Abstract
Iron is an essential cofactor in a number of critical biochemical reactions, and as such, its acquisition, storage, and metabolism is highly regulated in most organisms. The obligate intracellular bacterium, Chlamydia trachomatis experiences a developmental arrest when iron within the host is depleted. The nature of the iron starvation response in Chlamydia is relatively uncharacterized because of the likely inefficient method of iron depletion, which currently relies on the compound deferoxamine mesylate (DFO). Inefficient induction of the iron starvation response precludes the identification of iron-regulated genes. This report evaluated DFO with another iron chelator, 2,2′-bipyridyl (Bpdl) and presented a systematic comparison of the two across a range of criteria. We demonstrate that the membrane permeable Bpdl was superior to DFO in the inhibition of chlamydia development, the induction of aberrant morphology, and the induction of an iron starvation transcriptional response in both host and bacteria. Furthermore, iron starvation using Bpdl identified the periplasmic iron-binding protein-encoding ytgA gene as iron-responsive. Overall, the data present a compelling argument for the use of Bpdl, rather than DFO, in future iron starvation studies of chlamydia and other intracellular bacteria.
Keywords: Chlamydia, iron, deferoxamine, bipyridyl, transcription, persistence
Introduction
As an obligate, intracellular pathogen, Chlamydia has an absolute requirement of the host for essential nutrients. The chlamydial requirement for the biometal, iron, has been well established (Raulston, 1997; Al-Younes et al., 2001). Iron is an essential nutrient for nearly all organisms, as it is utilized as an electron intermediate, cofactor, or prosthetic group in a number of conserved biochemical cellular processes. Its biological importance can be attributed to its role as a transition metal with the ability to accept and donate single electrons as it alternates between the ferric (Fe3+) and ferrous (Fe2+) oxidative states (Hentze et al., 2004). When deprived of iron via long-term treatment using the hydrophilic chelator, deferoxamine mesylate (DFO), Chlamydiae enter an alternative growth mode, termed persistence, which was defined as a “viable but non-cultivatable state” of growth (Beatty et al., 1994). Persistence induced by iron starvation is phenotypically similar to those induced by other well-characterized mediators, which include treatment of host cells with IFN-g (Beatty et al., 1993), amino acid limitation (Coles et al., 1993), and B-lactam antibiotic exposure (Matsumoto and Manire, 1970; Lambden et al., 2006), with all exhibiting the accumulation of aberrantly enlarged RBs within the inclusion and the loss of infectious progeny (Beatty et al., 1994; Hogan et al., 2004; Mpiga and Ravaoarinoro, 2006; Wyrick, 2010). Noteworthy is the reversibility of this aberrant phenotype removal of the stress condition (Beatty et al., 1994).
Intracellular pathogens, like Chlamydia, are naturally buffered by the host cell from severe fluctuation of iron levels, given the ability of the host cell to maintain intracellular iron homeostasis. This raises the question of whether chlamydia requires, or even possesses iron-dependent regulatory mechanisms of gene expression. Analysis of the chlamydial genome reveals evidence to support the existence of an iron-dependent gene expression – the best studied being dcrA, which encodes for a protein that has homology to the prototypical ferric uptake regulator (Fur) protein that regulate transcription of target genes. The exact function of this protein is not known, but it has been demonstrated by Wyllie and Raulston (2001) that it harbors a DNA-binding activity, and that it can bind the same palindromic sequence recognized by E. coli Fur. However, further attempts to identify the cis-acting sequence recognized by DcrA in Chlamydia was unsuccessful (Rau et al., 2005). Moreover, the lack of a genetically tractable system in Chlamydia has precluded the knock-out-mediated identification of the “DcrA-regulon.” This lack of further insight is not isolated to DcrA function in Chlamydia, but to the entire iron-dependent regulation of gene expression in general. We hypothesize that this paucity in our knowledge of this process could be attributed to a lack of an efficient iron starvation mechanism that overcomes the buffering ability of the cell in relation to the chlamydial organisms it harbors.
The only iron-chelating agent tested against members of the Chlamydiacea has been DFO, which binds Fe3+ well, but Fe2+ poorly (Martell and Smith, 1977). In addition, DFO is membrane impermeable, and thus its chelating ability is restricted to the extracellular media, and perhaps the lumen of the endosomal vesicles (Lloyd et al., 1991; Cable and Lloyd, 1999; Persson et al., 2003; Glickstein et al., 2005). Of the published iron-restriction protocols for the Chlamydiacea, the overwhelming majority have utilized DFO with some auxiliary mechanism for iron depletion, including combination with cyclohexamide to block host up-regulation of the transferrin pathway (Raulston, 1997; Dill et al., 2009; Wehrl et al., 2004), or long-term pre-treatment of the host cells prior to infection (Al-Younes et al., 2001; Peters et al., 2005; Mukhopadhyay et al., 2006; Dill et al., 2009; Timms et al., 2009). Taken together, these published protocols imply that DFO, by itself, may be an unsuitable or inefficient method for the iron starvation of the intracellular pathogen, Chlamydia trachomatis. Moreover, the need for additional manipulations raises caveats in data interpretations. Therefore, we sought to identify an alternative means of starving Chlamydia of iron with minimal manipulation of the experimental system. We discovered that a single treatment at 100 μM dose at the time of infection with the compound, 2,2′-bipyridyl (Bpdl), which was previously identified as a membrane permeable iron-chelating agent capable of depleting the cytosolic labile iron pool (Breuer et al., 1995; Romeo et al., 2001), proved effective in inducing an iron starvation response in both the host cell and chlamydia. We present evidence that the compound Bpdl is more potent than DFO in inducing an iron-starved, persistent phenotype in C. trachomatis. Direct comparison with DFO also revealed a more robust and consistent induction of the transcription of previously reported iron-responsive genes. As final evidence of the superiority of Bpdl over DFO, ytgA, which encodes for an iron-binding periplasmic protein was found to be induced in iron-limiting conditions. Thus, we present an iron starvation protocol that requires minimal manipulation, and possesses a potential broader applicability to Chlamydia and other intracellular bacteria.
Methods
Cell culture and chlamydial infections
Human endothelial HEp2 cells (CCL-23, ATCC) were grown at 37°C with 5% CO2 in Iscove's Modified Dulbecco's Medium (IMDM, Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS), 20 mM l-glutamine (Gibco), and 10 μg/ml gentamicin. C. trachomatis UW-3/CX (serovar D) was originally obtained from Dr. Ted Hackstadt (Rocky Mountain Laboratory, NIAID), and propagated as described (Caldwell et al., 1981).
For infections, HEp2 or HeLa cells were plated to 85–90% confluency in six-well tissue culture treated plates and incubated overnight. Next day, monolayers were treated with 45 μg/ml DEAE-Dextran (Sigma) in HBSS for 10–15 min at 37°C prior to inoculation with C. trachomatis. Plates were centrifuged at 1500 rpm at 4°C prior to a 30-min invasion step at 37°C. Inoculum was decanted, cells were washed with PBS, and cultured in complete IMDM (plus or minus iron-chelating agents).
Iron-chelating agents
Deferoxamine mesylate (Sigma, CAS: 138-14-7) was diluted to a stock concentration of 50 mM in ddH2O, filter sterilized and stored at 4°C for no more than 2 months. Bpdl (Sigma, CAS: 366-18-7) was diluted in ethanol to a stock concentration of 100 mM, aliquoted, and stored at −80°C.
Quantification of recoverable infectious progeny
HEp2 cells were infected and treated as described. At 24 h post-infection (p.i.; unless noted), infected monolayers were washed prior to physical dislodgement into 1 ml SPG buffer. Suspension was transferred to sterile eppendorf tubes containing three glass beads (3 mm, VWR) and vortexed for 60 s. Samples were stored at −80°C. For quantification, samples were diluted serially and appropriate doses were used to re-infect fresh, confluent monolayers of HEp2 cells in triplicate. The next day, samples were fixed and permeabilized with MeOH and stained with convalescent human sera. Inclusions were visualized after secondary staining with Alexa-594 conjugated mouse mAb anti-human IgG, and counted in five fields per well for the calculation of infectious titer.
Immunofluorescent analysis and microscopy
Infected samples were fixed and permeabilized with MeOH, and immunolabeled with convalescent human sera. Alexa-594 conjugated mouse mAb anti-human IgG was used to visualize chlamydial morphology. Images were captured using Leica SP2 upright confocal microscope. Bars represent 10 μm in each image.
Nucleic acid preparation
Infected monolayers were trypsinized, pelleted, and resuspended in 200 μl PBS for genomic DNA extraction, using the DNeasy Blood and Tissue Kit (QIAGEN) following the instructions of the manufacturer. Samples were eluted in a volume of 100 μl TE buffer.
RNA was extracted using Trizol Reagent (Invitrogen) using the instructions of the manufacturer for cells grown in a monolayer. Extracted RNA was DNAsed using Turbo DNase Kit (Applied Biosystems), precipitated in 80% ethanol overnight, dried, and resuspended in 20 μl nuclease free water (Applied Biosystems). RNA concentrations were measured using Nanodrop ND-1000 spectrophotometer. Equal amounts of total RNA (2 μg) were converted to cDNA using Superscript III reverse transcriptase kit (Invitrogen) according to the instructions of the manufacturer. Reverse transcription was primed using random nonamers (New England Biolabs). cDNA mixture was diluted 1:8 with 10 mM Tris (pH 7.5), for a final volume of 160 μl and stored at −80°C.
Real time quantitative PCR (RT-qPCR)
Applied Biosystems 7300 Real time PCR System was used to carry out quantification of cDNA or gDNA sequences. Primers were designed using the Applied Biosystems Primer Express 3.0 Software and are listed in Table 1. Primer sets were tested for amplification efficiency against gDNA extracted from purified EBs, and for the generation of specific products using 7300 system post-run Tm function. Samples were assayed in triplicate wells of a 96 well plate; each reaction contained 5 μl DNA (complimentary or genomic), 25 μl 2X SYBR green master mix (Applied Biosystems), 0.45 pmol forward and reverse primers, and 19.8 μl RNase/DNase free water (Applied Biosystems). No RT and no template controls were performed regularly.
Table 1.
Primers used for quantitative PCR.
| Gene | Organism | Fwd primer (5′–3′) | Rev primer (5′–3′) |
|---|---|---|---|
| gapdh | H. sapiens | ctgctcctcctgttcgacagt | accttccccatggtgtctga |
| tfr | H. sapiens | cattctttggacatgctcatctg | tgatgaccgagatggtggaa |
| dmt1 | H. sapiens | ggagtactcttgttttagctttcgtaaa | ccagactgcaaatcggattca |
| fer-H | H. sapiens | tgaagctgcagaaccaacga | cgctctcccagtcatcacagt |
| fer-L | H. sapiens | accgtttttgtggttagctcctt | caggtcggtggaataattctga |
| euo | Ctr serovar D | gctgttcctgttacttcgcaaa | aacatagatagcctgacgagtcaca |
| omcB | Ctr serovar D | ccaaagcgaaagacaacacttct | aaccggagcaacctctttacg |
| ahpC | Ctr serovar D | ccagttagctggacaaaccattccg | cgttccattgacgaggaattgcgt |
| devB | Ctr serovar D | acgaagatgtagaagctggaagta | tgcggtatccatacgaaagatttg |
| ytgA | Ctr serovar D | ctcttgtgtttagcaggctgtttc | tgcgattcatagacaagacatagatg |
| tyrP-1 | Ctr serovar D | tcgcaggaacaaccattgg | gtaacgtcgtaggcaggaatcc |
Relative expression of eukaryotic iron-responsive transcripts was quantified using the Pfaffl method (2ΔΔCt method; Pfaffl, 2001). Target iron-responsive transcripts were normalized to gapdh transcript levels and calibrated to the untreated sample. Absolute expression of Chlamydia transcripts was quantified using serially diluted C. trachomatis genomic DNA, which was extracted from EBs, as a standard. Levels of mRNA were normalized to the number of Chlamydia-specific genomes, which were derived from parallel samples.
Graphs and statistical analysis
Generation of all graphs and statistical tests were performed in GraphPad Prism v5.0 Software. Specific statistical tests are described in the Section “Results” and figure legends. Unless noted, graphs represent mean averages ± SD.
Results
Eukaryotic iron-responsive transcription after iron-chelator treatment
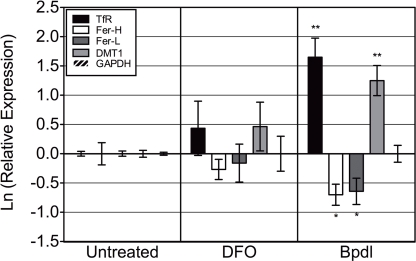
Intracellular chlamydia are naturally protected from severe fluctuations of extracellular environmental conditions, and thus eliciting a response for the bacterium would first require that the host cell be affected. Two different iron chelators, DFO and Bpdl (shown in Figure 1) were evaluated for their ability to induce an iron response from the host cell, with the underlying assumption that iron starvation of the host would lead to iron starvation of intracellular chlamydiae. To assess the level of host iron starvation, we chose to examine the differential regulation of a handful of iron-related transcripts that are known to respond to changes in iron levels. We utilized the mRNA levels of specific iron-responsive transcripts from uninfected HEp2 cells that had been treated with DFO or Bpdl for 10 h (Figure 2). The mRNA of two genes coding for iron-acquisitional proteins, transferrin receptor (TfR) and divalent metal transport protein 1 (DMT1) are normally stabilized under conditions of low cytoplasmic iron availability (Rao et al., 1986; Kato et al., 2007), resulting in the net increase in the levels of these transcripts. Conversely, ferritin heavy/light chain transcripts, which code for proteins that form a complex for the storage of excess iron, are normally degraded under the same low-iron conditions (Kato et al., 2007). Based on previous reports of intracellular iron-chelation, we expected Bpdl to cause an iron-starved transcriptional state of eukaryotic cells. Indeed, Bpdl treatment caused an elevated expression of tfr and dmt-1, which reached 5.2- and 3.5-fold over the untreated sample, respectively (unpaired, one-tailed t-test, p = 0.0054 and 0.0065, respectively). Moreover, treatment with Bpdl elicited an approximate twofold reduction in ferritin heavy/light chain (fer-H/L) transcripts (p = 0.0255 and 0.0458, respectively). The ability for Bpdl to cause an “iron-starved” transcriptional response from uninfected HEp2 cells, in a time frame where DFO could not, raised the possibility that Bpdl could induce a relatively robust iron starvation response and/or growth inhibition in C. trachomatis.
Figure 1.
Chemical structure of iron-chelating agents. Deferoxamine mesylate (DFO) is a hydrophilic compound with a high affinity for ferric iron (Martell and Smith, 1977). It binds ferric iron in a 1:1 (hexadentate) complex (Gaeta and Hider, 2005). 2,2′-bipyridyl (Bpdl) is a membrane permeable compound with similar moderate affinities for Fe2+ and Fe3+ (Smith and Martell, 1975), and binds both species of these molecules in a 3:1 molar ratio (bidentate; Gaeta and Hider, 2005).
Figure 2.
Bipyridyl treatment modulates eukaryotic iron-responsive transcription. Human epithelial HEp2 cells were incubated in complete medium containing 100 μM DFO or Bpdl for 10 h before select transcript levels were measured via RT-qPCR. Reported iron-responsive genes, transferrin receptor (TfR) and divalent metal transporter 1 (DMT1), and iron-repressed genes, ferritin heavy and light chain (Fer-H/L) were normalized to endogenous glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA levels and then calibrated to the untreated control (ΔΔCt method; Pfaffl, 2001). The y-axis presents the natural log of the target gene relative expression. Bars represent the natural log of mean relative expressions ± SD in one representative experiment containing two biological replicates per treatment, each assayed in triplicate. Experiment was performed independently two times and obtained similar transcriptional profiles in each case (i.e., total of four biological replicates were observed). Statistical significance from the untreated was determined by unpaired, one-tailed t-test (*p < 0.05, **p < 0.01).
The effect of bipyridyl on acute c. trachomatis infection
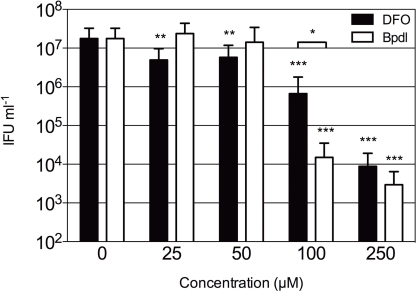
The observation that Bpdl was able to induce a greater iron starvation response from the host cell highly suggested that a similar response could be elicited from intracellular chlamydiae. Therefore we tested both Bpdl and DFO for effects on chlamydial development and inclusion morphology, with an emphasis on the simplest possible iron starvation protocol, which involves a single treatment at the time of infection. In order to test the effect of Bpdl on C. trachomatis development, a dose-response analysis of recoverable inclusion forming units (IFU) was performed (Figure 3). HEp2 cells were infected with C. trachomatis serovar D at an MOI of 0.5 prior to culture in normal (iron-replete) medium containing various doses of DFO or Bpdl for 24 h. Infectious progeny were harvested and replated onto fresh monolayers for quantification. C. trachomatis growing in iron-replete medium consistently produced over 107 IFUs/ml. No significant inhibition of recoverable IFUs was observed upon addition of low doses (25 or 50 μM) of Bpdl to the culture medium. The same doses of DFO (25 and 50 μM) each caused an approximate 70% decrease in IFU output that was significantly lower than the untreated control (unpaired, two-tailed t-test; p = 0.0046 and p = 0.0072, respectively). However, at concentrations of 100 and 250 μM, Bpdl was more effective relative to the untreated sample. At 100 μM, Bpdl caused a 2.5-log reduction of recoverable IFUs from the untreated control (p = 0.0001). Moreover, inhibition of infectious progeny was highly reproducible with the percent recovery ranging from just 0 to 0.33% of the untreated control over three independent experiments. At 100 μM concentration administered at the time of infection, DFO yielded a 95% reduction from the untreated control (p = 0.0002). However, this result was highly variable with a percent recovery that ranged from 0 (complete arrest) to 12.5% of the untreated control. Compared directly, treatment with Bpdl was 26-fold more effective than DFO in limiting the development of infectious progeny (p = 0.0408). While both compounds caused similar reduction in recoverable IFU at 250 μM, some cells were lost indicating possible cytotoxicity (data not shown). Therefore, 100 μM was deemed a suitable concentration for both iron-chelating agents for subsequent experiments.
Figure 3.
Infectious progeny of C. trachomatis samples cultured with iron-chelating agents. Various concentrations of DFO (black bars) or Bpdl (white bars) were included in the culture medium added immediately post-chlamydial invasion. Infectious titers were quantified after 24 h of treatment and presented as inclusion forming units (IFU) per milliliter of inoculum (y-axis). Graph represents the mean ± SD derived from at least two independent experiments, assayed in triplicate. Statistical significance determined with unpaired, two-tailed t-test in conjunction with F-test (*p < 0.05, **p < 0.01, ***p < 0.001). Asterisks without brackets represent comparison to the untreated control; brackets indicate the comparison of DFO to Bpdl at the same concentration.
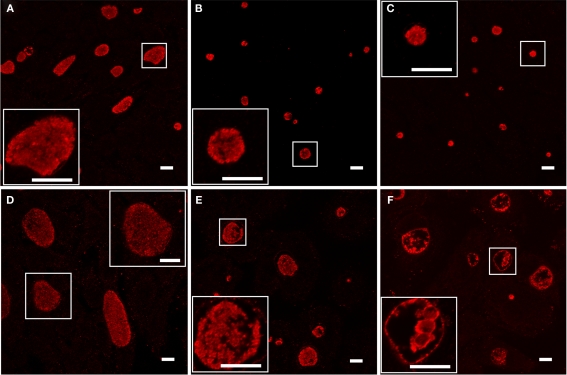
As the loss of recoverable infectious progeny due to developmental arrest is a hallmark of the chlamydial persistent phenotype (Beatty et al., 1994), we examined C. trachomatis samples cultured with Bpdl for another characteristic of persistence, the formation of aberrantly enlarged RBs. Infected cell cultures were treated for 24 or 48 h with 100 μM concentrations of Bpdl or DFO and visualized by immunofluorescent confocal microscopy (Figure 4). Compared to the untreated control, culture with both DFO and Bpdl (100 μM, administered at the time of infection) caused a qualitatively similar reduction in the size of chlamydial inclusions at 24 h p.i. (Figures 4A–C). Enlarged, aberrant RBs, characteristic of the persistent phenotype, were only observed after 48 h of culture with Bpdl (Figure 4F). Interestingly, the same 48-h length of treatment with DFO (Figure 4E) yielded a population of chlamydial inclusions, the majority of which resembled that of the untreated, 24-h p.i. sample (Figure 4A), suggesting that DFO may not be suitable for time-points of this length. It should be noted that growth media was neither replaced nor additional chelators added, consistent with our intention of minimizing manipulation of the experimental system. Under such conditions, Bpdl appeared to be more efficient in initiating and maintaining a developmental arrest in C. trachomatis.
Figure 4.
Chlamydial morphology after long-term incubation with inhibitory doses of Deferoxamine or Bipyridyl. C. trachomatis-infected cell cultures were incubated with 100 μM DFO (B, E) or Bpdl (C,F) in conjunction with untreated controls (A,D). Images from the top panel row (A–C) were fixed after 24 h p.i. and images from the bottom panel row (D–F) were fixed after 48 h p.i. Samples were stained immunofluorescently using convalescent human sera. Aberrant RB morphology and a lack of EB particles within the inclusional lumen was only observed upon 48 h of Bpdl treatment (F). Samples treated with DFO for 48 h (E) exhibited similar characteristics of untreated 24 h growth (A). Images were captured using Leica Confocal microscope. Bars represent 10 μM in each panel/inset. Images selected represent the overall trends observed in four independent experiments.
Chlamydial viability upon bipyridyl treatment
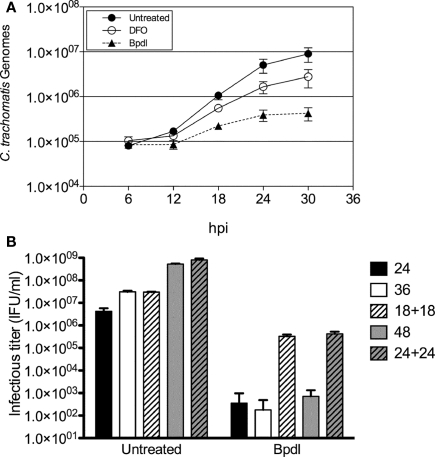
The term “persistence,” refers to the ability of non-cultivatable chlamydiae to re-enter the acute life-cycle and cause disease upon the removal of the condition of stress. We chose to examine the reversibility of the persistent phenotype of chlamydia exposed to Bpdl in two ways. First, persistent Chlamydiae often exhibit continued genomic replication, albeit at a reduced rate, despite a lack of re-differentiation into the infectious EB form (Belland et al., 2003; Lambden et al., 2006). Therefore Chlamydia-specific genomic DNA (gDNA) was monitored longitudinally during iron-chelation treatment using real time qPCR. Incubation with DFO and Bpdl (100 μM) retarded chlamydial DNA replication, compared to the untreated control. However, consistent with the recoverable IFU and IFA data, Bpdl treatment caused an even greater growth defect than that of DFO-treatment (Figure 5A). During the exponential phase of growth (between 12 and 24 h post-infection), Bpdl-treated C. trachomatis gDNA doubled once every ∼5.5 h, compared to approximately each 2.5 and 3.3 h for untreated and DFO-treated cultures, respectively, over the same time period. Importantly, the number of genomes increased during each time period tested for Bpdl-cultured samples, indicating active genomic replication, and thus viable organisms, despite the lack of infectious EB re-differentiation.
Figure 5.
Viability of C. trachomatis cultured with Bipyridyl. (A) Chlamydia-specific DNA was quantified via RT-qPCR from total genomic DNA extracted each 6 h. The numbers of chlamydial genomes (y-axis) were extrapolated from a standard curve of genomic DNA obtained from purified chlamydial EBs. Data points represent the mean ± SD of two independent experiments, which were representative of greater than five independent time course experiments. (B) Bpdl-cultured C. trachomatis were reactivated by washing and incubation with normal, iron-replete medium (“18 + 18” and “24 + 24”) and compared to cultures treated with Bpdl throughout the entire experiment (“24,” “36,” “48”). C. trachomatis treated with Bpdl for 18 and 24 h were still viable when cultured with normal media. Graph represents data combined from two independent experiments, each assayed in triplicate. Bars represent mean ± SD.
Reactivation of developmentally arrested chlamydiae upon removal of the chelator was monitored using recoverable IFUs as a measure of viability. Infected HEp2 cells were incubated with Bpdl for 18 or 24 h, and then “reactivated” by the replacement of Bpdl-supplemented media with normal (iron-replete) medium for another 18 to 24 h of incubation. The infectious titer of each sample was quantified and compared to samples incubated with Bpdl for the entire course of infection (Figure 5B). A near complete arrest of infectious EB re-differentiation was observed at 24, 36, and 48 h of continuous treatment with Bpdl, which was consistent with the aberrant morphology observed by immunofluorescent analysis at 48 h p.i. (Figure 4F). In contrast, “reactivated” samples exhibited an increase in recoverable infectious progeny of 2–3 logs over continuously treated samples. Although reactivated chlamydial samples were not completely rescued to the untreated output levels, a majority of chlamydiae were able to re-enter the acute life-cycle and re-differentiate into the infectious EB form.
Exogenous iron abrogates chlamydial inhibition caused by bipyridyl and deferoxamine
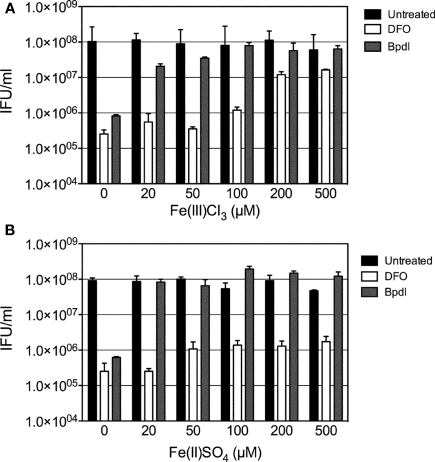
To ensure that effects elicited by culture with Bpdl resulted from the restriction of iron, an iron-titration experiment was performed, in which increasing concentrations of Fe3+ or Fe2+ were added to an inhibitory dose (100 μM) of the two iron-chelating agents, prior to their incubation with infected cell cultures. To confirm the binding capabilities of Bpdl, we utilized species of both ferric (FeCl3) and ferrous (FeSO4) oxidative states of iron. Based on reported affinities (Smith and Martell, 1975; Martell and Smith, 1977), it was not surprising that both Bpdl and DFO-mediated inhibition was abrogated by the addition of exogenous ferric chloride (Figure 6A). In contrast, the addition of ferrous sulfate was not able to rescue DFO-mediated inhibition of recoverable IFU output (Figure 6B), which was expected, based on its reported specific affinity for ferric iron (Martell and Smith, 1977). The lack of recovery exhibited by ferrous iron supplementation illustrated the inability of DFO to bind the divalent metal in the extracellular medium, coupled with the inability of the host cell to efficiently import the supplemented ferrous iron. The end result was the unaltered sequestration of ferric iron in the extracellular media, which, of course, is the main source of host cell iron acquisition. This result was not the case for Bpdl-mediated inhibition, as recoverable IFU output was completely reversed at 20 μM of FeSO4, which was the lowest dose tested, reflecting its ability to bind the divalent oxidation state of the metal. Taken together, the addition of exogenous ferric and ferrous iron abrogated the inhibitory effects of Bpdl, while only ferric iron restored IFU yield in DFO-treated samples.
Figure 6.
The effect of iron on iron-chelator mediated recoverable IFU inhibition. Increasing concentrations of ferric (A) or ferrous (B) iron were used to titrate out the inhibition of C. trachomatis infectious progeny caused by treatment with (100 μM) Bpdl or DFO. Exogenous iron was added to medium containing chelators immediately prior to its use in culture. Samples were harvested at 24 h p.i. for infectious titer quantification. Bars represent the mean ± SD of one experiment representative of two independent trials in which similar trends were obtained.
The effect of bipyridyl treatment on chlamydial transcription
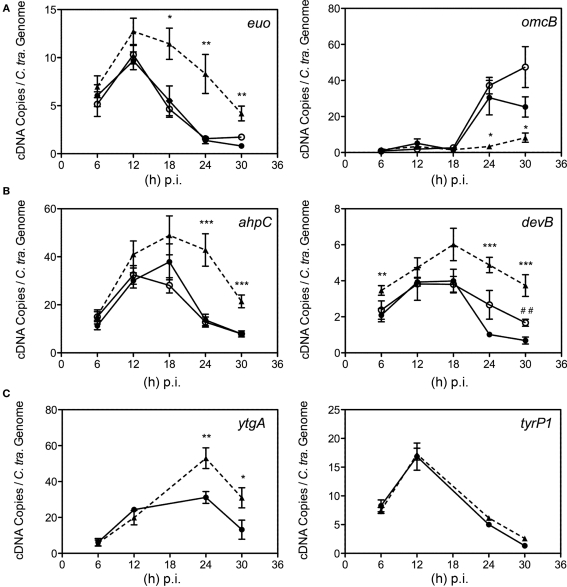
Having validated Bpdl treatment as an efficient mediator of the chlamydial persistent response, we decided to test its effects on chlamydial transcription of genes associated with persistence or known to be induced under iron-limiting conditions. We monitored the expression of known molecular markers of persistence – the continued expression of the early gene euo (Belland et al., 2003; Ouellette et al., 2006; Timms et al., 2009) and the failure to induce transcription of the late gene omcB (Iliffe-Lee and McClarty, 2000; Belland et al., 2003; Ouellette et al., 2006; Timms et al., 2009). Therefore the expression of persistence markers, euo and omcB, was measured longitudinally from DFO, Bpdl, and untreated cultures via RT-qPCR and normalized by the genome copy number derived from a parallel sample. As expected, appreciable omcB expression was not observed until 18 to 24 h p.i. in the untreated control, which correlates to the onset of EB re-differentiation for C. trachomatis. Alternatively, the expression of euo in the same samples increased during the earliest time-points measured and then decreased to near absent levels during the late stages of infection (Figure 7A). Culture with 100 μM DFO from the time of infection did not alter either of these transcriptional profiles, suggesting that a persistent phenotype could not be induced efficiently by this iron chelator when administered at the time of infection. In contrast, incubation with Bpdl caused the extended repression of omcB transcript expression throughout the entire time course. Concomitantly, expression of euo remained elevated until the late stages of infection. The altered expression of these two genes indicated that culture with Bpdl (100 μM), from the time of infection, elicited a persistent chlamydial phenotype.
Figure 7.
Time course analysis of selected transcript expression of C. trachomatis. Samples were cultured in normal iron-replete medium (--•--), or in conjunction with deferoxamine (--○--) or bipyridyl (--Δ--). Total RNA was extracted longitudinally and reverse transcribed to cDNA for quantification using specific primer sets (Table 1). cDNA levels were adjusted to reflect the number of chlamydial organisms by using the number of chlamydial genomes determined from parallel samples. (A) Transcripts whose expression were altered under multiple models of persistence (Iliffe-Lee and McClarty, 2000; Belland et al., 2003; Ouellette et al., 2006; Timms et al., 2009). (B) Transcripts from C. trachomatis whose expression were altered under iron-restriction. (C) ytgA, a transcript whose expression was hypothesized to be responsive to fluctuations in iron availability, along with tyrP1 whose transcription was altered under IFN-g treatment, but not iron-restriction in C. pneumoniae (Timms et al., 2009). Statistical significance relative to the untreated control was determined using two-tailed t-test (Bpdl-treated samples: *p < 0.05, **p < 0.01, ***p < 0.001; DFO-treated samples: ##p < 0.01).
We sought to validate the Bpdl model by monitoring its effects on the transcription of two previously reported chlamydial iron-responsive genes, ahpC and devB (Dill et al., 2009), in parallel with untreated and DFO-treated cultures. When included immediately post-invasion, a 100-μM concentration of DFO, noticeable increases in the levels of devB transcript were only observed at 24 (2.6-fold) and 30 (2.5-fold) h p.i. (unpaired, two-tailed t-test; p = 0.0740, p = 0.0023, respectively), while no differential transcription of ahpC was observed (Figure 7B). In contrast, culture with Bpdl caused statistically significant elevation over the untreated control at multiple time-points of both ahpC and devB (Figure 7B), with a statistically significant increase observed as early as 6 h post-treatment for devB. Maximum differential expression occurred at 24 h p.i. for ahpC, and reached 3.1-fold over the untreated levels (p = 0.0008). For devB, maximum differential expression reached 5.6-fold at 30 h p.i. (p = 0.0009). Taken together, treatment with Bpdl caused a more robust induction ahpC and devB than treatment with DFO. Not only was the altered expression more pronounced, it also lasted longer, suggesting significant and sustained iron-deprivation.
We also analyzed the expression of ytgA. YtgA, a proposed periplasmic iron-binding protein, was reported to be iron-responsive at the protein level upon long-term treatment with DFO (Miller et al., 2009), yet its differential expression at the transcript level has never been shown, and thus served as an excellent candidate to further evaluate the efficiency of iron starvation mediated by Bpdl. Using the Bpdl model, expression of ytgA was clearly elevated after 24 and 30 h p.i. (unpaired, two-tailed t-test; p = 0.0083, p = 0.0433, respectively; Figure 7C), providing a mechanism for the elevated protein expression previously observed (Miller et al., 2009). Thus, the greater effectiveness of the new iron starvation method was validated by the discovery of ytgA being an iron-responsive gene at the level of transcription.
To ensure that the up-regulation of ahpC, devB, and ytgA were specific to iron-limitation, the expression of tyrP-1, which codes for an aromatic amino acid transporter, was monitored. Its transcript expression in C. pneumoniae was differentially expressed under persistence mediated by IFN-g treatment, but not iron-restriction (Timms et al., 2009). IFN-g is thought to induce expression of the host enzyme, indoleamine dioxygenase (IDO), which effectively limits the available amount of tryptophan within the cell (Wood et al., 2004). Therefore its elevated expression under IFN-g mediated persistence presumably functions to increase acquisition of the limited amino acid. Because Timms et al. (2009) reported that it was not differentially expressed under conditions of low-iron availability, its transcript expression was monitored to distinguish the effects of Bpdl-mediated iron starvation from general persistence. As expected, transcription of tyrP1 was not altered under Bpdl treatment at any time-point tested (Figure 7C). The fact that this mid-stage gene was not differentially expressed under Bpdl-mediated persistence indicated that the elevated transcription published in this study was specific to iron-restriction. Taken together, the data indicate collectively that a single-dose Bpdl treatment is an efficient iron-restriction model for the growth of C. trachomatis, and is able to elicit a more uniform, and consequently, a more robust response from Chlamydiae than DFO under identical treatment conditions.
Discussion
We present a re-evaluation of the currently available iron starvation protocol for intracellular bacteria by directly comparing DFO and Bpdl for their abilities to induce iron starvation-related phenotypes in the obligate intracellular pathogen C. trachomatis. The goal was to identify an alternative means of starving Chlamydia of iron with minimal manipulations, such as pre-starving the host cell, or preventing it from synthesizing proteins involved in iron acquisition. Our motivation for simplifying iron starvation of Chlamydia by excluding such manipulations was to minimize potential contributory factors that may confound data interpretation. To this end, we identified Bpdl to be a better iron chelator than DFO under single-treatment experimental conditions. In a number of criteria, specifically IFU yield, aberrant RB formation, and transcriptional modulation of previously reported persistence indicating genes, Bpdl was consistently more potent than DFO. Moreover, the increased differential transcription of iron-responsive genes, ahpC and devB, in the Bpdl protocol relative to the DFO starvation method validated the alternative iron starvation system. Finally, the Bpdl model allowed the novel observation that ytgA, which codes for an iron-binding periplasmic protein thought to be involved in iron acquisition/transport into Chlamydia, is iron-responsive at the transcript level. The use of this Bpdl model could allow additional insights into the global iron-restricted regulon of C. trachomatis.
Our study was initiated due to the inefficient ability of DFO to elicit a uniform, persistent phenotype of C. trachomatis for analysis of differential gene expression. We hypothesized that DFO, which is not membrane permeable and can only bind ferric iron, could only be efficient in the iron-restriction of the obligate intracellular pathogen if the only chlamydial source of iron was the direct acquisition of ferric iron as it entered the host cell via the transferrin pathway. On the other hand, if Chlamydia were able to acquire iron from another source, e.g., from the cytosolic labile iron pool or the slow recycling pathway (Ouellette and Carabeo, 2010), then use of DFO would require longer treatment to deplete intracellular iron stores. In the literature, DFO has rarely been used without auxiliary steps or methods for iron-restriction. For instance, the combination of DFO with cyclohexamide (which is a general eukaryotic protein synthesis inhibitor) has been employed to prevent the host cell up-regulation of the transferrin iron-acquisition pathway (Raulston, 1997; Wehrl et al., 2004; Dill et al., 2009). Yet the modulation of host protein synthesis results in pleiotropic effects that likely influence chlamydial growth via means independent of iron starvation. Others have eliminated this need for cyclohexamide through the use of long-term DFO pre-treatment protocols in order to deplete host cells of intracellular iron stores prior to chlamydial invasion (Al-Younes et al., 2001; Peters et al., 2005; Mukhopadhyay et al., 2006; Dill et al., 2009; Timms et al., 2009). While the effects of this protocol could presumably be attributed solely to iron-deprivation, it is tedious and requires the infection of a compromised host cell population. One particularly illustrative example was published by Dill et al. (2009), in which both pre-treatment and combination with cyclohexamide was employed to observe transient and marginal (less than twofold) elevation of a handful of iron-responsive C. trachomatis transcripts. Meanwhile, transcripts of euo and omcB were not modulated (Dill et al., 2009), suggesting that the chlamydial persistent phenotype was not elicited by this treatment combination.
The salient properties of Bpdl are its membrane permeability and its high affinity for the ferric and ferrous forms of iron. Membrane permeability allows it access to the different iron reservoir inside the cell, as well as within the chlamydia organisms, while its ability to bind both forms of iron would result in a more complete iron depletion. Both properties combine to rapidly and efficiently starve chlamydia and its host of iron. An iron chelator with similar properties, salicylaldehyde isonicotinoyl hydrazone (SIH) was initially the compound of choice. However, its structural similarity to a family of molecules known to inhibit the function of type III secretion apparatus in a number of bacterial pathogens precluded accurate interpretations of its growth effects on chlamydia. Indeed, we have observed the inhibition of the cytosolic translocation of the Salmonella effector PipB2 in infected cells treated with SIH for 8 h (Thompson and Carabeo, unpublished data), and hence, investigations of the functions of type III secretion in Chlamydia using these SIH-related compounds must be taken with caution.
In summary, the detailed characterization of DFO and Bpdl with regards to their effects on C. trachomatis when supplemented at the time of infection, revealed the superiority of Bpdl. Moreover, this model is easier to apply and eliminated many of the caveats of other DFO- based iron-restriction protocols. As such, efforts to identify bona fide primary iron-responsive regulons may now be compatible with microarray approaches, given the availability of a faster acting iron chelator that, in addition to the extracellular iron pool, targets the pools in the DFO-inaccessible cytosolic, organellar, and chlamydial environment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Dr. Scot Ouellette and the rest of the Carabeo lab for their critiques and helpful suggestions. This research was supported by a grant from the Medical Research Council (G0900213) to Rey A. Carabeo.
References
- Al-Younes H. M., Rudel T., Brinkmann V., Szczepek A. J., Meyer T. F. (2001). Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell. Microbiol. 3, 427–437 [DOI] [PubMed] [Google Scholar]
- Beatty W. L., Byrne G. I., Morrison R. P. (1993). Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 90, 3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty W. L., Morrison R. P., Byrne G. I. (1994). Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58, 686–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R. J., Nelson D. E., Virok D., Crane D. D., Hogan D., Sturdevant D. E., Beatty W. L., Caldwell H. D. (2003). Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U.S.A. 100, 15971–15976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer W., Epsztejn S., Cabantchik Z. I. (1995). Iron acquired from transferrin by K562 cells is delivered into a cytoplasmic pool of chelatable iron(II). J. Biol. Chem. 270, 24209–24215 [DOI] [PubMed] [Google Scholar]
- Cable H., Lloyd J. B. (1999). Cellular uptake and release of two contrasting iron chelators. J. Pharm. Pharmacol. 51, 131–134 10.1211/0022357991772231 [DOI] [PubMed] [Google Scholar]
- Caldwell H. D., Kromhout J., Schachter J. (1981). Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31, 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles A. M., Reynolds D. J., Harper A., Devitt A., Pearce J. H. (1993). Low-nutrient induction of abnormal chlamydial development: a novel component of chlamydial pathogenesis? FEMS Microbiol. Lett. 106, 193–200 [DOI] [PubMed] [Google Scholar]
- Dill B. D., Dessus-Babus S., Raulston J. E. (2009). Identification of iron-responsive proteins expressed by Chlamydia trachomatis reticulate bodies during intracellular growth. Microbiology 155(Pt 1), 210–219 [DOI] [PubMed] [Google Scholar]
- Gaeta A., Hider R. C. (2005). The crucial role of metal ions in neurodegeneration: the basis for a promising therapeutic strategy. Br. J. Pharmacol. 146, 1041–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickstein H., El R. B., Shvartsman M., Cabantchik Z. I. (2005). Intracellular labile iron pools as direct targets of iron chelators: a fluorescence study of chelator action in living cells. Blood 106, 3242–3250 10.1182/blood-2005-02-0460 [DOI] [PubMed] [Google Scholar]
- Hentze M. W., Muckenthaler M. U., Andrews N. C. (2004). Balancing acts: molecular control of mammalian iron metabolism. Cell 117, 285–297 10.1016/S0092-8674(04)00343-5 [DOI] [PubMed] [Google Scholar]
- Hogan R. J., Mathews S. A., Mukhopadhyay S., Summersgill J. T., Timms P. (2004). Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72, 1843–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliffe-Lee E. R., McClarty G. (2000). Regulation of carbon metabolism in Chlamydia trachomatis. Mol. Microbiol. 38, 20–30 [DOI] [PubMed] [Google Scholar]
- Kato J., Kobune M., Ohkubo S., Fujikawa K., Tanaka M., Takimoto R., Takada K., Takahari D., Kawano Y., Kohgo Y., Niitsu Y. (2007). Iron/IRP-1-dependent regulation of mRNA expression for transferrin receptor, DMT1 and ferritin during human erythroid differentiation, 35, 879–887 10.1016/j.exphem.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Lambden P. R., Pickett M. A., Clarke I. N. (2006). The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology 152(Pt 9), 2573–2578 10.1099/mic.0.29032-0 [DOI] [PubMed] [Google Scholar]
- Lloyd J. B., Cable H., Rice-Evans C. (1991). Evidence that desferrioxamine cannot enter cells by passive diffusion. Biochem. Pharmacol. 41, 1361–1363 [DOI] [PubMed] [Google Scholar]
- Martell A., Smith R. M. (1977). Critical Stability Constants/Vol. 3 Other Organic Ligands. New York: Plenum Press, 303–304 [Google Scholar]
- Matsumoto A., Manire G. P. (1970). Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J. Bacteriol. 101, 278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. D., Sal M. S., Schell M., Whittimore J. D., Raulston J. E. (2009). Chlamydia trachomatis YtgA is an iron-binding periplasmic protein induced by iron restriction. Microbiology 155(Pt 9), 2884–2894 10.1099/mic.0.030247-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpiga P., Ravaoarinoro M. (2006). Chlamydia trachomatis persistence: an update. Microbiol. Res. 161, 9–19 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Miller R. D., Sullivan E. D., Theodoropoulos C., Mathews S. A., Timms P., Summersgill J. T. (2006). Protein expression profiles of Chlamydia pneumoniae in models of persistence versus those of heat shock stress response. Infect. Immun. 74, 3853–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette S. P., Carabeo R. A. (2010). A functional slow recycling pathway of transferrin is required for growth of chlamydia. Front. Microbiol. 1:112. 10.3389/fmicb.2010.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette S. P., Hatch T. P., AbdelRahman Y. M., Rose L. A., Belland R. J., Byrne G. I. (2006). Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNgamma-mediated host cell tryptophan starvation. Mol. Microbiol. 62, 1387–1401 [DOI] [PubMed] [Google Scholar]
- Persson H. L., Yu Z., Tirosh O., Eaton J. W., Brunk U. T. (2003). Prevention of oxidant-induced cell death by lysosomotropic iron chelators. Free Radic. Biol. Med. 34, 1295–1305 [DOI] [PubMed] [Google Scholar]
- Peters J., Hess S., Endlich K., Thalmann J., Holzberg D., Kracht M., Schaefer M., Bartling G., Klos A. (2005). Silencing or permanent activation: host-cell responses in models of persistent Chlamydia pneumoniae infection, 7, 1099–1108 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K. K., Harford J. B., Rouault T. A., McClelland A., Ruddle F. H., Klausner R. D. (1986). Transcriptional regulation by iron of the gene for the transferrin receptor. Mol. Cell. Biol. 6, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau A., Wyllie S., Whittimore J. D., Raulston J. E. (2005). Identification of Chlamydia trachomatis genomic sequences recognized by chlamydial divalent cation-dependent regulator A (DcrA). J. Bacteriol. 187, 443–448 10.1128/JB.187.2.443-448.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulston J. E. (1997). Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65, 4539–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo A. M., Christen L., Niles E. G., Kosman D. J. (2001). Intracellular chelation of iron by bipyridyl inhibits DNA virus replication: ribonucleotide reductase maturation as a probe of intracellular iron pools. J. Biol. Chem. 276, 24301–24308 [DOI] [PubMed] [Google Scholar]
- Smith R. M., Martell A. (1975). Critical Stability Constants/Vol. 2, Amines. New York: Plenum Press, 235–237 [Google Scholar]
- Timms P., Good D., Wan C., Theodoropoulos C., Mukhopadhyay S., Summersgill J. T., Mathews S. A. (2009). Differential transcriptional responses between the interferon-gamma-induction and iron-limitation models of persistence for Chlamydia pneumoniae. J. Microbiol. Immunol. Infect. 42, 27–37 [PubMed] [Google Scholar]
- Wehrl W., Meyer T. F., Jungblut P. R., Muller E. C., Szczepek A. J. (2004). Action and reaction: Chlamydophila pneumoniae proteome alteration in a persistent infection induced by iron deficiency, 4, 2969–2981 [DOI] [PubMed] [Google Scholar]
- Wood H., Roshick C., McClarty G. (2004). Tryptophan recycling is responsible for the interferon-gamma resistance of Chlamydia psittaci GPIC in indoleamine dioxygenase-expressing host cells. Mol. Microbiol. 52, 903–916 [DOI] [PubMed] [Google Scholar]
- Wyllie S., Raulston J. E. (2001). Identifying regulators of transcription in an obligate intracellular pathogen: a metal-dependent repressor in Chlamydia trachomatis. Mol. Microbiol. 40, 1027–1036 [DOI] [PubMed] [Google Scholar]
- Wyrick P. B. (2010). Chlamydia trachomatis persistence in vitro: an overview. J. Infect. Dis. 201, 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]