). F.H. received support from a SNF fellowship and M.B. received support from a DFG fellowship. Microarray data have been deposited at NCBI-GEO.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Information
Six figures, 1 table, Supplementary Discussion, and Experimental Procedures are available on line (http://www.current-biology.com).
Author contributions: T.-H.K. and J.I.S. designed the research; T.-H.K., F.H., T.H., S.X., M.B., N.N., K.H., S.L., and N.R. performed the experiments at UCSD and N.P. at MPIPZ. Experiments addressing reviewer comments were performed by S.M. (UCSD), B.-h.L. (SU) and T.-H.K. (DWU) and N.P. (MPIPZ). T.-H.K. and J.I.S. wrote the paper with contributions from J.E.P. Most of the work research completed when T.-H.K. was in J.I.S.’ laboratory at UCSD.
References
- 1.Mauch-Mani B, Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol. 2005;8:409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. Pathological hormone imbalances. Curr Opin Plant Biol. 2007;10:372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell. 2008;20:1678–1692. doi: 10.1105/tpc.107.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan J, Hill L, Crooks C, Doerner P, Lamb C. Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 2009;150:1750–1761. doi: 10.1104/pp.109.137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Metraux JP, Mauch-Mani B. Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell. 2005;17:987–999. doi: 10.1105/tpc.104.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y. Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3129–3134. doi: 10.1073/pnas.0500185102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christmann A, Hoffmann T, Teplova I, Grill E, Muller A. Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol. 2005;137:209–219. doi: 10.1104/pp.104.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci U S A. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S, Hua J. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004;16:1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci U S A. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- 17.Gray WM, Muskett PR, Chuang HW, Parker JE. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, Dangl JL. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 20.Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol. 2005;8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 24.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks DM, Hernandez-Guzman G, Kloek AP, Alarcon-Chaidez F, Sreedharan A, Rangaswamy V, Penaloza-Vazquez A, Bender CL, Kunkel BN. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2004;17:162–174. doi: 10.1094/MPMI.2004.17.2.162. [DOI] [PubMed] [Google Scholar]
- 31.Penaloza-Vazquez A, Preston GM, Collmer A, Bender CL. Regulatory interactions between the Hrp type III protein secretion system and coronatine biosynthesis in Pseudomonas syringae pv. tomato DC3000. Microbiology. 2000;146(Pt 10):2447–2456. doi: 10.1099/00221287-146-10-2447. [DOI] [PubMed] [Google Scholar]
- 32.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 34.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 36.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Wang GX, Xin M, Yang HM, Wu XJ, Li T. The parameters of guard cell calcium oscillation encodes stomatal oscillation and closure in Vicia faba. Plant Science. 2004;166:415–421. [Google Scholar]
- 43.Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 2006;4:e327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen GJ, Chu SP, Harrington CL, Schumacher K, Hoffmann T, Tang YY, Grill E, Schroeder JI. A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature. 2001;411:1053–1057. doi: 10.1038/35082575. [DOI] [PubMed] [Google Scholar]
- 45.Siegel RS, Xue S, Murata Y, Yang Y, Nishimura N, Wang A, Schroeder JI. Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K channels in Arabidopsis guard cells. Plant J. 2009;59:207–220. doi: 10.1111/j.1365-313X.2009.03872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Torres Zabala M, Bennett MH, Truman WH, Grant MR. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009;59:375–386. doi: 10.1111/j.1365-313X.2009.03875.x. [DOI] [PubMed] [Google Scholar]
- 47.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 48.Noel LD, Cagna G, Stuttmann J, Wirthmuller L, Betsuyaku S, Witte CP, Bhat R, Pochon N, Colby T, Parker JE. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell. 2007;19:4061–4076. doi: 10.1105/tpc.107.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou F, Mosher S, Tian M, Sassi G, Parker J, Klessig DF. The Arabidopsis gain-of-function mutant ssi4 requires RAR1 and SGT1b differentially for defense activation and morphological alterations. Mol Plant Microbe Interact. 2008;21:40–49. doi: 10.1094/MPMI-21-1-0040. [DOI] [PubMed] [Google Scholar]
- 50.Holt BF, 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 51.Romeis T, Piedras P, Jones JD. Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell. 2000;12:803–816. doi: 10.1105/tpc.12.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romeis T, Ludwig AA, Martin R, Jones JD. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001;20:5556–5567. doi: 10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy AS, Poovaiah BW. Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature. 2009;457:1154–1158. doi: 10.1038/nature07612. [DOI] [PubMed] [Google Scholar]
- 54.Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gelli A, Higgins VJ, Blumwald E. Activation of Plant Plasma Membrane Ca2+-Permeable Channels by Race-Specific Fungal Elicitors. Plant Physiol. 1997;113:269–279. doi: 10.1104/pp.113.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.