Abstract
Prior genetic and physical mapping has shown that the Naip gene cluster on mouse chromosome 13D1-D3 contains a gene, Lgn1, that is responsible for determining the permissivity of ex vivo macrophages to Legionella pneumophila replication. We have identified differences in the structure of the Naip array among commonly used inbred mouse strains, although these gross structural differences do not correlate with differences in L. pneumophila permissiveness. A physical map of the region employing clones of the C57BL/6J haplotype confirms that there are fewer copies of Naip in this strain than are in the physical map of the 129 haplotype. We have also refined the genetic location of Lgn1, leaving only Naip2 and Naip5 as candidates for Lgn1. Our genetic map suggests the presence of two hotspots of recombination within the Naip array, indicating that the 3′ portion of Naip may be involved in the genomic instability at this locus.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF240489–AF240530.]
Genetic variation in the ability of ex vivo mouse macrophages to support the intracellular replication of Legionella pneumophila has been mapped to mouse chromosome 13D1-D3 (Lgn1; Beckers et al. 1995; Dietrich et al. 1995). The human region orthologous to the Lgn1 locus on chromosome 5q11.2-q13.3 contains mutations responsible for a family of autosomal recessive neurodegenerative diseases termed spinal muscular atrophy (SMA; Gilliam et al. 1990). Distinct genes present in both intervals have been attributed to the primary effects of each phenotype. While one or more closely related Naip (neuronal apoptosis inhibitory protein) paralogs has been identified as a candidate gene for Lgn1 (Endrizzi et al. 1999; Growney et al. 2000), SMN is responsible for the majority of SMA cases (Lefebvre et al. 1995; Rodrigues et al. 1995). However, Naip remains a candidate modifying gene for SMA disease severity (Roy et al. 1995; Morrison 1996; Chang et al. 1997). Correlation between disease severity and the extent of genomic deletions has provided additional support for the role of Smn flanking genes in SMA progression (DiDonato et al. 1994; Wirth et al. 1995; Burlet et al. 1996). Therefore, continued study of the Naip gene family will likely provide insight into L. pneumophila susceptibility and SMA.
Both the mouse and human Lgn1/SMA intervals contain multiple copies of large segments of DNA (Carpten et al. 1994; Burglen et al. 1996, 1997; Carter et al. 1997). We have previously demonstrated that the structure and number of the repeated segments in these regions is different in mouse and human, indicating an independent origin for the genomic amplifications in these species (Growney et al. 2000). For example, the SMA locus resides within a large inverted duplication that includes SERF1, SMN, NAIP, and GTF2H2, while the mouse Lgn1 region contains a direct repeat of a variable number of Naip genes. To date, Naip is the only identified gene common to the amplifications in both species (Scharf et al. 1996, 1998; Schrank et al. 1997; Viollet et al. 1997).
The differences in the structure of the array between mouse and human, as well as the complexity of the mouse array, suggest that this region of the genome is subject to various types of rearrangements. Furthermore, differences in SMN and NAIP gene copy numbers in human SMA patients (Rajcan-Separovic et al. 1996) imply that understanding the mechanisms of genomic instability of these loci may play a crucial role in predicting SMA outcome. Continued comparative genomic analysis may identify additional common sequence elements that will further our understanding of the instability of these regions.
We have previously constructed a detailed physical map of the Naip gene array in the 129 mouse haplotype (Growney et al. 2000). This map contains a direct repeat of seven complete Naip genes with three 5′ truncated Naip loci interspersed within the array. An important character of the 129 structure is that the central portion of the Naip array contains several copies of Naip genes (known as Naip3–Naip7 and ΔNaip1–3), which are much more highly related to each other than they are to the Naip genes on the proximal and distal borders (which are known as Naip1 and Naip2). The central Naip genes share such extensive homology that copies of microsatellite loci in their introns are simultaneously amplified by the same pair of primers, whereas Naip1 and Naip2 contain markers within their introns that map uniquely in the mouse genome. Based on the marker content and exon sequence homology of the different Naip paralogs, we have proposed a model for the origins of the 129 Naip array (Growney et al. 2000).
Our previous genetic and physical maps of the region have suggested that one or more copies of Naip are responsible for the Lgn1 phenotype. However, polymorphisms between the strains used in our genetic mapping (A/J and C57BL/6J) and the strains used in our physical mapping (129) have prevented us from refining the Lgn1 genetic interval within the central portions of the Naip array (Dietrich et al. 1995; Scharf et al. 1996; Endrizzi et al. 1999; Growney et al. 2000). Because we have several animals from our cross that are recombinant within the Naip array, it seemed likely that additional attempts to physically map the location of recombinant genetic markers contained in the central repetitive Naip array would allow us to improve our genetic mapping of Lgn1.
Our interest in understanding both the mechanisms involved in the apparent genomic instability of this region and in identifying the Lgn1 gene have led us to compare the structure of the Naip array among commonly used inbred mouse strains. Here, we report significant differences in the structure of the array among commonly used inbred mouse strains that do not obviously correlate with permissiveness to L. pneumophila replication. We have also constructed a new physical map of the Lgn1 region using clones of the C57BL/6J haplotype. Using this physical map to localize the position of polymorphic but repetitive markers, we have refined the genetic interval for Lgn1 to the highest resolution possible with our current backcross (Scharf et al. 1996). Because we have identified apparent recombination hotspots within the Naip array, it is possible that sequences within the 3′ portion of Naip may be responsible for the instability within the Naip array.
RESULTS
Comparison of Naip Repeat Structure among Inbred Mouse Strains
The sequence of BAC 149m19 (AF131205; Endrizzi et al. 1999), BAC 26f17 (AF242431, AF242432), and P1–9045 (AF242433, AF242434, AF242435; Endrizzi et al. 2000) together encompass six complete Naip genes. These include the “unique” Naip genes on the borders, Naip2 and Naip1, as well as four of the five Naip genes that lie in the central amplification, namely Naip5, Naip7, Naip6, and Naip3 (Fig. 1; Growney et al. 2000). Our analysis of these genomic sequences indicates that intron 13, which is present in all copies of Naip, exhibits gene-specific polymorphisms and can be grouped into two classes. We call one group “type I” loci, present in Naip2, Naip5, Naip3, ΔNaip, and Naip1, and which are 337, 304, 318, 318, and 289 bp, respectively. We call the second group “type II” loci, present in Naip7 and Naip6. Type II loci are 6,545 bp due to an insertion of an L1 repeat unit (Endrizzi et al. 2000).
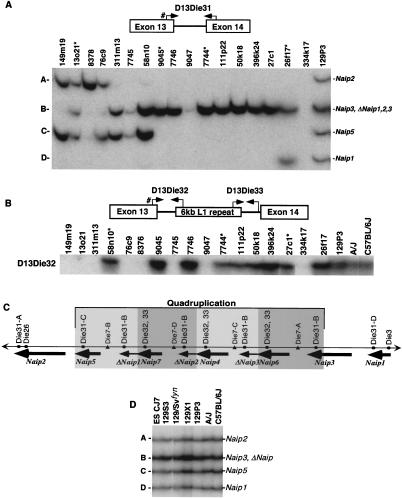
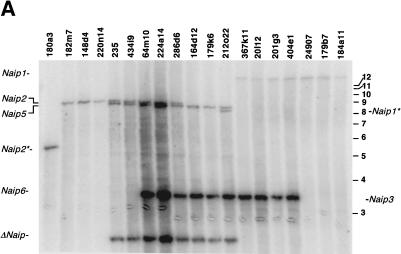
Figure 1.
Intron 13 identifies conserved Naip elements. (A) Autoradiography of 6% denaturing polyacrylamide gel of PCR products amplified from the indicated 129 mouse Lgn1 BACs using STS D13Die31 (see Methods). A schematic of D13Die31 is indicated to show primer orientation. Pound sign indicates common primer with D13Die32. Band sizes are labeled A–D from largest to smallest. Asterisk indicates nonoverlapping clones in 129 physical map of the Naip array that contain D13Die31-B, indicating four distinct D13Die31-B loci. (B) Autoradiography of 6% denaturing polyacrylamide gel of PCR products amplified from the indicated 129 mouse Lgn1 BACs using STS D13Die32 (see Methods). A schematic of D13Die32 and D13Die33 is indicated to show primer orientation. Pound sign indicates common primer with D13Die31. Asterisk indicates nonoverlapping clones in 129 physical map of the Naip array that contain D13Die32, indicating three distinct D13Die32 loci. Identical patterns of amplification were obtained with D13Die32 (data not shown). (C) Schematic of the structure of the Naip array in mice of the 129 haplotype. Single-copy markers D13Die26 and D13Die3 are indicated, identifying “unique” Naip loci. The four copies of the repeated marker D13Die7 are indicated. Gray shading indicates four subunits of the quadruplication of Naip sequence. Positions of Naip intron 13 loci identified with D13Die31, D13Die32 and D13Die33 are shown. (D) Autoradiography of 6% denaturing polyacrylamide gel of PCR products amplified with D13Die31 on genomic DNA from cell line ES CJ7 and several inbred mouse lines. Band sizes are labeled as in A. Identification of Naip loci are as determined from 129 physical map (Growney et al. 2000).
The presence of these polymorphisms led us to use intron 13 as a marker for comparison of the structure of the Naip array among inbred mouse strains. To do this, we devised a PCR assay, D13Die31, which amplifies all type I intron 13 loci and two PCR assays, D13Die32 and D13Die33, which amplify the proximal and distal ends of the type II intron 13 (Table 1). As expected, D13Die31 yields four bands from a panel of 129 BAC and P1 clones spanning the entire Lgn1 interval (Fig. 1A). Three of these bands were unique to specific Naip loci. Notably, band A maps to Naip2, band C maps to Naip5, and band D maps to Naip1. Band B, on the other hand, is present in multiple nonoverlapping clones, mapping them to multiple loci, including Naip3 and the three ΔNaip loci. This finding is in agreement with our previous prediction that the ΔNaip loci are closely related to Naip3 (Growney et al. 2000). We also found that D13Die32 (Fig. 1B) and D13Die33 (data not shown) are present in multiple copies throughout the map, mapping intron 13 type II to Naip6, Naip4, and Naip7. The positions of these markers within the Naip array from the 129 haplotype are indicated in Figure 1C.
Table 1.
Primers for STSs Used in This Work
| Locus name | Gene location | F-primera | R-primera | Size (bp)c | Reference |
|---|---|---|---|---|---|
| Naip-RS | Naip exon 3 | AGCTCATGGATACCACAGGAGATG | ATATCCCTTGGACATAAAATGGC | 263 | (Scharf et al. 1996) |
| Ocln | Unmapped exon | ATTATGCACCAAGCAATGACA | ATGCACATCACAATAATGAGCA | 169 | (Growney et al. 2000) |
| Smn | Smn exon 4intron 4 | TGGAAAGTTGGTGACAAGTG | CGTCTGTGGAAACTTGACTTC | 435 | This work |
| D13Die3 | Naip1 intron 7 | ACGACAGCGTCTTCGCTAAT | GGGCAGCATTGCAGACAT | 209 | (Scharf et al. 1996) |
| D13Die6 | Smn upstream region | CACAGTGCACAAAACCATCC | AACTTCAACAAGGCCACACC | 126 | (Scharf et al. 1996) |
| D13Die7 | Naip-ΔNaip intergenic region | AATTTCTGTGTGGTTGTGTG | ATAAACCATCCCTAGTTGCCTG | 125–136 | (Scharf et al; 1996) |
| D13Die24 | Naip2 intron 4 | CGGTGAGAGGCTCAGAGTTC | TGGTGTAGCCCTTTTCTGCT | 154 | (Endrizzi et al. 1999) |
| D13Die25 | Naip5, 7, 4, 6 intron 8 | TGCAGAGGACTTCAAGGTTT | CAACACTGACAGAATCACTGGA | 216–250 | (Endrizzi et al. 1999) |
| D13Die26 | Naip2 intron 12 | TGGAGAACAATTCTGAAACACA | AACAGATTTCCCCCAAAGCT | 164 | (Endrizzi et al. 1999) |
| D13Die27 | Naip2–Naip5 intergenic region | TGGGGTTCTCTAAGGTAACAGA | GGCCTTTGTGAGAAGGAACA | 151 | (Endrizzi et al. 1999) |
| D13Die30 | Naip exon 16 ΔNaip exon 8 junction | AGACACCCCCAGTCCAAAC | CTTGTTGTGCTCTTGTATTGGG | 1479 | (Growney et al. 2000) |
| D13Die30.2 | Naip6-ΔNaip junction | CTGTAGAGCTGAACAGAGACTGA | GACTTCTTGGTGTCTGCTTCC | 2904 | This work |
| D13Die31 | Naip intron 13 type 1 | ATGCTTTGAAGCCATGACCTb | AAAACAAAATTTGGCAAAATGT | A = 385 B = 366 C = 353 D = 337 | This work |
| D13Die32 | Naip intron 13 type II | ATGCTTTGAAGCCATGACCTb | GGCCCATTAGACTTGCAAAC | 410 | This work |
| D13Die33 | Naip intron 13 type II | TGACAGCTTCTGGGACAGG | GAAGATGAAGGCCCAAAACA | 272 | This work |
| D13Die34 | Naip exon 11 | GAAGCTGGCAGTGGAAAGAC | GTATAGGCGGATGTCTCTGACC | 366 | (Growney et al. 2000) |
| D13Die35 | Naip5/Naip 6 intron 11 | AAGGACATGAGAGAAAGAAGCC | ACTTGCTAGTGAGAGGGAGAGC | 172–270 | This work |
| D13Die36 | Naip3/ΔNaip intron 14 | AGGACAACTGTACACAGCTTGC | TCTCCTCCCATTGATGACCT | 215–230 | This work |
| D13Die37 | Naip2 intron 8 | GAGAGATCACTCATGCTCATGA | CAAGGATGGTGATTTCCCC | 357 | This work |
| D13Die38 | 180a3 SP6 | GTGCACAAGTACACACATGGG | GAGGATTTGCTCGAATATTTGG | 272 | This work |
| D13Die39 | 182m7 T7 | TAGCCATGTAACGGGCTACC | TGTGGGGCCTTAGGTTAGTG | 193 | This work |
| D13Die40 | 220n14 SP6 | AAGAAGAGTCGGCTCCACAA | TGTCTTAACGTCCGTTTGCA | 322 | This work |
| D13Die41 | 434i9 T7 64m10 T7 | CCCTGTAAACCCAGCACCTA | CAAGCTTGGTGTGTTAGTGCA | 205 | This work |
| D13Die42 | 404e1 T7 | CTTTTTTAGTTCGGGAGGGG | TGTTATATACCAGAAACCCCGG | 202 | This work |
| D13Die43 | 20l12 SP6 | TCAGAGCAGGGACATGAGC | AAAGGCACCTTACATCTGAAGG | 291 | This work |
| D13Die44 | 480i6 T7 477p2 SP6 201g3 SP6 | ACAGCATAAGGACCTTCACTCC | AGGCAGAGTCTCCCACTGAA | 278 | This work |
| D13Die45 | 180a11 T7 | AGACTACCCACACAGGGGC | CCCCTGACCCAGTTGCTATA | 148 | This work |
| D13Die46 | 179b7 SP6 | TCCTGGCATCTGCCTCTC | CCCTCCCCAGCTAAGAGAAC | 192 | This work |
Novel STS were developed using Primer 0.5 (http://www.genome.wi.mit.edu/). Primers for all STS except D13Die30.2 were chosen to be 18–22 bp in length, with an optimal melting temperature of 60°C (range 57°C–63°C). Primers for STS D13Die30.2 were developed for long PCR. Primers were chosen to be 20–23 bp in length, with an optimal melting temperature of 64°C (range 60°C–68°C).
D13Die31 and D13Die32 share a common forward primer mapping to Naip exon 13.
PCR product sizes predicted from 129 genomic sequence.
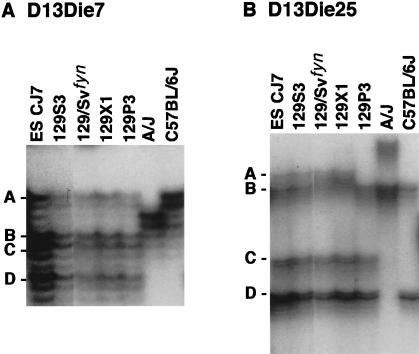
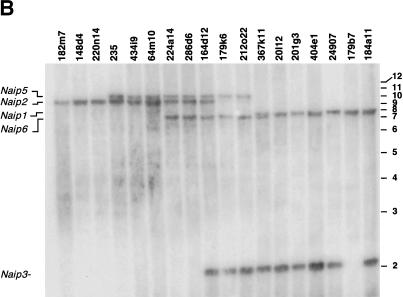
We took several approaches to investigate whether the structure of the Naip array in strains C57BL/6J and A/J, which are the parents of the cross we used to map Lgn1, was the same as that observed in the 129 haplotype. First, we found that the intron 13 type I marker D13Die31 yields an identical pattern of bands when amplified on genomic DNA from the A/J, C57BL/6J and 129 strains, suggesting that all the basic elements of the Naip array are present in these strains (Fig. 1D). Second, we analyzed genomic DNA with microsatellite markers that map to the central portion of the Naip array (Growney et al. 2000). As can be seen in Figure 2, the number of polymorphic bands amplified by D13Die7 and D13Die25 varies between mouse strains, with 129 strains consistently yielding more bands than A/J and C57BL/6J.
Figure 2.
Amplification of polymorphic microsatellite markers from mouse genomic DNA identifies differences in the Naip repeat structure among inbred mouse strains. Autoradiography of 6% denaturing polyacrylamide gel of PCR products amplified from genomic DNA of the indicated mouse strains using primers for microsatellite markers D13Die7 (A) and D13Die25 (B). Four bands representing four distinct loci for each marker identified in the 129 haplotype are labeled A–D, from largest to smallest. A, D13Die7: all 129 substrains analyzed demonstrate four amplified loci. B, D13Die25: band A is polymorphic in 129P3. These markers amplify two distinct bands in A/J and C57BL/6J.
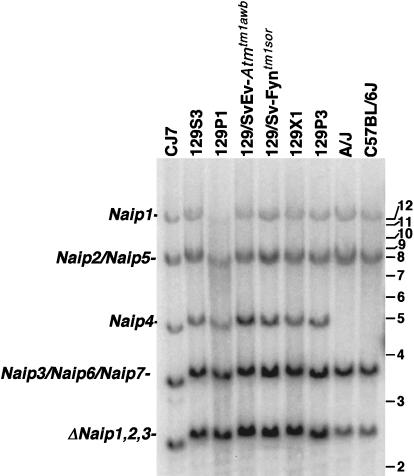
Because this data suggests the possibility that inbred mouse strains differ in the number of Naip gene subunits contained within the central repeat, we performed hybridization experiments in an attempt to compare the total number of Naip genes in different mouse strains. We observed significant differences in the hybridization pattern of a Naip exon 11 probe to BamHI genomic Southern Blots (Fig. 3). We have previously mapped ten Naip exon 11 loci in mice of the 129 haplotype (Growney et al. 2000). Consistent with differences in the number of copies of repetitive microsatellite markers, the A/J and C57BL/6J strains demonstrate hybridization patterns distinct from 129 strains and consistent with differences in the number of copies of repetitive microsatellite markers. Notably, these strains lack the Naip4 specific 5-kb band. In addition, the 3.5-kb triplet band, mapping to Naip3, Naip6, and Naip7, and the 2.5-kb fragment mapping to the three ΔNaip loci are consistently less intense in A/J and C57BL/6J. Together, these data suggest that the central amplification within the Naip array is larger in 129 strains than in A/J and C57BL/6J.
Figure 3.
Southern blot analysis of BamHI-digested genomic DNA identifies differences in Naip copy number among inbred mouse strains. Correlation of the Naip exon 11 hybridizing BamHI fragments of genomic DNA from several 129 strains with Naip loci was done by comparison of the fragments in BAC and P1 clones from the 129 haplotype that make up a complete map of the region (data not shown) and from completed genomic sequence (Endrizzi et al. 1999, 2000). Horizontal bars and numbers indicate position and size (kb) of fragments in 1-kb ladder molecular weight marker (GIBCO). A single band of 12 kb mapping to Naip1 and an 8.5-kb doublet fragment mapping to Naip2 and Naip5 are well conserved among all strains. In contrast, the 5-kb fragment mapping to Naip4 is unique to mice of the 129 lineage. A 3.8-kb band mapping to Naip3, Naip6, and Naip7 in the 129 haplotype is polymorphic in other strains. This fragment is less intense in A/J and C57BL/6J. The 2.5-kb fragment mapping to the three ΔNaip loci in the 129 haplotype is significantly decreased in intensity in A/J and C57BL/6J.
Construction of C57BL/6J Physical Map
To determine the structure of the Naip array in C57BL/6J, we constructed a physical map of the region using clones derived from this strain. We identified 21 clones from two distinct BAC libraries (see Methods; Table 2). We ordered the clones into a complete physical map of the C57BL/6J Naip array using markers developed in mapping the 129 interval (Table 1) (Dietrich et al. 1994, 1995, 1996; Scharf et al. 1996, 1998; Endrizzi et al. 1999; Growney et al. 2000). Our reasoning behind our ability to order the clones and markers is outlined as follows. First, all clones were typed for the presence of single-copy markers. These include D13Die3, D13Die6, D13Die24, D13Die26, D13Die27, D13Die37, and Ocln (data not shown, Table 1). These markers identified clones that contain single-copy sequence on the proximal and distal borders of the region. Second, clones containing Naip2, Naip5, and Naip1 were identified by D13Die31 loci A, C, and D (Fig. 4A). In addition, clones that extend into the repeat were identified by the presence of D13Die31-B.
Table 2.
C57BL/6J BAC Clone Summary
| Clonea | Size (kb)b | Cen end | Cen end seq.accession # | Cen end map position | Tel end | Tel end seq.accession # | Tel end map position |
|---|---|---|---|---|---|---|---|
| 180a3 | 180 | SP6 | AF240489 | 83 kb proximal to 149m19–D13Die38 | T7 | AF240490 | Naip2 intron 8 |
| 182m7 | 190 | T7 | AF240491 | 56 kb proximal to 149m19–D13Die39 | SP6 | AF240492 | Naip2 intron 2 |
| 220n14 | 177 | SP6 | AF240493 | 52 kb proximal to 149m19–D13Die40 | T7 | AF240494 | Naip2 intron 2 |
| 148d4 | 191 | T7 | AF240495 | 38 kb proximal to 149m19 | SP6 | AF240496 | Naip5 intron 12 |
| 434i9 | 214 | T7 | AF240497 | Tfnrc–D13Die41 | SP6 | AF240498 | Naip6 intron 13 |
| 64m10 | 220 | T7 | AF240499 | Tfnrc–D13Die41 | SP6 | AF240500 | Naip6 intron 6 |
| 224a14 | 194 | SP6 | AF240501 | Smn 5′ region | T7 | AF240502 | Naip6–Naip3 intergenic region |
| 235 | 150 | T7 | AF240503 | Smn exon 1 | SP6 | AF240504 | ΔNaip exon 7 |
| 286d6 | 180 | T7 | AF240505 | Smn intron 1 | SP6 | AF240506 | Naip6–Naip3 intergenic region |
| 164d12 | 220 | SP6 | AF240507 | Naip2 intron 5 | T7 | AF240508 | Naip1 exon 16 |
| 179k6 | 194 | T7 | AF240509 | Naip2 intron 2 | SP6 | AF240510 | Naip1–Naip3 intergenic region |
| 212o22 | 194 | T7 | AF240511 | Naip5 intron 15 | SP6 | AF240512 | Naip1 intron 9 |
| 367k11 | 190 | T7 | AF240513 | Naip6 exon 16 | SP6 | AF240514 | Gtf2h2d |
| 20112 | 218 | T7 | AF240515 | Naip5–Naip6 intergenic region | SP6 | AF240516 | 56 kb distal to 26f17; D13Die43 |
| 201g3 | 218 | T7 | AF240517 | Naip3 intron 14 | SP6 | AF240518 | 70 kb distal to 26f17; D13Die44 |
| 404e1 | 190 | SP6 | AF240519 | Naip3 intron 14 | T7 | AF240520 | 48 kb distal to 26f17; D13Die42 |
| 24907 | 125 | SP6 | AF240521 | Naip3 intron 9 | T7 | AF240522 | Gtf2h2d |
| 477p2 | 170 | T7 | AF240523 | Naip3 intron 4 | SP6 | AF240524 | 55 kb distal to 26f17; D13Die44 |
| 184a11 | 194 | SP6 | AF240525 | Naip3 intron 4 | T7 | AF240526 | 79 kb distal to 26f17; D13Die45 |
| 480i6 | 180 | SP6 | AF240527 | Naip3 intron 1 | T7 | AF240528 | 73 kb distal to 26f17; D13Die44 |
| 179b7 | 210 | T7 | AF240529 | Naip3 intron 1 | SP6 | AF240530 | 113 kb distal to 26f17; D13Die46 |
Clones 235 and 24907 are derived from the Genome Systems St. Louis, MO C57BL/6J BAC library. All remaining clones are derived from the RPCl-23 C57BL/6J BAC library.
Sizes determined from pulsed field gel analysis of Notl digested DNA (Methods).
No cDNA for mouse Tfnr has been described, so we cannot define exon nomenclature.
No cDNA for mouse Gtf2h2 has been described, so we cannot define exon nomenclature.
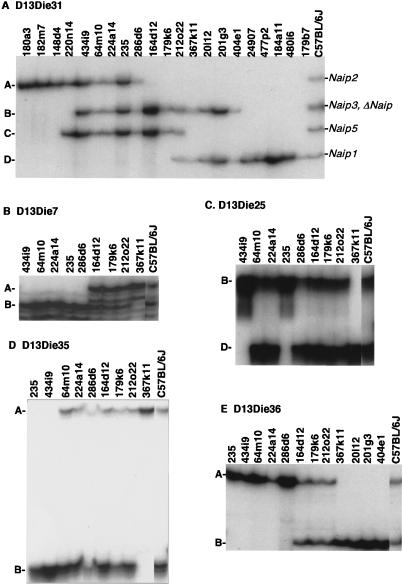
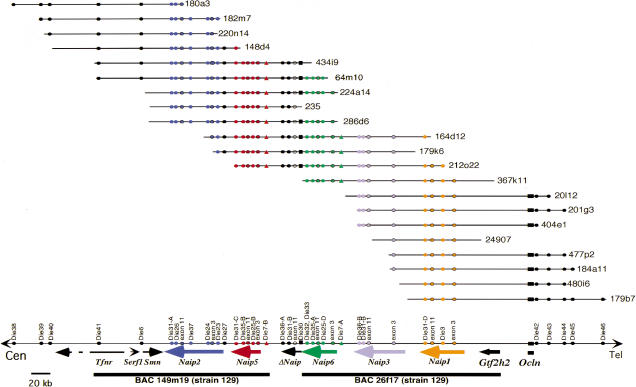
Figure 4.
Amplification of repetitive polymorphic microsatellite markers mapping within the Naip array of the C57BL/6J haplotype. Autoradiography of 6% denaturing polyacrylamide gel of PCR products amplified from genomic DNA and genomic clones using primers for STS D13Die31 (A) and microsatellite markers D13Die7 (B), D13Die25 (C), D13Die35 (D), and D13Die36 (E). (A) Four bands representing four distinct D13Die31 loci are labeled A–D, from largest to smallest. A single genomic clone, 212o22, which includes Naip5 (band C) and Naip1 (band D) spans the entire central repeat region of the Naip array. (B–E) Microsatellite repeat markers identify and separate two loci for each marker present in C57BL/6J genomic DNA.
Finally, those clones that extend into the central Naip repeat were ordered by typing microsatellite markers D13Die7 (Fig. 4B), D13Die25 (Fig. 4C), D13Die35 (Fig. 4D), and D13Die36 (Fig. 4E). For each marker, two distinct loci were amplified from genomic DNA. At least one clone was identified for all markers that contains only one locus of each repeat marker, allowing duplicated loci to be ordered relative to each other. For example, clones 434i9 and 235 each contain a single copy of D13Die25, band B (for nomenclature, see Methods). Both clones contain the single-copy marker D13Die27 (data not shown), indicating that D13Die25-B maps on the proximal side of the repeat. Clone 367k11 contains only one copy of D13Die25, band D. This clone also contains the distal single-copy marker D13Die3, placing D13Die25-D distal to D13Die25-B. While we were able to order both loci amplified with a given marker relative to each other, the order of distinct markers in two regions of the map was ambiguous from the marker content of the clones. The first region, located in the interval from Naip5 to ΔNaip, includes D13Die35-B, D13Die25-B, D13Die7-B, and D13Die36-A. The second region, within Naip6, includes D13Die35-A and D13Die25-D. Because these markers have been previously mapped to specific intronic or intergenic regions, they were ordered relative to each other based on our prior knowledge of genomic sequence (Endrizzi et al. 1999, 2000).
These data allowed us to order all the clones into a contiguous map of the Naip region. Additional Naip sequence content was identified by Southern blot analysis. We probed each clone for the presence of Naip exon 3 (Fig. 5A) and Naip exon 11 (Fig. 5B). We were able to map six copies of Naip exon 11 and five copies of Naip exon 3. Specific Naip loci were identified based on the relative position of restriction fragments within the contig (proximity to single-copy markers) and comparison of restriction fragments to Southern blot analysis of 129 BACs (data not shown) (Growney et al. 2000). Consistent with the identification of six Naip exon 11 loci and five Naip exon 3 loci, only a single locus for D13Die30, a marker for the junction of a complete Naip gene and a 5′ truncated Naip locus, and intron 13 type II (D13Die32 and D13Die33) was identified (data not shown). Finally, the end sequence of all clones in our map were compared to the sequenced 129 clones BAC 149m19 (AF131205), BAC 26f17 (AF242431, AF242432), and P1 9045 (AF242433, AF242434, AF242435) and the strain B10.A-derived Naip cDNA clones (Huang et al. 1999; Table 2). The positions of all clone ends within the map are consistent with the marker content of each clone.
Figure 5.
Southern blot analysis of BamHI- and EcoRI-digested BAC DNA identifies six copies of Naip exon 11 and five copies of Naip exon 3. Correlation of the restriction fragments with specific Naip loci was done by comparison of the bands on the BAC blot with predicted fragments from genomic sequence and our previous physical map of the 129 Naip array (data not shown) (Growney et al. 2000). Horizontal bars and numbers indicate position and size (kb) of fragments in 1-kb ladder molecular weight marker (GIBCO). (A) BamHI-digested DNA probed with Naip exon 11 identifies six Naip exon 11 loci. 129 haplotype genomic sequence predicts the observed 14.3-kb fragment mapping to Naip1, a 9-kb fragment mapping to Naip2, an 8.6-kb fragment mapping to Naip5, and a doublet of 3.5 kb and 3.6 kb mapping to Naip6 and Naip3. The remaining 2.2-kb band maps to ΔNaip, as observed in the 129 haplotype (data not shown). Asterisk indicates vector junction fragments mapping to Naip1 and Naip2. (B) EcoRI-digested DNA probed with Naip exon 3 identifies five Naip exon 3 loci. 129 haplotype genomic sequence predicts the observed 10.2-kb fragment mapping to Naip5, an 8.5-kb fragment mapping to Naip2, a 7.5-kb fragment mapping to Naip1, a 7.2-kb fragment mapping to Naip6, and 2.1-kb mapping to Naip3.
Taken together, this data identifies six Naip loci in C57BL/6J. Beginning at the proximal edge of the array, these loci are Naip2, Naip5, ΔNaip, Naip6, Naip3, and Naip1 (Fig. 6). We are also now able to definitively map all known Naip cDNAs to specific Naip loci (Table 3). Our completed map, together with our completed genomic sequence of the proximal and distal aspects of the Naip array, enables us to estimate the size of the Naip array in the C57BL/6J haplotype from Naip2 to Naip1 to be 313 kb, while it is ca. 471 kb in strains of the 129 lineage because of the presence of two additional 79-kb repeat subunits, each containing one complete Naip gene and one ΔNaip locus.
Figure 6.
Complete physical map of the Lgn1 region in the C57BL/6J haplotype identifies five complete Naip genes and one 5′ truncated Naip locus. A physical map is indicated that has been constructed by ordering the clones and loci based on marker content of D13Die7, D13Die25, D13Die35, and D13Die36 (Fig. 5B–E) and several indicated single-copy markers (data not shown; Table 1). Naip gene content was determined by amplification with D13Die31 (Fig. 4A), D13Die32, D13Die33, and D13Die30 (data not shown), as well as Southern blot analysis with Naip exon 3 and Naip exon 11 (Fig. 5). Distal mouse chromosome 13 is indicated by a single horizontal line at the bottom. Genomic clones are indicated by the named horizontal lines. The relative positions of two sequenced 129 BAC clones, 149m19 (AF131205) and 26f17 (AF242431, AF242432) , are indicated with bold horizontal lines. Colored arrows indicate the position and orientation of genes present in the interval. Single-copy marker content is indicated by black circles. Markers within Naip loci are indicated with colored circles. Naip exon 3 and Naip exon 11 loci are indicated by colored circles with black trim. D13Die7 is indicated with filled colored triangles. The position of the single-copy D13Die30 is indicated with black squares. The map is drawn approximately to scale, based on genomic sequence and estimated sizes of C57BL/6J BAC clones. The direction of transcription of Ocln is unknown. The 3′ portion of Tfnr has not been mapped and is indicated with a dashed arrow. RFLP data suggests that C57BL/6J Naip6 has elements of 129 Naip6 and Naip7 (data not shown). Because microsatellite marker data and sequence analysis suggests that these two loci are very similar (Growney et al. 2000; Endrizzi et al. 2000), we designated this locus Naip6 to simplify nomenclature. Accordingly, we have designated specific loci of repeat markers to be consistent with their relative position in the 129 haplotype (for example, D13Die25; see Methods).
Table 3.
Mouse Naip Nomenclature
| Naip locusa | Accession No. of cDNAsb |
|---|---|
| Naip2c | AF135490 |
| AF102871 | |
| Naip5c | AF135492d |
| AF135493d | |
| Naip6 | AF135494 |
| Naip3 | NAe |
| Naip1 | AF135491 |
| AF007769 |
Naip nomenclature previously defined (Yaraghi et al. 1998).
cDNA sequences were reported in the following papers: Yaraghi et al. 1998; Yaraghi et al. 1999; and Huang et al. 1999.
Included within Lgn1 critical interval.
Alternative splice variants of this locus. We have previously reported two Naip5 cDNA clones (clones 11 and 22) that contain a single base insertion within Naip5 exon 11 resulting in a frameshift (Huang et al. 1999). Direct sequencing of Naip5 exon 11 PCR products derived from genomic DNA indicate that the two cDNA clones containing frameshift mutations are artifacts of cloning (data not shown).
We have not identified a cDNA clone corresponding to this locus (Endrizzi et al. 2000).
Improved Genetic Mapping of Lgn1
Our genomic mapping data (Figs. 1, 2, 3; unpublished observations) demonstrate that a permissive strain (A/J) and a nonpermissive strain (C57BL/6J) have similar gene arrangements and copy numbers within the Naip array. This indicates that differences in Naip gene copy number are not responsible for the Lgn1 phenotypic differences observed in our A/J and C57BL/6J cross. Therefore, the Lgn1 mutation is unlikely to result from a gross alteration of the structure of the Naip array, at least for these two strains. This suggests that it should be possible to narrow the genetic interval of Lgn1 to a small subset of genes.
We previously reported a 466 animal backcross ([A/J × C57BL/6J] × A/J), which was used to map Lgn1 to chromosome 13 (Dietrich et al. 1995; Scharf et al. 1996). Five animals from this backcross are recombinant across the remaining Lgn1 interval (between D13Die26 and D13Die3). Therefore, we used our completed map of the C57BL/6J Naip array to finely map the recombination breakpoints in these animals relative to the position of the genes in the interval.
We previously isolated several markers from the central Naip array (e.g., D13Die7) that amplify one or more loci that recombine on the distal side of Lgn1. However, up to this point, we have not been able precisely to precisely physically map these recombinant loci relative to the genes in the Naip array (Scharf et al. 1996; Endrizzi et al. 1999; Growney et al. 2000). The C57BL/6J map allowed us to physically localize each recombinant locus in the Naip array. In particular, analysis of D13Die30 for single-nucleotide polymorphisms allowed us to very narrowly define the Lgn1 critical interval (Fig. 7). This new critical interval only contains the Naip2, Naip5, and 3′ portion of ΔNaip genes. In addition, our genetic and physical map analysis allowed us to define two small regions within the Lgn1 interval that contain all five recombination events from our cross. The recombination event for three animals occurs within a 17.1-kb interval flanked by D13Die26 and D13Die37, located within Naip2, spanning exons 9–12. The breakpoint for the remaining two recombinant animals lies within a 17.7-kb interval between D13Die30 and D13Die36-A, located within ΔNaip, spanning exons 7–14. Further refinement of the recombination break points will not exclude additional Naip loci from the Lgn1 critical interval with our current 466 animal backcross.
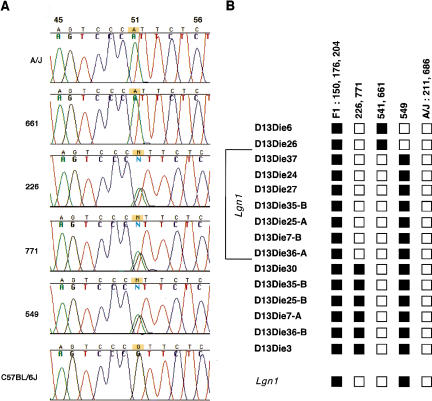
Figure 7.
Fine genetic mapping of Lgn1. (A) Sequence chromatograms of PCR products amplified with D13Die30.2 from genomic DNA of A/J, C57BL/6J, and selected backcross animals. Extensive sequencing of D13Die30.2 PCR products from A/J and C57BL/6J genomic DNA (see Methods) detected a single nucleotide polymorphism at position 51 in the 1479 bp corresponding to D13Die30 (A/J, A; C57BL/6J, G). D13Die30.2 PCR products from 10 backcross animals were sequenced with D13Die30-F and Die30-seq3. These animals include two A/J-like controls (211, 686), three heterozygous controls (150, 176, and 204) and five animals previously identified as recombinants within the Naip array (226, 771, 541, 549, and 661). Partial sequence tracings using primer D13Die30-F spanning the SNP at position 51 in D13Die30 from selected animals are shown. Heterozygous animals 226, 771, and 549 have two equivalent peaks at this position. (B) Polymorphic markers spanning the Naip array were assayed on genomic DNA from 10 animals in our Lgn1 cross (see Methods). Marker positions are indicated as ordered in the C57BL/6J haplotype (Fig. 6). Black squares indicate heterozygous loci, open circles indicate A/J-like homozygous loci. Three recombination events (541, 661, and 549) were mapped between D13Die26 and D13Die37, located within Naip2. Two recombination events (226 and 771) were mapped between D13Die30 and D13Die36-A, located within ΔNaip. The genetic interval for Lgn1, including the 5′ portion of Naip2, all of Naip5, and the 3′ portion of ΔNaip are indicated. The Lgn1 phenotype is indicated at the bottom (C57BL/6J nonpermissive; A/J permissive).
DISCUSSION
The Lgn1/SMA region of the mouse and human genomes continues to be a rich locus for comparative genomic studies. Previous comparative genomic mapping of this region has identified differences in the structure of the amplified segments present in the mouse and human genomes. Specifically, the human SMA disease interval on chromosome 5 in most individuals contains a large (∼500 kb) inverted duplication with each subunit of the duplication containing at least one copy of SMN, NAIP, GTF2H2 (previously known as BTF2P44), and SERF1 (previously known as H4F5; Melki et al. 1994; Roy et al. 1995; Lefebvre et al. 1995; Burglen et al. 1997; Carter et al. 1997; Scharf et al. 1998, Growney et al. 2000). The mouse genome, however, contains only a single copy of Smn, Serf1, and Gtf2h2 (Scharf et al. 1996; Viollet et al. 1997; Schrank et al. 1997; Bergin et al. 1997; DiDonato et al. 1997, 1999). In contrast to human, the mouse Lgn1 interval contains a direct repeat of six to ten complete and partial copies of Naip (Scharf et al. 1996; Huang et al. 1999). While the differences in structure have so far precluded the identification of common unstable elements, such studies have been useful for identifying candidate phenotypic modifying genes for SMA (Scharf et al. 1998). Here, our comparative genomic analysis has allowed us to identify significant differences in the structure of the Naip array among commonly used inbred mouse strains and to refine the Lgn1 genetic interval.
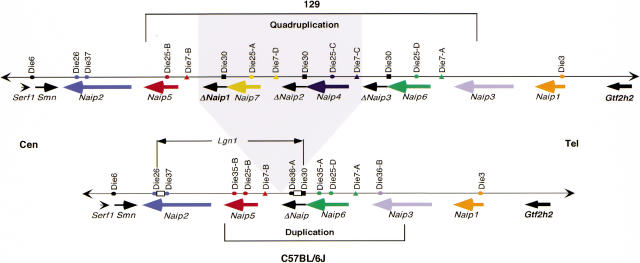
Our studies of the Naip array employing microsatellite marker analysis and hybridization studies suggest that several features of the Naip array are conserved among inbred mouse strains (Fig 8). Notably, Naip2 and Naip1 form the proximal and distal aspects of the array, respectively. These loci flank a centrally located repeat of more closely related Naip loci. Naip5 and a ΔNaip form the proximal side of the repeat, while Naip6 and Naip3 form the distal end of the repeat. However, 129 strains contain four additional Naip loci, namely ΔNaip1, Naip7, ΔNaip2, and Naip4 between Naip5 and ΔNaip3. In Figure 8, we have indicated that Naip7, Naip4, and two ΔNaip loci have been inserted between Naip5 and the single ΔNaip locus present in the C57BL/6J genome. However, our designation of Naip6 in the C57BL/6J haplotype is arbitrary and is done to simplify nomenclature of genes and marker loci. The insertions resulting in the current Naip array within the 129 haplotype may have occurred on the proximal side of what we have designated Naip6 in the C57BL/6J haplotype or in any position between Naip3 and Naip5 (Fig. 8). Future genomic sequencing of Naip6 from C57BL/6J may determine if it is more closely related to Naip7, Naip4, or Naip6 in the 129 haplotype.
Figure 8.
Comparative map of Naip array in mice of the 129 and C57BL/6J haplotypes. A summary of physical maps constructed in two different inbred mouse strains is indicated. Conserved gene loci are indicated by common colors. Repeat microsatellite markers identify a quadruplication in mice of the 129 lineage and a duplication in the C57BL/6J haplotype. Gray shading indicates the two complete and two partial Naip loci specific to the 129 haplotype. A hypothetical insertion point between Naip5 and ΔNaip in the C57BL/6J haplotype is indicated (see Discussion). The genetically mapped Lgn1 interval is indicated in the C57BL/6J haplotype. Two open boxes indicate the positions of two recombination hot spots identified in our Lgn1 backcross.
Our complete map allows us to definitively identify the transcriptionally active Naip loci and correlate them with previously identified Naip cDNAs. We have not identified any cDNAs mapping to the Naip3 locus, which genomic sequencing suggests is likely to be nonfunctional (Endrizzi et al. 2000). Thus, we have mapped all identified Naip cDNAs to four loci (Table 3; Yaraghi et al. 1998, 1999; Huang et al. 1999). Our cDNA clones indicate that Naip6 transcription extends through ΔNaip exons 7 and 8 and then is polyadenylated. For that reason, the ΔNaip locus likely does not encode a functional transcript. While the Naip6 locus has been shown to be transcriptionally active, no information concerning the transcription of Naip7 or Naip4 in the 129 haplotype is available. It is interesting to speculate that amplification of the Naip6 locus may indicate a selection for specific functions of this Naip locus.
A principal goal in mapping this region was to identify the physical location of mapped polymorphic markers from the interval to improve the resolution of our genetic mapping of Lgn1. As a result of our work, the current critical region for Lgn1 has been shortened to the interval from D13Die26 to D13Die30. This critical region includes Naip2, Naip5, and the 3′ portion of ΔNaip. Because ΔNaip is unlikely to be functional, Naip2 and Naip5 are the remaining candidates for Lgn1. This is further supported by our findings that mouse strains 129S3 and 129P3, which contain additional copies of Naip6 and ΔNaip, are permissive for Legionella pneumophila replication, while F1 progeny of these strains with C57BL/6J are nonpermissive (data not shown).
It is difficult to speculate about which of the two remaining candidate genes is the most likely to contain the Lgn1 mutation. Both Naip2 and Naip5 are expressed in macrophages (Huang et al. 1999). Because so little is known about the molecular function of the Naip proteins, even the presence of considerable variation in the amino acid sequences of the different Naip products do not allow for confident predictions of which gene will encode the unique function implicated by the genetics.
Recent studies provide a preliminary analysis of the expression profiles of the various Naips. Consistent with our exclusion of Naip1 from the Lgn1 critical interval, Naip1 has been shown to be more highly expressed in the brain than in the macrophage-rich spleen (Yaraghi et al. 1999). Furthermore, Naip1 has been shown to play a role in hippocampus cell survival following limbic seizures (Holcik et al. 2000). However, Naip1 expression is not limited to the brain, as transcripts corresponding to this locus have been identified from a macrophage cDNA library (Huang et al. 1999). In contrast, Naip2 has been shown to be highly expressed in spleen and bone marrow macrophages (Huang et al. 1999; Yaraghi et al. 1999). While cDNA cloning suggests that Naip2 is likely the most strongly expressed Naip locus in macrophages, data for the expression of Naip5 is not currently available.
Differences in the relative expression of Naip between A/J- (permissive) and C57BL/6J- (nonpermissive) derived peritoneal macrophages have recently been identified (Diez et al. 2000). C57BL/6J-derived macrophages expressed stronger levels of Naip than A/J-derived macrophages, suggesting differences in transcription or mRNA stability as a possible cause of the Lgn1 phenotype. However, the levels of Naip increased in both strains in response to L. pneumophila infection. Furthermore, this upregulation of Naip expression was not specific to L. pneumophila infection, as infection with Salmonella typhimurium and feeding with latex beads also induced this response. Unfortunately, these studies were performed with probes that do not distinguish between specific Naip loci, making it impossible to tell if the upregulation is from Naip2, Naip5, or some other Naip gene unrelated to the Lgn1 phenotype. Our complete physical map and precise mapping of Naip cDNAs will allow us to generate probes likely to be specific for individual Naip loci. These studies are currently underway in our laboratory.
Our data identify the 3′ portion of Naip as a variable region of this gene family that may contribute to the instability of the Naip array. Interestingly, analysis of recombinant animals in our ([A/J × C57BL/6J] × A/J) backcross employed to map Lgn1 has identified two recombination hotspots within the Naip array. Of five animals with recombination events within the Naip interval, three recombination events occur within 17.1-kb interval between D13Die26 and D13Die37 in Naip2, while the remaining recombinations take place within a 17.7-kb interval between D13Die30 and D13Die36-A in ΔNaip. Both intervals map to the 3′ portion of Naip genes. While we have not identified any particular sequence elements in these regions that suggest a mechanism for the high frequency of recombination in these intervals, several interesting observations can be made. First, this position correlates with the likely position of an ancient amplification that may have given rise to Naip5 and ΔNaip, as well as a potential breakpoint for recombination resulting in the more recent expansion of the array in the 129 haplotype. This suggests misalignment of homologous sequences, for example, between Naip3 on one strand and ΔNaip on the other, followed by subsequent unequal recombination as a possible mechanism resulting in the expansion of the array. Second, the BIR domains are encoded by Naip exons 3–9, suggesting that these hotspots also correspond to the position within Naip where it diverges from other members of the IAP family of proteins (Huang et al. 1999). Third, the junction between the conserved BIR domains and the divergent carboxyl terminus of Naip appears to be a variable region within this family of proteins. Specifically, Naip exon 10 is absent from Naip3 and the ΔNaips, while Naip2, Naip3, and ΔNaip contain a unique 150-bp extension at the 5′ end of exon 11. Finally, the identification of a human NAIP cDNA containing portions of OCLN within the 3′ UTR suggest a genomic deletion within the 3′ end of NAIP resulting in the fusion of NAIP and OCLN. These observations suggest that the 3′ portion of NAIP may play a role in the genomic instability resulting in the generation of SMA alleles in humans.
Our mapping studies that identify Naip2 and Naip5 as the remaining Lgn1 candidate genes suggest several future lines of study. Continued comparative genomics likely will entail sequencing of the Lgn1 critical interval from A/J and C57BL/6J to identify candidate mutations. While several C57BL/6J-derived BAC clones identified in this study will facilitate analysis of this haplotype, our detailed physical maps of the region will enable a PCR-based sequence analysis of the Lgn1 critical interval in the A/J haplotype, for which no genomic clones are currently available.
Assuming that the permissiveness of A/J strains to L. pneumophila replication is not due to a haploinsufficient function that is lacking in the C57BL/6J haplotype, functional complementation of the permissive phenotype using genomic (this article) or cDNA (Huang et al. 1999) clones that we have identified remains a tantalizing possibility. However, these approaches have proven technically challenging because of the difficulty of using A/J as a background for transgenic embryos and because of the refractory nature of myeloid cells to transfection (data not shown; Chisholm and Symonds 1988; Rupprecht and Coleman 1991; Wright and Farber 1991; Stacey et al. 1993; Nicolet and Paulnock 1994). The use of a substitute permissive transgenic background or the use of viral vectors may facilitate future analysis of Naip2 and Naip5 by functional complementation (Strair et al. 1990; Haddada et al. 1993).
METHODS
BAC/P1 Clones
All of the 129-haplotype-derived BAC and P1 clones have been previously described (Diez et al. 1997; Yaraghi et al. 1998; Growney et al. 2000). C57BL/6J clones derive from two BAC libraries. Clones 24907 and 235 were identified by Genome Systems. Briefly, library pools were screened by PCR with primers for Naip exon 3. Candidate clones were confirmed by PCR and Southern blot analysis. All remaining C57BL/6J BAC clones derive from the RPCI-23 C57BL/6J BAC library (http://bacpac.med.buffalo.edu). Briefly, six high-density filter arrays were screened by Southern blot analysis with a Naip exon 11 (D13Die34) probe as per the manufacturer's instructions. Twenty-five candidate clones were identified and confirmed by PCR with D13Die34. Nineteen positive clones were chosen for further study (Table 2).
Markers Used in This Study
Primers used in PCR assays to amplify all STSs (sequence tagged sites) in this study are indicated in Table 1. Novel PCR assays were developed as follows. First, STS D13Die31 was developed to amplify all intron 13 loci type I. Primers were chosen to reside in conserved regions of Naip exons 13 and 14 based on the sequence of BAC 149m19 (accession AF131205), BAC 26f17 (accession AF242431 and AF242432), and P1 9045 (accession AF242433, AF242434, and AF242435; Endrizzi et al. 1999, 2000). D13Die32 and D13Die33 were devised to amplify the distal and proximal borders of Naip intron 13 type II, based on the sequence of BAC 26f17.
Second, microsatellite markers D13Die35, D13Die36, and D13Die37 were designed to amplify novel microsatellites identified from the sequence of BACs 149m19 (AF131205) and 26f17 (AF242431, AF242432).
Third, STSs D13Die38–D13Die46 were developed from the end sequence of selected C57BL/6J BAC clones that were not homologous to any sequence in 149m19 (AF131205) or 26f17 (AF242431, AF242432).
Finally, for D13Die30.2, novel primers that flank and encompass locus D13Die30 were developed to improve amplification of this locus.
PCR
Except where noted, all STSs were amplified as previously described (Growney et al. 2000). All amplifications were performed in an MJ Research thermal cycler (PTC-225). Amplifications of BAC clones employed 5μL of a bacterial suspension. Amplifications of genomic DNA employed 5–25 ng of DNA. Primers for microsatellite markers were end labeled with (γ32P)-ATP, 6000 Ci/mmol (NEN), and PCR products amplified as previously described (Dietrich et al. 1992). Amplification products were resolved on 6% denaturing polyacrylamide gel (National Diagnostics) at 120 W for 2–3 hr.
D13Die30.2 was amplified using Expand Long Template PCR System (Boehringer, Mannheim). Briefly, 25 ng of genomic DNA was amplified in a 50-μL reaction containing 300 nM each of forward and reverse primer, 350 μM dNTPs in 1× buffer system 1. Reactions were heated to 94°C for 2 min and cooled to 88°C before 0.75 μL of Expand enzyme mix was added. After addition of enzyme, 10 cycles of 94°C for 10 s, 65°C for 30 s, and 68°C for 10 min was performed, followed by 20 cycles of 94°C for 10 s, 65°C for 30 s, and 68°C for 10 min + 20 s/cycle. A final extension at 68°C for 7 min was performed. PCR products were resolved in a 1% SeaPlaque agarose (FMC) gel and purified using QIAquick Gel Extraction Kit (Qiagen), following the manufacturer's directions.
Genomic DNAs
Mouse genomic DNAs for A/J (stock number JR0646), C57BL/6J (JR0664), and 129 strains (as previously described) were obtained from Jackson Labs (Growney et al. 2000). Where applicable, revised strain nomenclature is presented (Festing et al. 1999). Genomic DNA for ES cell line CJ7 was a gift from E-Chiang Lee (Mammalian Genetics Laboratory, NCI-FCRDC, Fort Detrick, Frederick, Md.).
Genomic DNA from animals in our backcross was prepared using a high–molecular weight DNA prep method (Strauss 1998). Briefly, 10 mL of ice-cold PBS was added to one-quarter of a frozen (−80°C) mouse liver in a 50-mL conical tube. Tissue was homogenized for 20 s with a Polytron (PT 1200) tissue homogenizer (Kinematica AG) on setting 5. Homogenates were centrifuged at 1,000 rpm at 4°C for 10 min in a Beckman GS-6KR rotor. The supernatant was aspirated, and the nuclear pellet was resuspended in 4.5 mL of ice-cold STE (10 mM Tris-Cl pH 7.5 : 10 mM NaCl : 1 mM EDTA). To each sample, 25 μL of 20 mg/ml proteinase K, 25 μL of 10 mg/mL RNAse A, and 125 μL 10% SDS was added. Samples were incubated at 50°C for 15 hr, followed by two extractions with phenol : chloroform : isoamyl alcohol (25 : 24 : 1; Boehringer Mannheim). The aqueous phase was dialyzed against TE pH 8.0 at 4°C with two buffer changes over 24 hr.
BAC DNA for Southern blot analysis was prepared by a modified PEG prep method as previously described (Growney et al. 2000). BAC DNA for Pulsed Field Gel mapping was prepared using the Large-Construct Kit (Qiagen) as per the manufacturer's directions.
Pulsed Field Gel Mapping
150 ng of BAC DNA was digested overnight with NotI. Fragments were resolved in 1% LE agarose (FMC)/0.5 × TBE at 4°C with a BioRad Chef Mapper. Samples were run into the gel using the following Chef Mapper settings: forward gradient = 6.0 V/cm (180° FIGE), initial switch time = 20 min, final switch time = 20 min, reverse gradient = 0, initial switch time = 20 min, final switch time = 20 min, total run time = 20 min, ramp = linear. Fragments 3–100 kb were resolved with the following settings: run time = 15 hr 16 min, voltage gradient = 6.0 V/cm (two state), initial switch time = 0.22 s, final switch time = 8.53 s, angle = 120°, ramp = linear. Fragments 72–250 kb were resolved with the following settings. Run time = 30 hr 4 min; voltage gradient = 6.0 V/cm (two state); initial switch time = 9.29 s; final switch time = 21.79 s, angle = 120°, ramp = linear.
Sequencing
PCR Products
Purified D13Die30.2 PCR products were sequenced with ABI dye terminator chemistry (ABI prism DNA sequencing kit, PE Applied BioSystems). We sequenced 1,479 bp of the 2,904 bp D13Die30.2 PCR product corresponding to D13Die30 from A/J and C57BL/6J DNA with five primers, D13Die30-F, D13Die30-R, D13Die30-seq2 (GCTGGACACTAAAGGCACTATG), D13Die30-seq3 (GCGAAGAAGCTGTCGTTG), and D13Die30-seq4 (CATGCATGCATGTGCAAG). We amplified 290 ng of purified DNA in a 20 μL reaction containing 4 μL Big Dye, 20 pM primer and 2 μL 5× CSA SEQ Buffer (Perkin Elmer). Products were amplified with 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Reactions were resolved on a Perkin Elmer 3700 Capillary Sequencer. Sequences were imported into Sequencher (Gene Codes) for analysis.
BAC Ends
The SP6 and T7 ends of purified BAC DNA were sequenced using ABI dye terminator chemistry (ABI prism DNA sequencing kit, PE Applied BioSystems). 1–2 μg of PEG prep BAC DNA was amplified in a 40-μL reaction containing 20 pM SP6 or T7 primer, 8 μL Big dye, and 4 μL 5× CSA SEQ Buffer (Perkin Elmer). Following an initial denaturation at 95°C for 5 min, products were amplified with 50 cycles of 95°C for 30 s, 55°C for 20 s, and 60°C for 4 min. The entire reaction was loaded onto a Perkin Elmer 377 slab gel sequencer.
Southern Blotting
Naip exon 3 and exon 11 probes were as previously described (Growney et al. 2000). An STS for Smn exon 4-intron 4 was amplified from A/J genomic DNA and labeled as described.
Genomic Blots
In a 0.7 % SeaKem LE agarose (FMC) gel, 10 μg of total genomic DNA (Jackson Labs) was resolved after having been digested overnight with BamHI (NEB). DNAs were transferred to Gene Screen Plus hybridization membrane (NEN) and hybridized with 1 × 106-cpm probe/mL overnight at 68°C in Church's Buffer (Church and Gilbert 1984; Growney et al. 2000).
BAC Blots
In 0.7% SeaKem LE (FMC) agarose gel, fragments of 5 μg of BAC DNA that had been digested overnight with EcoRI or BamHI (NEB) were resolved for 20 hours at 1.2 volts/cm. Blots were performed as previously described (Growney et al. 2000). Blots were imaged on a Storm 860 Phosphoimager (Molecular Dynamics).
Nomenclature
Repetitive microsatellite loci within the Naip array were originally named based on the size of PCR products, from largest to smallest, as amplified from 129 genomic DNA (Growney et al. 2000). C57BL/6J loci names correlate to 129 loci based on the finding that Naip7 and Naip4 are not present in this haplotype (see Results). For example, Naip5 in the physical maps for C57BL/6J and 129 presented here and elsewhere contains D13Die25-B and is flanked by D13Die7-B, while Naip6 in both strains contains D13Die25-D and is flanked by D13Die7-A. Exceptions include markers D13Die35 and D13Die36, which have not been tested in 129 strains and are thus labeled by size in C57BL/6J.
Acknowledgments
We thank Matthew Endrizzi and Vey Hadinoto for technical assistance with sequencing described in this work. We thank Victor Boyartchuk and James Watters for critical reading of this manuscript. This work was partially supported by a grant to W.F.D. from the Muscular Dystrophy Association. W.F.D. is an assistant investigator at Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dietrich@rascal.med.harvard.edu; FAX (617)432-3993.
REFERENCES
- Beckers M C, Yoshida S, Morgan K, Skamene E, Gros P. Natural resistance to infection with Legionella pneumophila: chromosomal localization of the Lgn1 susceptibility gene. Mamm Genome. 1995;6:540–545. doi: 10.1007/BF00356173. [DOI] [PubMed] [Google Scholar]
- Bergin A, Kim G, Price D L, Sisodia S S, Lee M K, Rabin B A. Identification and characterization of a mouse homologue of the spinal muscular atrophy–determining gene, survival motor neuron. Gene. 1997;204:47–53. doi: 10.1016/s0378-1119(97)00510-6. [DOI] [PubMed] [Google Scholar]
- Burglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, Munnich A, Melki J. Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996;32:479–482. doi: 10.1006/geno.1996.0147. [DOI] [PubMed] [Google Scholar]
- Burglen L, Seroz T, Miniou P, Lefebvre S, Burlet P, Munnich A, Pequignot E V, Egly J M, Melki J. The gene encoding p44, a subunit of the transcription factor TFIIH, is involved in large-scale deletions associated with Werdnig-Hoffmann disease. Am J Hum Genet. 1997;60:72–79. [PMC free article] [PubMed] [Google Scholar]
- Burlet P, Burglen L, Clermont O, Lefebvre S, Viollet L, Munnich A, Melki J. Large scale deletions of the 5q13 region are specific to Werdnig- Hoffmann disease. J Med Genet. 1996;33:281–283. doi: 10.1136/jmg.33.4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpten J D, DiDonato C J, Ingraham S E, Wagner-McPherson C, Nieuwenhuijsen B W, Wasmuth J J, Burghes A H. A YAC contig of the region containing the spinal muscular atrophy gene (SMA): identification of an unstable region. Genomics. 1994;24:351–356. doi: 10.1006/geno.1994.1626. [DOI] [PubMed] [Google Scholar]
- Carter T A, Bonnemann C G, Wang C H, Obici S, Parano E, De Fatima Bonaldo M, Ross B M, Penchaszadeh G K, Mackenzie A, Soares M B, Kunkel L M, Gilliam T C. A multicopy transcription-repair gene, BTF2p44, maps to the SMA region and demonstrates SMA associated deletions. Hum Mol Genet. 1997;6:229–236. doi: 10.1093/hmg/6.2.229. [DOI] [PubMed] [Google Scholar]
- Chang J G, Jong Y J, Lin S P, Soong B W, Tsai C H, Yang T Y, Chang C P, Wang W S. Molecular analysis of survival motor neuron (SMN) and neuronal apoptosis inhibitory protein (NAIP) genes of spinal muscular atrophy patients and their parents. Hum Genet. 1997;100:577–581. doi: 10.1007/s004390050555. [DOI] [PubMed] [Google Scholar]
- Chisholm O, Symonds G. Transfection of myeloid cell lines using polybrene/DMSO. Nucleic Acids Res. 1988;16:2352. doi: 10.1093/nar/16.5.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato C J, Brun T, Simard L R. Complete nucleotide sequence, genomic organization, and promoter analysis of the murine survival motor neuron gene (Smn) Mamm Genome. 1999;10:638–641. doi: 10.1007/s003359901061. [DOI] [PubMed] [Google Scholar]
- DiDonato C J, Chen X N, Noya D, Korenberg J R, Nadeau J H, Simard L R. Cloning, characterization, and copy number of the murine survival motor neuron gene: homolog of the spinal muscular atrophy–determining gene. Genome Res. 1997;7:339–352. doi: 10.1101/gr.7.4.339. [DOI] [PubMed] [Google Scholar]
- DiDonato C J, Morgan K, Carpten J D, Fuerst P, Ingraham S E, Prescott G, McPherson J D, Wirth B, Zerres K, Hurko O, et al. Association between Ag1-CA alleles and severity of autosomal recessive proximal spinal muscular atrophy. Am J Hum Genet. 1994;55:1218–1229. [PMC free article] [PubMed] [Google Scholar]
- Dietrich W, Katz H, Lincoln S E, Shin H S, Friedman J, Dracopoli N C, Lander E S. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W F, Damron D M, Isberg R R, Lander E S, Swanson M S. Lgn1, a gene that determines susceptibility to Legionella pneumophila, maps to mouse chromosome 13. Genomics. 1995;26:443–450. doi: 10.1016/0888-7543(95)80161-e. [DOI] [PubMed] [Google Scholar]
- Dietrich W F, Miller J, Steen R, Merchant M A, Damron-Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O'Connor T J, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Dietrich W F, Miller J C, Steen R G, Merchant M, Damron D, Nahf R, Gross A, Joyce D C, Wessel M, Dredge R D, et al. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- Diez E, Beckers M C, Ernst E, DiDonato C J, Simard L R, Morissette C, Gervais F, Yoshida S I, Gros P. Genetic and physical mapping of the mouse host resistance locus Lgn1. Mamm Genome. 1997;8:682–685. doi: 10.1007/s003359900536. [DOI] [PubMed] [Google Scholar]
- Diez E, Yaraghi Z, MacKenzie A, Gros P. The neuronal apoptosis inhibitory protein (Naip) is expressed in macrophages and is modulated after phagocytosis and during intracellular infection with Legionella pneumophila. J Immunol. 2000;164:1470–1477. doi: 10.4049/jimmunol.164.3.1470. [DOI] [PubMed] [Google Scholar]
- Endrizzi M, Huang S, Scharf J M, Kelter A-R, Wirth B, Kunkel L M, Miller W, Dietrich W F. Comparative sequence analysis of the mouse and human Lgn1/SMA interval. Genomics. 1999;60:137–151. doi: 10.1006/geno.1999.5910. [DOI] [PubMed] [Google Scholar]
- Endrizzi, M. G., Hadinoto, V., Growney, J. D., Scharf, J. M., Miller, W., and Dietrich, W. F. 2000. Genomic sequence analysis of the nouse Naip gene array. Genome Res.. (This issue). [DOI] [PMC free article] [PubMed]
- Festing M F, Simpson E M, Davisson M T, Mobraaten L E. Revised nomenclature for strain 129 mice. Mamm Genome. 1999;10:836. doi: 10.1007/s003359901099. [DOI] [PubMed] [Google Scholar]
- Gilliam T C, Brzustowicz L M, Castilla L H, Lehner T, Penchaszadeh G K, Daniels R J, Byth B C, Knowles J, Hislop J E, Shapira Y, et al. Genetic homogeneity between acute and chronic forms of spinal muscular atrophy. Nature. 1990;345:823–825. doi: 10.1038/345823a0. [DOI] [PubMed] [Google Scholar]
- Growney J D, Scharf J M, Kunkel L M, Dietrich W F. Evolutionary divergence of the mouse and human Lgn1/SMA repeat structures. Genomics. 2000;64:62–81. doi: 10.1006/geno.1999.6111. [DOI] [PubMed] [Google Scholar]
- Haddada H, Lopez M, Martinache C, Ragot T, Abina M A, Perricaudet M. Efficient adenovirus-mediated gene transfer into human blood monocyte-derived macrophages. Biochem Biophys Res Commun. 1993;195:1174–1183. doi: 10.1006/bbrc.1993.2168. [DOI] [PubMed] [Google Scholar]
- Holcik M, Thompson C S, Yaraghi Z, Lefebvre C A, MacKenzie A E, Korneluk R G. The hippocampal neurons of neuronal apoptosis inhibitory protein 1 (NAIP1)-deleted mice display increased vulnerability to kainic acid- induced injury. Proc Natl Acad Sci. 2000;97:2286–2290. doi: 10.1073/pnas.040469797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Scharf J M, Growney JD, G J, Endrizzi M G, Dietrich W F. The mouse Naip gene cluster on Chromosome 13 encodes several distinct functional transcripts. Mamm Genome. 1999;10:1032–1035. doi: 10.1007/s003359901155. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy–determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Melki J, Lefebvre S, Burglen L, Burlet P, Clermont O, Millasseau P, Reboullet S, Benichou B, Zeviani M, Le Paslier D, et al. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science. 1994;264:1474–1477. doi: 10.1126/science.7910982. [DOI] [PubMed] [Google Scholar]
- Morrison K E. Advances in SMA research: review of gene deletions. Neuromuscular Disorders. 1996;6:397–408. doi: 10.1016/s0960-8966(96)00368-9. [DOI] [PubMed] [Google Scholar]
- Nicolet C M, Paulnock D M. Promoter analysis of an interferon-inducible gene associated with macrophage activation. J Immunol. 1994;152:153–162. [PubMed] [Google Scholar]
- Rajcan-Separovic E, Mahadevan M S, Lefebvre C, Besner-Johnston A, Ikeda J E, Korneluk R G, MacKenzie A. FISH detection of chromosome polymorphism and deletions in the spinal muscular atrophy (SMA) region of 5q13. Cytogenet Cell Genet. 1996;75:243–247. doi: 10.1159/000134493. [DOI] [PubMed] [Google Scholar]
- Rodrigues N R, Owen N, Talbot K, Ignatius J, Dubowitz V, Davies K E. Deletions in the survival motor neuron gene on 5q13 in autosomal recessive spinal muscular atrophy. Hum Mol Genet. 1995;4:631–634. doi: 10.1093/hmg/4.4.631. [DOI] [PubMed] [Google Scholar]
- Roy N, Mahadevan M S, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- Rupprecht A P, Coleman D L. Transfection of adherent murine peritoneal macrophages with a reporter gene using DEAE-dextran. J Immunol Methods. 1991;144:157–163. doi: 10.1016/0022-1759(91)90082-q. [DOI] [PubMed] [Google Scholar]
- Scharf J M, Damron D, Frisella A, Bruno S, Beggs A H, Kunkel L M, Dietrich W F. The mouse region syntenic for human spinal muscular atrophy lies within the Lgn1 critical interval and contains multiple copies of Naip exon 5. Genomics. 1996;38:405–417. doi: 10.1006/geno.1996.0644. [DOI] [PubMed] [Google Scholar]
- Scharf J M, Endrizzi M G, Wetter A, Huang S, Thompson T G, Zerres K, Dietrich W F, Wirth B, Kunkel L M. Identification of a candidate modifying gene for spinal muscular atrophy by comparative genomics. Nat Genet. 1998;20:83–86. doi: 10.1038/1753. [DOI] [PubMed] [Google Scholar]
- Schrank B, Gotz R, Gunnersen J M, Ure J M, Toyka K V, Smith A G, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey K J, Ross I L, Hume D A. Electroporation and DNA-dependent cell death in murine macrophages. Immunol Cell Biol. 1993;71:75–85. doi: 10.1038/icb.1993.8. [DOI] [PubMed] [Google Scholar]
- Strair R K, Towle M, Smith B R. Retroviral mediated gene transfer into bone marrow progenitor cells: use of beta-galactosidase as a selectable marker. Nucleic Acids Res. 1990;18:4759–4762. doi: 10.1093/nar/18.16.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss W M. Preparation of genomic DNA from mammalian tissue. In: Ausubel F M, et al., editors. Current Protocols in Molecular Biology. Boston: Wiley; 1998. pp. 2.2.1–2.2.3. [DOI] [PubMed] [Google Scholar]
- Viollet L, Bertrandy S, Bueno Brunalti A L, Lefebvre S, Burlet P, Clermont O, Cruaud C, Guenet J L, Munnich A, Melki J. cDNA isolation, expression, and chromosomal localization of the mouse survival motor neuron gene (Smn) Genomics. 1997;40:185–188. doi: 10.1006/geno.1996.4551. [DOI] [PubMed] [Google Scholar]
- Wirth B, Hahnen E, Morgan K, DiDonato C J, Dadze A, Rudnik-Schoneborn S, Simard L R, Zerres K, Burghes A H. Allelic association and deletions in autosomal recessive proximal spinal muscular atrophy: association of marker genotype with disease severity and candidate cDNAs. Hum Mol Genet. 1995;4:1273–1284. doi: 10.1093/hmg/4.8.1273. [DOI] [PubMed] [Google Scholar]
- Wright T M, Farber J M. 5′ regulatory region of a novel cytokine gene mediates selective activation by interferon gamma. J Exp Med. 1991;173:417–422. doi: 10.1084/jem.173.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaraghi Z, Diez E, Gros P, MacKenzie A. cDNA cloning and the 5′ genomic organization of Naip2, a candidate gene for murine Legionella resistance. Mamm Genome. 1999;10:761–763. doi: 10.1007/s003359901087. [DOI] [PubMed] [Google Scholar]
- Yaraghi Z, Korneluk R G, MacKenzie A. Cloning and characterization of the multiple murine homologues of NAIP (neuronal apoptosis inhibitory protein) Genomics. 1998;51:107–113. doi: 10.1006/geno.1998.5378. [DOI] [PubMed] [Google Scholar]