Abstract
Escherichia coli O157:H7 (O157) causes human diarrheal disease and healthy cattle are its primary reservoir. O157 colonize the bovine epithelial mucosa at the recto-anal junction (RAJ). Previous studies show that O157 at this site are not eliminated by aggressive interventions including applications of O157-specific lytic bacteriophages and other bactericidal agents. We hypothesize that some O157 at the RAJ mucosa are protected from these killing agents by host cell internalization. To test this hypothesis, rectal biopsies from O157 culture positive and negative cattle were analyzed by fluorescent microscopy and subjected to gentamicin protection assays. GFP-labeled bacteria were found located deep within the tissue crypts and a small number of O157 were recovered from rectal biopsies after gentamicin treatment. Primary bovine rectal epithelial (PBRE) cell cultures were incubated with O157 and subjected to gentamicin protection assays. Strains ATCC 43895, 43894, Sakai, and WSU180 entered the PBRE cells with different levels of efficiency ranging from 0.18 to 19.38% of the inocula. Intracellular bacteria were confirmed to be within membrane-bounded vacuoles by electron microscopy. Cytochalasin D curtailed internalization of O157 indicating internalization was dependent on eukaryotic microfilament assembly. Strain ATCC 43895 exhibited the highest efficiency of internalization and survived for at least 24 h within PBRE cells. Deletion mutation of intimin or its receptor in ATCC 43895 did not reduce bacterial internalization. This strain produced more biofilm than the others tested. Retrospective analysis of cattle challenged with two O157 strains, showed ATCC 43895, the most efficient at host cell internalization, was most persistent.
Keywords: E. coli O157:H7, cattle, internalization, epithelial cells, EHEC, bovine
Introduction
Enterohemorrhagic Escherichia coli (EHEC) cause human disease ranging from self-limited watery diarrhea to the life-threatening condition of hemorrhagic colitis and its sequelae – the hemolytic uremic syndrome (HUS; Karmali et al., 1983; Paton and Paton, 1998). E. coli O157:H7 is the predominate EHEC serotype isolated from disease outbreaks in North America, the United Kingdom, and Japan (Griffin and Tauxe, 1991; Smith, 1998; Mead et al., 1999; Michino et al., 1999). Healthy cattle are the major reservoir for E. coli O157:H7, the most common source for human infections (Hancock et al., 1994; Zhao et al., 1995; Chapman et al., 2001), and carry the bacteria with no apparent disease symptoms (Blanco et al., 1996; Besser et al., 1997; Hancock et al., 1997). Previous studies demonstrate that the recto-anal junction (RAJ) mucosa is the primary site of E. coli O157:H7 colonization in cattle (Grauke et al., 2002; Naylor et al., 2003; Cobbold et al., 2007; Dean-Nystrom et al., 2008), and the carriage of E. coli O157:H7 at this site is correlated to high level fecal shedding (Low et al., 2005). Rectal administration of E. coli O157:H7 results in efficient bacterial colonization at the RAJ mucosa that is maintained for >1 month (Sheng et al., 2004, 2006b). Because the terminal rectal mucosa may be the ideal target for controlling E. coli O157:H7 in cattle, we and others have applied O157-specific lytic bacteriophages (Sheng et al., 2006a; Rivas et al., 2010) or chemical chlorhexidine (Naylor et al., 2007) directly to this location. However, when these treatments are effective they reduce the level of the E. coli O157:H7 carriage, but never completely eliminate E. coli O157:H7 from the rectal mucosa.
Enterohemorrhagic E. coli and enteropathogenic E. coli (EPEC) belong to the “attaching and effacing (A/E) pathogen” category because of their ability to induce A/E lesions on intestinal epithelial cells. The characteristics of the A/E lesions include the localized effacement of the brush border microvilli, intimate bacterial attachment to the host epithelium, and the formation of cytoskeleton-rich pedestal-like structures beneath the adherent bacteria (Moon et al., 1983). The A/E pathogens share a highly homologous mobile genetic element called the locus of enterocyte effacement (LEE) pathogenicity island that is responsible for A/E lesion formation (McDaniel and Kaper, 1997; Hueck, 1998; Elliott et al., 1999, 2000). The LEE encodes a type III secretion system (T3SS), secreted proteins, chaperone, and regulatory molecules, and an outer membrane adhesion protein called intimin (Perna et al., 1998; Elliott et al., 1999). The T3SS is responsible for translocating into infected cells both LEE- and non-LEE-encoded (Nle)-effectors, including translocated intimin receptor (Tir), EspA, EspB, and EspD (Kenny et al., 1997; Knutton et al., 1998; Kenny, 2001). Tir is inserted into the host cell plasma membrane with Tir–intimin interaction triggering signaling events leading to pedestal formation (Rosenshine et al., 1992a; Kenny et al., 1997). There are a number of studies that show the LEE encoding effector proteins play a role in ruminant intestinal colonization in vivo (Dean-Nystrom et al., 1998; Cornick et al., 2002; Dziva et al., 2004; Naylor et al., 2005; Sheng et al., 2006b; Vlisidou et al., 2006).
Enteropathogenic E. coli is usually considered an extracellular organism but under in vitro conditions bacteria can be seen intracellularly and this phenotype is dependent on a functional T3SS (Francis et al., 1991; Rosenshine et al., 1992a; Jepson et al., 2003). The effecter EspT facilitates EPEC internalization into non-phagocytic cells in a process involving Rac1 and Wave2 (Bulgin et al., 2009). In fact, in addition to the generation of intimate adherence and the A/E lesions, EHEC strains, like EPEC, are capable of efficient entry into a variety of epithelial cell types (Dibb-Fuller et al., 2001; Martinez and Hultgren, 2002; Uhlich et al., 2002, 2009; Abu-Ali et al., 2010), though they are also regarded as extracellular pathogens. Internalization of EHEC O157:H7 by human epithelial cells is also known (Oelschlaeger et al., 1994) and Dibb-Fuller et al. (2001) demonstrate E. coli O157:H7 is internalized in several distinct bovine cell lines. The interaction of E. coli O157:H7 with bovine host cells and the mechanism(s) that lead to the specific tropism and persistence at the rectal mucosa are not fully understood. We speculated that the bovine epithelial cells at the terminal rectal mucosa may internalize a subpopulation of E. coli O157:H7 and these intracellular bacteria may contribute to the persistence of E. coli O157:H7 at this colonization site. The aims of this study were to (1) describe E. coli O157:H7 in biopsy tissue taken from E. coli O157:H7 culture positive cattle; and (2) investigate the internalization of E. coli O157:H7 using primary bovine rectal epithelial (PBRE) cells.
Materials and Methods
Bacteria and culture
All strains used in this study are listed in Table 1. The clinical isolates were chosen because they are well characterized human pathogens and two have full DNA sequence known (ATCC 43895, also referred to as EDL933, and Sakai). All have been used previously by our laboratory and others in cattle trails of E. coli O157:H7 carriage. E. coli O157:H7 and other E. coli strains were grown in Luria–Bertani (LB) broth or agar. Recombinant E. coli O157:H7 43894 GFPuv was grown in LB with 100 μg/ml ampicillin.
Table 1.
Bacterial strains used in this study.
| Strains | Description | Source or reference | ||
|---|---|---|---|---|
| 43895 (EDL933) | E. coli O157:H7 ATCC 43895, a clinical isolate from ground beef, stx1+/stx2+ | ATCC | ||
| 43894 | E. coli O157:H7 ATCC 43894, a clinical isolate from human stool, stx1+/stx2+ | ATCC | ||
| Sakai | E. coli O157:H7, a clinical isolate, stx1+/stx2+ | Lim et al. (2010), Tatsuno et al. (2001 | ||
| 43894 GFPuv | 43894 carry pCRII-GFPuv and produces strong fluorescence when excited by UV light. AmpR | Laboratory stock | ||
| 43895Δeae | 43895 with intimin gene deletion | Sheng et al. (2006b) | ||
| 43895Δtir | 43895 with Tir gene deletion | Sheng et al. (2006b) | ||
| 43894Δeae | 43894 with intimin gene deletion | This work | ||
| 43894Δtir | 43894 with Tir gene deletion | This work | ||
| WSU180 | E. coli O157:H7, a bovine isolate, stx1+/stx2+ | Rice et al. (2003) | ||
| K-12 | E. coli MG1655 strain | Laboratory stock |
ATCC, American type culture collection, Manassas, VA, USA.
Gene manipulation
To make a mutant deficient in expression of intimin or Tir in E. coli O157:H7 43894, the λ-Red recombinase system (Datsenko and Wanner, 2000) was used for eae or tir gene deletion, respectively. The plasmid pKD4 was used as a template to amplify a kanamycin resistance gene, and the same primers were used as described, previously (Sheng et al., 2006b).
Cattle challenge
All animal procedures were approved by the Institutional Animal Care and Use and Biosafety Committees and were strictly followed in this study. Seven to 10-month-old Holstein steers were housed in a quarantined facility at the University of Idaho Agriculture Experiment Station as previous described (Sheng et al., 2006b). Steers were given a single rectal application of 107 CFU of E. coli O157:H7 strains 43895, 43894, or 43894 GFPuv as previously described (Sheng et al., 2004). Retrospective analysis of strain persistence used previously published and unpublished cattle rectal application challenge culture data collected by our laboratory since 2004 (Kudva et al., 2006; Sheng et al., 2006b; and unpublished data).
Preparation of rectal biopsy tissue
Feces were manually removed from the cattle rectum and mucosal biopsies were taken from the rectal mucosa 10–15 cm proximal to the anus using a standard bone curette. The wet weights of biopsy tissues ranged from 30 to 60 mg. The biopsy tissues were rinsed with cold phosphate buffered saline (PBS) and held in cold Hanks’ balanced salt solution (HBSS) on ice for transport to the laboratory.
Tissue-embedded E. coli O157:H7 recovery assay
The terminal rectal tissue biopsies from E. coli O157:H7 culture positive cattle were transferred into sterile 10-cm Petri dishes with 15 ml PBS, fecal debris, and clotted blood was trimmed away and the remaining tissue was cut into fine pieces with sterile scissors. The minced tissue was recovered by centrifugation at 500×g for 5 min, the supernatant was removed, 100 μg/ml gentamicin in HBSS was added, and the tissue was incubated with shaking (150 rpm) at 37°C for 2 h to kill extracellular bacteria. The biopsy tissues were then washed three times with PBS, lysed with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) in PBS by vigorous vortexing. Serial dilutions of the mixture were plated on sorbitol MacConkey agar (SMAC) supplemented with cefixime (50 μg/l; Lederle Laboratories), potassium tellurite (2.5 mg/l; Sigma), vancomycin (40 mg/l; Sigma), and 4-methylumbelliferyl-β-d-glucuronide (0.1 μg/ml, Biosynth AG, Staad, Switzerland). The plates were incubated at 37°C overnight. E. coli O157:H7 colonies were identified and confirmed as O157 by latex agglutination as previous described (Sheng et al., 2008).
Preparation of tissue biopsies for microscopy
To prepare cryostat sections, biopsy tissues from the terminal rectum of 43894 GFPuv culture positive and negative control cattle were fixed with 4% (w/v) paraformaldehyde for 30 min, and infiltrated with up to 25% (w/v) sucrose in PBS (7.5, 15, 25% step gradient). Tissue was oriented to obtain optimal apical to basolateral cryostat cross sections (5–10 μm) that were placed on polylysine-coated slides, air dried, and observed using fluorescence microscopy.
Primary bovine epithelial cell isolation and cell culture
Primary bovine rectal epithelial cells were isolated from rectal biopsies as described by others with some modifications (Follmann et al., 2000; Dziva et al., 2007; Stamm et al., 2008). Briefly, rectal biopsies were taken from E. coli O157:H7 negative steers, cleaned, and cut as described above. The treated tissues were digested in Dulbecco's modified Eagle medium (DMEM) containing 1% (v/v) heat-inactivated fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin (Invitrogen), 30 mg/ml streptomycin (Invitrogen), 25 mg/ml gentamicin (Invitrogen), 75 U/ml collagenase (Sigma), and 20 mg/ml accutase (Millipore, Billerica, MA, USA) with gentle shaking at 37°C until isolated crypts were observed under microscopy. A series of differential centrifugation steps with DMEM containing 2% (w/v) sorbitol was used to enrich the disassociated crypts. The crypt cell pellet was resuspended in cell culture medium [DMEM, 2.5% (v/v) FBS, 0.25 U/ml insulin (Sigma), 10 ng/ml epidermal growth factor (EGF; Sigma), 100 U/ml penicillin, 30 mg/ml streptomycin, and 30 mg/ml gentamicin]. Approximately 400 crypts are seeded per well into 24-well culture plates precoated with collagen (Becton Dickinson Labware, Bedford, MA, USA). Media was supplemented with 100 μg/ml cis-OH-proline (Sigma) to selectively eliminate fibroblasts from the cultures (Willis et al., 2005). Confluence of the cultured cells was reached within 6–7 days and contained approximately 1.8 × 105 cells/well. The characteristics of the epithelial cells were confirmed by immunocytochemistry as described previously (Hoey et al., 2003).
Adherence and internalization assays
The bacterial internalization by the PBRE cells was measured using a standard gentamicin protection assay (Small et al., 1987; Matthews et al., 1997). Monolayers of the PBRE cells were washed twice with HBSS and infected with 1 ml of the cell culture medium without antibiotics and FBS (internalization medium) at a MOI of 100 bacteria/epithelial cell. After 4 h incubation at 37°C with 5% CO2, unattached extracellular bacteria were removed using a serological pipette and the infected monolayers were washed three times with HBSS. The total number of cell-associated bacteria (adhered and internalized) were measured by disrupting the monolayers for 10 min with 0.1% Triton X-100 in 0.1 M PBS (pH 7.2) and gentle vortexing. Serial dilutions were plated on LB agar to determine bacterial numbers. A standard Giemsa stain procedure was used in replicate sample wells to view the interaction of the bacteria with PBEC cells, microscopically. For measurement of internalization, fresh internalization medium containing 100 μg/ml of gentamicin was added to kill adhered extracellular bacteria. After incubation for an additional 2 h, the monolayers were washed three times with HBSS without Ca2+ or Mg2+ and epithelial cells were lysed by addition of 100 μl of 0.5% trypsin–EDTA (Sigma) and 900 μl of 0.05% Triton X-100 for 5 min. Samples were removed, serially diluted, and plated on LB agar to determine the number of CFU. Internalization levels were expressed as the number of CFU recovered/well or as a percentage of the original inoculum resisting gentamicin treatment.
Internalization inhibition assay
To assess the effect of eukaryotic cell inhibitors on bacterial internalization, the PBRE cell monolayers were incubated with cytochalasin D (1 μg/ml, Sigma), colchicine (1.25 μM, Sigma), compactin (25 μM, Sigma), or genistein (50 μM, Sigma) prior to addition of bacteria as described previously (Matthews et al., 1997). Cytochalasin D and colchicine were added to the monolayers 30 min prior to inoculation, genistein 15 min prior to inoculation, and compactin 18 h prior to inoculation. All of the inhibitors were present throughout the 4-h bacterial incubation period. Controls verified that inhibitors did not affect eukaryotic cell viability by trypan blue exclusion and bacterial viability by plate count. Data are expressed as means of the averages obtained from three experiments.
Immunofluorescence microscopy
To visualize the location of E. coli O157:H7 in the biopsy tissue sections, 43894 GFPuv which expresses green fluorescent protein was applied at the rectal mucosa of two steers. The GFP carry-plasmid was stable in 43894 without antibiotic supplement. The cryostat sections were examined by epifluorescence microscopy using an Olympus BX51 microscope with a 100× oil immersion objective (Olympus, Tokyo, Japan). Digital images were acquired using an Olympus DP-70 digital camera and merged using DP manager software version 1.1.1.71.
Electron microscopy
The PBRE cells were infected with 43894 or 43895 for 4 h and fixed in 2.5% glutaraldehyde 2% paraformaldehyde in 0.1 M cacodylate buffer, followed by post-fixation in 2% osmium tetroxide in 0.1 M in 0.1 M cacodylate buffer for 1 h. The cells were dehydrated in an ethanol series, followed twice by 100% acetone, infiltrated with Spurr's resin and polymerized overnight at 70°C. Thin (90 nm) sections were stained with 4% uranyl acetate and Reynolds lead before viewing with a JEOL 1200 EX JEM transmission electron microscope located at the Franceschi Microscopy and Imaging Center (FMIC), Washington State University; Pullman, WA, USA.
Biofilm formation assay
The biofilm formation assay was performed using a microtiter plate method as described previously (Djordjevic et al., 2002) with some modifications. Briefly, a single colony of each E. coli O157:H7 strain was inoculated into 4 ml LB broth and incubated at 37°C overnight with aeration. Then, 5 μl overnight culture was inoculated into 96 well microtiter plates (Corning, Inc., New York, NY, USA) containing 160 μl minimal salt medium (MSM) supplemented with 1 mg/l of yeast extract and 0.04% glucose and incubated at 37°C for 24 h under stationary conditions. Each strain was inoculated in triplicate. Following incubation the plates were washed vigorously with DI water and stained for 15 min with crystal violet (1%) and rinsed vigorously with DI water again to remove unattached cells and residual dye. Biofilm formation was evaluated by measuring the absorbance of solubilized dye in 95% ethanol at 595 nm using a PowerWave XS plate reader (Bio-Tek, Winooski, VT, USA). The biofilm formation experiments were performed in triplicate.
Statistical analysis
Graphs were drawn using Microsoft Excel and GraphPad Prism software 4.0 (San Diego, CA, USA). The numbers of E. coli O157:H7 recovered from gentamicin protection assay were transformed to log10 values. The data were analyzed by GraphPad Prism software 4.0. The data were analyzed by the chi-square test unless the variables needed the Statistical Analysis's t test procedure. For retrospective analysis of strain persistence, survival analysis was used to compare the duration of E. coli O157:H7 infections of two groups of cattle rectally inoculated with 43895 or 43894. Survival time was defined as the number of days a rectal swab sample was continuously cultured positive for E. coli O157:H7. The Log-rank (Mantel–Cox) test was conducted to evaluate the statistical significance of the difference between durations of carriage caused by 43895 and 43994.
Results
E. coli O157:H7 in biopsy tissue survived gentamicin treatment and was deep in the mucosal crypts
To determine if E. coli O157:H7 were internalized by bovine epithelial cells at the terminal rectum, rectal tissue biopsies from culture positive cattle were analyzed using a gentamicin protection assay. Two groups of cattle received rectal application of E. coli O157:H7 strain 43895 or 43894 at day 0, and rectal tissue was biopsied on days 1, 4, and 7 post-challenge. The average tissue sample was 47 mg and contained 12 crypts (data not shown). Table 2 shows that the number of E. coli O157 recovered from rectal biopsy tissues after 2 h gentamicin treatment. On each sampling time (day 1, 4, and 7), higher numbers of the bacteria were recovered after gentamicin treatment from the rectal biopsies of the 43895-infected cattle compared to the 43894-infected cattle (P < 0.05). The data indicated that more 43895 were protected from gentamicin killing than 43894, and suggest the two strains were different in their interaction with bovine host cells.
Table 2.
Escherichia coli O157:H7 from bovine rectal tissue biopsy.
| Strain | ATCC 43894a | ATCC 43895a | ||
|---|---|---|---|---|
| Days post- challenge |
CFUb/cryptc (mean ± SD) |
CFUb/cryptc (mean ± SD) |
||
| 1 | 34 ± 15 | 278 ± 114* | ||
| 4 | 241 ± 99 | 1788 ± 865* | ||
| 7 | 30 ± 21 | 523 ± 102* |
aApproximately 107 CFU E. coli O157:H7 ATCC 43894 or 43895 were inoculated at the terminal rectal mucosa in two separate groups of four steers on day 0.
bCFU recovered after gentamicin treatment.
cA minimum of three biopsies were taken from each animal on each sampling day and each biopsy contained an average of 12 crypts.
*Indicates a higher number of 43895 recovered compared to 43894 on the respective sampling days (P < 0.05).
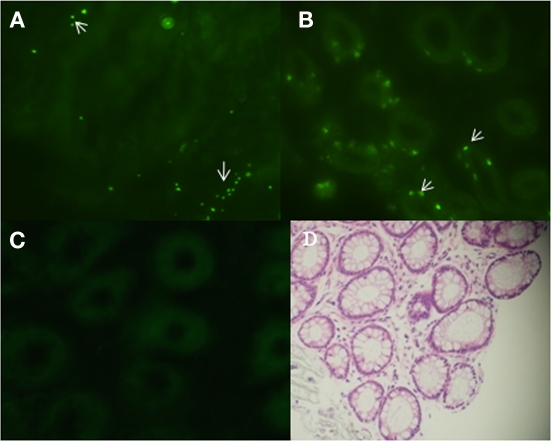
To localize E. coli O157:H7 in biopsy tissue, 43894 GFPuv was applied to the terminal rectal mucosa of steers and 4 days after challenge, biopsies of rectal tissue were taken and frozen sections prepared for fluorescence microscopy. Tissue sections of the first 10 μm from the lumen side showed green fluorescent bacteria adhered to the surface of the rectal mucosa (Figure 1A). Interestingly, 43894 GFPuv were visible in sections 30–40 μm proximal to the lumen surface, deep within the crypt (Figure 1B). No fluorescent bacteria were seen in samples from culture-negative cattle (Figure 1C). The cellular architecture and crypt bodies were visible in tissue sections from culture-negative steers due to weak tissue autofluorescence (Figure 1C) and in rectal tissue biopsies stained with H and E (Figure 1D).
Figure 1.
Escherichia coli O157:H7 penetrated into the crypts of the bovine terminal rectal mucosa. Cattle were challenged with E. coli O157:H7 43894 GFPuv by standard rectal application of bacteria and negative control animals received no bacteria. Rectal mucosal biopsies were taken using a curette 4 days post-challenge. Tissue was processed, cryosectioned, and observed by light or fluorescent microscopy. Representative biopsy sections from O157 culture-negative and -positive cattle are shown. The sections were cut sequentially from the lumen side. (A) E. coli O157:H7 43894 GFPuv (arrow) visible 10 μm from the lumen surface and (B) around crypts at 30–40 μm from the lumen surface. (C) Biopsy tissue section from an O157 culture-negative animal. (D) Hematoxylin and eosin-stained section showing crypts of the terminal rectal mucosa. Crypts show autofluorescence with the fluorescence microscopy (original magnification ×200).
PBRE cell culture was established and characterized
To test the internalization of E. coli O157:H7 by bovine rectal epithelial cells, PBRE cells were obtained from rectal tissue biopsies. After mechanical disruption and enzymatic digestion of the tissue, crypt bodies were obtained (Figure 2A). With appropriate culture conditions and cell densities, a confluent epithelial cell layer derived from the crypts was obtained after ∼6 days (Figure 2B). The epithelial characteristics of the cultured cells were confirmed using pan-cytokeratin antibody specific for the intermediate filaments present in epithelial cells and anti-vimentin labeled V79 fibroblasts served as a negative control (data not shown).
Figure 2.
Establishment of primary bovine rectal epithelial (PBRE) cells. (A) Crypt bodies released from biopsies of the terminal rectal mucosa after 2 h enzymatic digestion at 37°C. Incubation at 37°C in supplemented DMEM medium for 6 days gave rise to (B) a confluence monolayer of epithelial cells. Both representative images taken by inverted microscopy (original magnification, ×100).
E. coli O157:H7 adhered to and were internalized by the PBRE cells
Cell association and the gentamicin protection assays were employed to quantitatively assess adherence and internalization of four different E. coli O157:H7 strains by PBRE cells. The bacterial strains included three E. coli O157:H7 isolates from outbreaks of human disease: 43895, 43894, Sakai, a bovine isolate:WSU180, and a non-pathogenic laboratory strain of E. coli: K-12 MG1655. All strains grew at indistinguishable rates both in LB and minimal medium. Table 3 shows the numbers of cell-associated and internalized bacteria. The numbers of cell-associated and internalized bacteria varied significantly among the E. coli O157:H7 strains with 43895 values significantly higher compared with to 43894, Sakai, and WSU 180 (P < 0.001). Not only were high numbers of 43895 internalized by the PBRE cells, but a mean of 19.38% of the original number of 43895 survived after 2 h gentamicin treatment. This higher internalization level of 43895 in the PBEC cells compared to 43894 is in agreement with the biopsy data that showed higher numbers of 43895 than 43894 were recovered from the biopsy tissues after gentamicin treatment. Cell association and internalization of E. coli K-12 MG1655, used as a non-pathogenic E. coli control, was significantly lower than all tested E. coli O157:H7 strains (P < 0.05).
Table 3.
Internalization of E. coli O157:H7 and K-12 by primary bovine rectal epithelial cells (n = 3).
| Strains | Cell-associateda (CFU × 107/well) |
Internalizedb(CFU × 104/well) | % Internalizationc | ||
|---|---|---|---|---|---|
| ATCC 43895 | 11.88 ± 3.75* | 547.12 ± 110.13* | 19.38 ± 3.91* | ||
| ATCC 43894 | 2.09 ± 0.82 | 5.32 ± 2.29 | 0.20 ± 0.09 | ||
| Sakai | 2.45 ± 0.73 | 5.41 ± 2.21 | 0.19 ± 0.08 | ||
| WSU180 | 2.23 ± 0.77 | 4.89 ± 3.05 | 0.18 ± 0.12 | ||
| E. coli K-12 | 1.02 ± 0.39 | 1.40 ± 0.49* | 0.07 ± 0.02* |
aCFU recovered from washed, lysed monolayers after 4 h incubation with bacteria.
bCFU recovered from lysed monolayers after 4 h incubation with bacteria and 2 h gentamicin treatment.
cPercentage of inoculae surviving gentamicin treatment.
*Indicates a difference (P > 0.05) compared with the other E. coli O157:H7 strains.
E. coli O157:H7 43895 were associated with PBRE cells in an aggregative pattern and internalized bacteria were in vacuoles
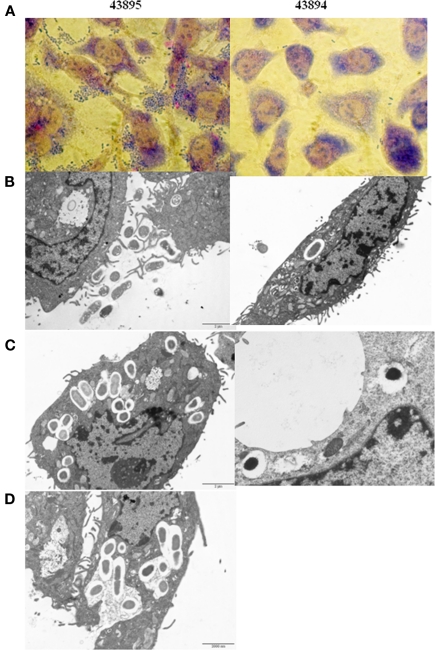
Giemsa staining showed 43895 were associated with PBRE cells with numerous bacteria aggregated together. Other tested E. coli O157:H7 strains, 43894 (Figure 3), Sakai (data not shown), and WSU180 (data not shown) had fewer PBRE cell-associated bacteria and were mostly as single bacterial cells. PBRE cells were examined by TEM 4 h post-incubation with bacteria and intracellular E. coli O157:H7 were observed in membrane-bound vacuoles (Figures 3B,C). The PBRE cells frequently internalized ten to twenty 43895 (Figure 3) with >80% of PBEC cell monolayer having engulfed bacteria (data not shown), while only one to three 43894 cells were internalized (Figure 3) by approximately 15% of the PBEC cell monolayer (data not shown). These TEM results were consistence with the internalization assay results (Table 3) in which a 100-fold higher recovery of 43895, compared to 43894, occurred. It also appeared that 43895 may be dividing in the PBRE vacuoles (Figure 3D), although this was not tested further. These data confirmed that 43895 had an exceptional ability to be internalized by bovine host cells compared to other three tested E. coli O157:H7 strains.
Figure 3.
Escherichia coli O15:H7 adheres to and enters the PBRE cells. E. coli O15:H7 43894 or 43895 were incubated with the monolayer of PBRE cells at 100 MOI for 4 h. More 43895 bacteria adhered to and were internalized by the PBRE cells compared to 43894. (A) Giemsa staining showing 43895 aggregative adherence pattern (inverted microscope, original magnification, ×1000). (B,C) Transmission electronic microscope showing extracellular and intracellular bacteria. (D) 43895 appeared to divide within the vacuoles of the PBRE cells (Original magnification, ×10000).
E. coli O157:H7 43895 survived within the PBRE cells
To investigate the survival of E. coli O157:H7 in the PBRE cells, the number of intracellular bacteria was determined at various times after gentamicin exposure and expressed as percentages of the total bacteria recovered. The number of 43895 or 43894 recovered after 6 h exposure to gentamicin decreased to 42.1 and 23.2% of the total bacteria recovered at 2 h, respectively (Figure 4). After 24 h exposure to gentamicin, the number of intracellular bacteria was dramatically decreased. The survival rate of 43895 was decreased to 25.9% while <1% of 43894 survived at 24 h (Figure 4). The number of PBRE cells attached to the wells and infected by 43895 decreased to 94% at 6 h and to 85% at 24 h of gentamicin treatment compared to the number at 2 h (data not shown). This decrease in 43895-infected cells suggested that some (6–15%) 43895-infected PBRE cells were destroyed over the course of the experiment and released bacteria that would be killed by gentamicin. Thus, the decreased number of 43895 recovered after 6 and 24 h exposure to gentamicin, may include these released cells. Almost all (>99%) attached PBRE cells incubated with 43895 or 43894 excluded the trypan blue dye. Similar and minimal tissue culture cellular debris/destruction was noted with 43894 or 43895 during the 4-h incubation period (data not shown). In addition, the culture media containing gentamicin was tested and remained sterile indicating that the bacteria recovered were intracellular.
Figure 4.
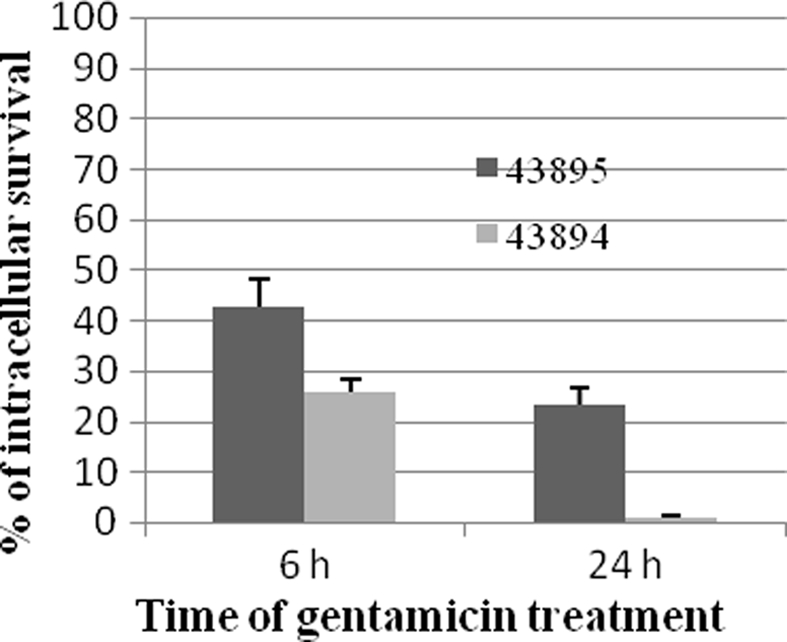
Survival of E. coli O157:H7 in PBRE cells. Bacteria were incubated with monolayers of the PBRE cells at 100 MOI for 4 h. Gentamicin (100 μg/ml) was added for 2, 6, or 24 h incubation. Cells were lysed and plated to LB agar to enumerate surviving bacteria. The bacterial CFU recovered after 2 h Gentamicin treatment was set at 100%. The percentage of intracellular bacteria surviving after 6 and 24 h are shown and each value is the mean of three separate experiments.
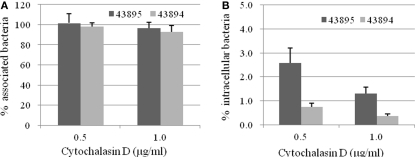
Cytochalasin D inhibited internalization of E. coli O157:H7 by the PBRE cells
To identify host cell factors involved in internalization of 43895 and 43894, PBRE cells were treated with eukaryotic cell inhibitors known to block bacterial invasion (Rosenshine et al., 1992b; Matthews et al., 1997; Martinez and Hultgren, 2002). Treatment with cytochalasin D, an inhibitor of actin microfilaments, did not modify the number of cell-associated bacteria (P > 0.05; Figure 5A). In contrast, its addition significantly inhibited the entry of both 43895 and 43894. The numbers of intracellular bacteria decreased by 97.4 and 99.2% at 0.5 μg/ml (P < 0.001) and by 98.7 and 99.6% at 1 μg/ml concentration (P < 0.001), for 43895 and 43894, respectively (Figure 5B). Inhibitory assays showed that colchicines, a eukaryotic microtubule formation inhibitor, compactin, a pan-Rho GTPase inhibitor, and Genistein, a tyrosine kinase inhibitor did not influence PBRE cell internalization of 43895 or 43894 (data now shown). The data indicated that the internalization process of E. coli O157:H7 was dependent on actin microfilament assembly.
Figure 5.
Effect of cytochalasin D on E. coli O157:H7 association with and internalization by the PBRE cells. The PBRE cell monolayers were pretreated with cytochalasin D for 30 min before the addition of E. coli O157:H7 at 100 MOI and incubated for 4 h in the presence of the inhibitor. (A) Total cell-associated bacteria were quantified after PBRE cell-lysis with 0.1% (v/v) Triton X-100. (B) Intracellular bacteria were quantified after gentamicin treatment for 2 h. Results are expressed as cell-associated or intracellular bacteria relative to those obtained without inhibitor, taken as 100%. Each value is the mean of three separate experiments.
Intimin and Tir are not required for E. coli O157:H7 internalization
Our previous study shows that intimin and its receptor-Tir are essential for efficient E. coli O157:H7 colonization at the RAJ mucosa (Sheng et al., 2006b). To test whether intimin or Tir had a role in internalization by the PBRE cells, we examined the ability of intimin or Tir deletion mutants of both 43894 and 43895 to enter the PBRE cells. Although internalization was slightly reduced for 43894Δeae, a mutant in which intimin was deleted, it was not significantly different from the wild-type. However, the slight reduction in PBRE cell internalization of the 43894Δtir mutants (log value 4.03 ± 0.11) compared to the wild-type (log value 4.73 ± 0.17) was significant (P < 0.05; Figure 6), although not dramatic. Similar trends were observed in Tir and intimin mutants of 43895. Both 43895Δeae and Δtir showed reduced levels of internalization (log value 5.95 ± 0.25, 6.03 ± 0.21, respectively); however, these decreased levels were not significant compared to wild-type 43895 (log value 6.33 ± 0.19; P > 0.05). Notably, the levels of 43895Δeae and Δtir internalization were significantly higher than the wild-type 43894, its corresponding mutants (43894Δeae and 43894Δtir; P < 0.001; Figure 6). The data showed strain variation in that deletion of Tir did not affect the high internalization properties of 43895, but did reduce the lower internalization properties of strain 43894 in PBRE cells.
Figure 6.
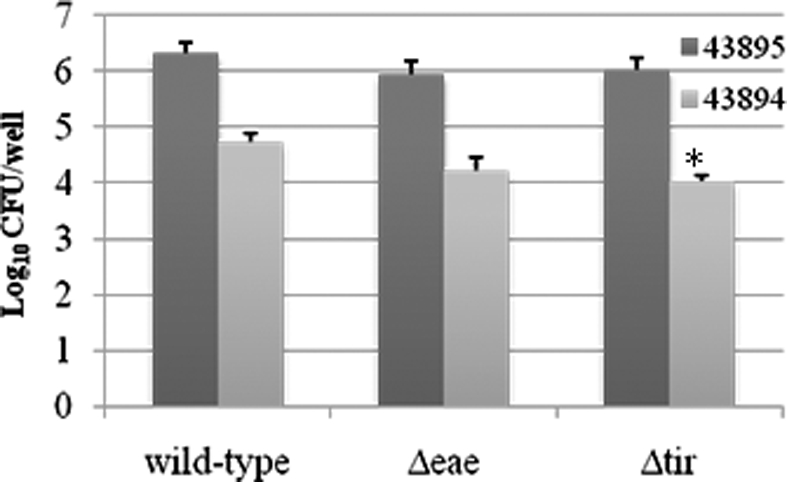
The effect of the deletion of intimin or Tir on PBRE cell internalization of E. coli O157:H7. E. coli O157:H7 43894 or 43895 and the derivatives lacking intimin (Δeae) or Tir (Δtir) with the monolayer of the PBRE cells at 100 MOI. The number of bacteria recovered after gentamicin treatment indicated the number internalized by the epithelial cells. The experiment was repeated three times with three replicated wells. Bars represent means ± SE. Significant difference compared with wild-type E. coli O157:H7 is indicated by the asterisk (P < 0.05).
43895 is most efficient in biofilm formation
Because the pattern of adherence appear aggregative for 43895, the E. coli O157:H7 strains used in this study were assessed for biofilm formation in a standard microtiter plate crystal violet biofilm assay in which formation of bacterial matrices attached to the plastic wells were quantified. 43895 formed more biofilm (P < 0.05), with mean optical densities of 0.55 ± 0.043, than 43894 (0.09 ± 0.02), Sakai (0.14 ± 0.05), or WSU180 (0.11 ± 0.03; Figure 7). The enhanced ability of 43895 biofilm formation correlated with its enhanced ability to adhere to and enter PBEC cells.
Figure 7.

Escherichia coli O157:H7 biofilm formation in crystal violet assays. The E. coli O157:H7 strains indicated were incubated at 37°C in MSM containing 4% glucose for 24 h in 96-well microtiter plate under stationary condition. Biofilm formation was measured by the absorbance of solubilized dye in 95% ethanol at 595 nm. The assays were performed six times for each isolate. All values are means and SE. Significant difference is indicated by the asterisk (P < 0.05).
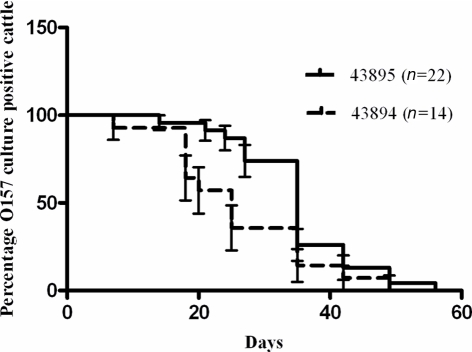
Strain 43895 persists longer than 43894 in cattle
Retrospective analysis of previous studies in which cattle were challenged with a single rectal application of 107 CFU of E. coli O157:H7 43895 or 43894 was done to compare durations of bacterial carriage. The proportion of animals that were culture positive for E. coli O157:H7 from each group on each sampling day was compared and shown in Figure 8. There was a difference (P < 0.05) between 43895 challenged animals (n = 22) and 43894 challenged animals (n = 14) determined using the Log-rank (Mantel–Cox) test. The median duration of carriage with 43895 or 43894 was 34 and 25 days, respectively. The detection limit for E. coli O157:H7 carriage, as previously described for enrichment culture, was 10 CFU/swab (Kudva et al., 2006; Sheng et al., 2006b). This result indicated that E. coli O157:H7 strain 43895 persisted in cattle longer than strain 43894 and correlated with its increased internalization by host cells.
Figure 8.
Differential persistence of E. coli O157:H7 in cattle. Cattle were challenged with 43895 (n = 22) or 43894 (n = 14) using a single rectal application of 107 CFU of bacteria. Rectal swab samples from each animal was taken twice a week and cultured for E. coli O157:H7 until animals cleared the carriage of the bacteria. Percent survival indicated the percent of the cattle that were O157 culture positive. Survival time (duration of carriage) was defined as the number of days animals were cultured positive with E. coli O157:H7 and the P value was calculated by the Log-rank (Mantel–Cox) test.
Discussion
The most important finding of this work was that a subpopulation of E. coli O157:H7 were internalized by epithelial cells at the bovine RAJ mucosa. This finding supports previous observations that not all E. coli O157:H7 are killed by phage lysis or by antimicrobials applied topically or fed. In addition, internalization of E. coli O157:H7 by cells at the RAJ mucosa may play a role in bacterial persistence in cattle. The conclusion that some bacteria are internalized and survive in bovine rectal epithelial cells was made both from tissue biopsies from culture positive cattle and from in vitro culture of bovine rectal epithelial cells. To accomplish the latter, primary cell culture techniques were adapted to culture epithelial cells from the RAJ mucosa of adult cattle.
The architecture of the bovine RAJ mucosa is comprised of epithelial cells lining crypts and the area is rich in lymphoid follicular tissue (Naylor et al., 2003). The intestinal epithelial cells at this site are rapidly dividing and originate from progenitor cells at the base of the crypt, migrate up and out of the crypt onto the luminal surface of the mucosa where they are shed (Magnuson et al., 2000). Estimates are that the epithelial cells lining the terminal rectum turnover every 4 days as older cells are sloughed and newly dividing cells emerge from the crypts (Falk et al., 1998; Sheng et al., 2006b). Also, the average duration individual animals carry E. coli O157:H7 is 1 month (Besser et al., 1997). Cattle carrying E. coli O157:H7 are colonized at this tissue site and bacterial cells have been characterized in close association with the mucosa, attached to the surface of the host cells (Naylor et al., 2003). The parameters that trigger clearance or persistence in the carriage of E. coli O157:H7 are not fully understood but likely involve a dynamic between bacterial attachment to host cells, bacterial division, host cell division, and host cell turnover. E. coli O157:H7 internalization and survival in epithelial cells likely adds to this host:microbe interaction. In this study, the bacteria were present deep in the crypts and were able to survive gentamicin treatment of biopsied tissue that kills all extracellular bacteria. Also, E. coli O157:H7 attached to, were internalized by, and survived in cultured cells (PBRE) from this site.
Most pathogenic types of E. coli involved in gastrointestinal infections are able to invade cultured epithelial cells (Nataro and Kaper, 1998). Our findings showed that for E. coli O157:H7, the efficiency of internalization differed among the strains we tested. The uptake of ATCC 43895 by the PBRE cells was 19.38% of the original inoculum. This level is ∼100-fold higher than the other E. coli O157:H7 strains tested (Table 3) and was at a level similar to previous studies done with a variety of cell lines (Matthews et al., 1997; Dibb-Fuller et al., 2001; Uhlich et al., 2002, 2009; Abu-Ali et al., 2010). Also, ATCC 43895 was more proficient at forming biofilm compared to the other strains tested. These two phenotypes of enhanced host cell internalization and biofilm formation may be related and may contribute to the duration bacterial strains are carried by cattle. This idea is supported by the retrospective analysis of cattle challenged with a single rectal application of E. coli O157:H7 43895 or 43894 that showed strain 43895 persisted in cattle longer than strain 43894.
Bacterial internalization is associated with the rearrangement of the host cell cytoskeletal structure resulting in endocytosis of the pathogen (Goosney et al., 1999; Donnenberg, 2000). Most invasive enteric bacteria including Salmonella, Shigella, Listeria, and Yersinia spp. (Falkow et al., 1992; Finlay and Cossart, 1997; Mengaud et al., 1996) trigger largely microfilament-dependent entry pathways. Invasion of EPEC cells in tissue culture is inhibited by cytochalasin D, colchicines, and by broad specificity Rho GTPase inhibitors (Donnenberg et al., 1990; Ben-Ami et al., 1998). In contrast, E. coli O157:H7 internalization was inhibited by cytochalasin D (Figure 5) but not by other eukaryotic cell inhibitors. This suggests that E. coli O157:H7 uptake by the bovine epithelial cells was through a microfilament-dependent pathway. These data are in agreement with previous studies using immortal cell lines (Matthews et al., 1997) and indicate that EPEC and EHEC exploit distinct host cell processes to stimulate their uptake.
In addition to the actin-dependent host cell microfilament activity required for E. coli O157:H7 internalization by epithelial cells, bacterial factors play a role because strain differences in the rate of internalization were found. Tir was not required for efficient uptake of the more efficiently internalized ATCC 43895 strain, but its deletion affected the less efficiently internalized ATCC 43894, although this could be related to the differences in the number of internalized cells or a unique change in the association with the PBRE cells that affected the internalization rate. Close examination of the electron microscopy images of O157-infected PBRE cells showed the interaction between 43895 and PBRE cells was associated with microvilli-like extensions projecting from the host cell membranes and surrounding the bacteria (Figure 3B). In addition, Transmission electron microscopy of O157-infected PBRE cells showed bacteria enclosed by membrane-bound vacuoles. This suggests that the localized membrane elongations, which may be induced when the bacteria come into contact with the epithelial cell surface, play a role in the uptake by the PBRE cells. Also, EM images of cross sectioned cells showed >10 intracellular 43895/epithelial cell. This number of intracellular bacteria is comparable with numbers previously reported for other enteric bacterial pathogens such as Shigella flexneri (Sansonetti et al., 1986) and Salmonella typhimurium (Leung and Finlay, 1991). Invasion of eukaryotic cells is known to be an important survival mechanism of these enteric pathogens (Finlay and Falkow, 1997). However, the internalization of O157 by RAJ epithelial cells is not associated with disease in cattle.
In conclusion, the internalization and survival of a subset of E. coli O157:H7 by epithelial cells at the bovine RAJ contributes to the host:microbe interaction. Further study will be required to determine the extent to which internalization contributes to persistence and/or the bovine immune response to this human pathogen and the mechanism of E. coli O157:H7 internalization by bovine epithelial cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Lonie Austin and Duane Bull for animal care and handling. We thank Linda Liou and Ann Norton for technique help with cell culture and fluorescence microscopy. This work was supported, in part, by the Idaho Agriculture Experiment Station, Public Health Service grants AI053067, U54-AI-57141, and P20-RR16454 from the National Institutes of Health, and by grants from the Idaho Beef Council.
References
- Abu-Ali G. S., Ouellette L. M., Henderson S. T., Whittam T. S., Manning S. D. (2010). Differences in adherence and virulence gene expression between two outbreak strains of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 156, 408–419 10.1099/mic.0.033126-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami G., Ozeri V., Hanski E., Hofmann F., Aktories K., Hahn K. M., Bokoch G. M., Rosenshine I. (1998). Agents that inhibit Rho, Rac, and Cdc42 do not block formation of actin pedestals in HeLa cells infected with enteropathogenic Escherichia coli. Infect. Immun. 66, 1755–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser T. E., Hancock D. D., Pritchett L. C., McRae E. M., Rice D. H., Tarr P. I. (1997). Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J. Infect. Dis. 175, 726–729 [DOI] [PubMed] [Google Scholar]
- Blanco M., Blanco J. E., Blanco J. E., Gonzalez A., Mora A., Prado C., Fernandez L., Rio M., Ramos J., Alonso M. P. (1996). Prevalence and characteristics of Escherichia coli serotype O157:H7 and other verotoxin-producing E. coli in healthy cattle. Epidemiol. Infect. 117, 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgin R., Arbeloa A., Goulding D., Dougan G., Crepin V. F., Raymond B., Frankel G. (2009). The T3SS effector EspT defines a new category of invasive enteropathogenic E. coli (EPEC) which form intracellular actin pedestals. PLoS Pathog. 5, e1000683. 10.1371/journal.ppat.1000683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P. A., CerdanMalo A. T., Ellin M., Ashton R. (2001). Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in South Yorkshire, UK. Int. J. Food Microbiol. 64, 139–150 10.1016/S0168-1605(00)00453-0 [DOI] [PubMed] [Google Scholar]
- Cobbold R. N., Hancock D. D., Rice D. H., Berg J., Stilborn R., Hovde C. J., Besser T. E. (2007). Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 73, 1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornick N. A., Booher S. L., Moon H. W. (2002). Intimin facilitates colonization by Escherichia coli O157:H7 in adult ruminants. Infect. Immun. 70, 2704–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean-Nystrom E. A., Bosworth B. T., Moon H. W., O'Brien A. D. (1998). Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect. Immun. 66, 4560–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean-Nystrom E. A., Stoffregen W. C., Bosworth B. T., Moon H. W., Pohlenz J. F. (2008). Early attachment sites for Shiga-toxigenic Escherichia coli O157:H7 in experimentally inoculated weaned calves. Appl. Environ. Microbiol. 74, 6378–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb-Fuller M. P., Best A., Stagg D. A., Cooley W. A., Woodward M. J. (2001). An in vitro model for studying the interaction of Escherichia coli O157:H7 and other enteropathogens with bovine primary cell cultures. J. Med. Microbiol. 50, 759–769 [DOI] [PubMed] [Google Scholar]
- Djordjevic D., Wiedmann M., McLandsborough L. A. (2002). Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68, 2950–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg M. S. (2000). Pathogenic strategies of enteric bacteria. Microbiology 153, 561–569 [DOI] [PubMed] [Google Scholar]
- Donnenberg M. S., Donohue-Rolfe A., Keusch G. T. (1990). A comparison of HEp-2 cell invasion by enteropathogenic and enteroinvasive Escherichia coli. FEMS Microbiol. Lett. 57, 83–86 [DOI] [PubMed] [Google Scholar]
- Dziva F., Mahajan A., Cameron P., Currie C., McKendrick I. J., Wallis T. S., Smith D. G. E., Stevens M. P. (2007). EspP, a Type V-secreted serine protease of enterohaemorrhagic Escherichia coli O157:H7, influences intestinal colonization of calves and adherence to bovine primary intestinal epithelial cells. FEMS Microbiol. Lett. 271, 258–264 [DOI] [PubMed] [Google Scholar]
- Dziva F., van Diemen P. M., Stevens M. P., Smith A. J., Wallis T. S. (2004). Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology 150, 3631–3645 10.1099/mic.0.27448-0 [DOI] [PubMed] [Google Scholar]
- Elliott S. J., Sperandio V., Giron J. A., Shin S., Mellies J. L., Wainwright L., Hutcheson S. W., McDaniel T. K., Kaper J. B. (2000). The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68, 6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S. J., Yu J., Kaper J. B. (1999). The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67, 4260–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk P. G., Hooper L. V., Midtvedt T., Gordon J. I. (1998). Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62, 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S., Isberg R. R., Portnoy D. A. (1992). The interaction of bacteria with mammalian cells. Annu. Rev. Cell Biol. 8, 333–363 10.1146/annurev.cb.08.110192.002001 [DOI] [PubMed] [Google Scholar]
- Finlay B. B., Cossart P. (1997). Exploitation of mammalian host cell functions by bacterial pathogens. Science 276, 718–725 10.1126/science.276.5313.718 [DOI] [PubMed] [Google Scholar]
- Finlay B. B., Falkow S. (1997). Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61, 136–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follmann W., Weber S., Birkner S. (2000). Primary cell cultures of bovine colon epithelium: isolation and cell culture of colonocytes. Toxicol. In Vitro 14, 435–445 10.1016/S0887-2333(00)00033-3 [DOI] [PubMed] [Google Scholar]
- Francis C. L., Jerse A. E., Kaper J. B., Falkow S. (1991). Characterization of interactions of enteropathogenic Escherichia coli O127:H6 with mammalian cells in vitro. J. Infect. Dis. 164, 693–703 [DOI] [PubMed] [Google Scholar]
- Goosney D. L., Knoechel D. G., Finlay B. B. (1999). Enteropathogenic E. coli, Salmonella, and Shigella: masters of host cell cytoskeletal exploitation. Emerging Infect. Dis. 5, 216–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauke L. J., Kudva I. T., Yoon J. W., Hunt C. W., Williams C. J., Hovde C. J. (2002). Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68, 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin P. M., Tauxe R. V. (1991). The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13, 60–98 [DOI] [PubMed] [Google Scholar]
- Hancock D. D., Besser T. E., Kinsel M. L., Tarr P. I., Rice D. H., Paros M. G. (1994). The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol. Infect. 113, 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock D. D., Besser T. E., Rice D. H., Herriott D. E., Tarr P. I. (1997). A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 118, 193–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey D. E. E., Sharp L., Currie C., Lingwood C. A., Gally D. L., Smith E. D. G. (2003). Verotoxin 1 binding to intestinal crypt epithelial cells results in localization to lysosomes and abrogation of toxicity. Cell. Microbiol. 5, 85–97 [DOI] [PubMed] [Google Scholar]
- Hueck C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson M. A., Pellegrin S., Peto L., Banbury D. N., Leard A. D., Mellor H., Kenny B. (2003). Synergistic roles for the Map and Tir effector molecules in mediating uptake of enteropathogenic Escherichia coli (EPEC) into non-phagocytic cells. Cell. Microbiol. 5, 773–783 [DOI] [PubMed] [Google Scholar]
- Karmali M. A., Steele B. T., Petric M., Lim C. (1983). Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1, 619–620 10.1016/S0140-6736(83)91795-6 [DOI] [PubMed] [Google Scholar]
- Kenny B. (2001). The enterohaemorrhagic Escherichia coli (serotype O157:H7) Tir molecule is not functionally interchangeable for its enteropathogenic E. coli (serotype O127:H6) homologue. Cell. Microbiol. 3, 499–510 [DOI] [PubMed] [Google Scholar]
- Kenny B., DeVinney R., Stein M., Reinscheid D. J., Frey E. A., Finlay B. B. (1997). Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520 10.1016/S0092-8674(00)80437-7 [DOI] [PubMed] [Google Scholar]
- Knutton S., Rosenshine I., Pallen M. J., Nisan I., Neves B. C., Bain C., Wolff C., Dougan G., Frankel G. (1998). A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17, 2166–2176 10.1093/emboj/17.8.2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudva I. T., Krastins B., Sheng H., Griffin R. W., Sarracino D. A., Tarr P. I., Hovde C. J., Calderwood S. B., John M. (2006). Proteomics-based expression library screening (PELS): a novel method for rapidly defining microbial immunoproteomes. Mol. Cell. Proteomics 5, 1514–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K. Y., Finlay B. B. (1991). Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 88, 11470–11474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. Y., La H. J., Sheng H., Forney L. J., Hovde C. J. (2010). Influence of plasmid pO157 on Escherichia coli O157:H7 Sakai biofilm formation. Appl. Environ. Microbiol. 76, 963–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J. C., McKendrick I. J., McKechnie C., Fenlon D., Naylor S. W., Currie C., Smith D. G. E., Allison L., Gally D. L. (2005). Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Appl. Environ. Microbiol. 71, 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B. A., Davis M., Hubele S., Austin P. R., Kudva I. T., Williams C. J., Hunt C. W., Hovde C. J. (2000). Ruminant gastrointestinal cell proliferation and clearance of Escherichia coli O157:H7. Infect. Immun. 68, 3808–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. J., Hultgren S. J. (2002). Requirement of Rho-family GTPases in the invasion of type 1-piliated uropathogenic Escherichia coli. Cell. Microbiol. 4, 19–28 [DOI] [PubMed] [Google Scholar]
- Matthews K. R., Murdough P. A., Bramley A. J. (1997). Invasion of bovine epithelial cells by verocytotoxin-producing Escherichia coli O157:H7. J. Appl. Microbiol. 82, 197–203 [DOI] [PubMed] [Google Scholar]
- McDaniel T. K., Kaper J. B. (1997). A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23, 399–407 [DOI] [PubMed] [Google Scholar]
- Mead P. S., Slutsker L., Dietz V., McCaig L. F., Bresee J. S., Shapiro C., Griffin P. M., Tauxe R. V. (1999). Food-related illness and death in the United States. Emerg. Infect. Dis. 5, 607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J., Ohayon H., Gounon P., Cossart P. (1996). E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84, 923–932 10.1016/S0092-8674(00)81070-3 [DOI] [PubMed] [Google Scholar]
- Michino H., Araki K., Minami S., Takaya S., Sakai N., Miyazaki M., Ono A., Yanagawa H. (1999). Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150, 787–796 [DOI] [PubMed] [Google Scholar]
- Moon H. W., Whipp S. C., Argenzio R. A., Levine M. M., Giannella R. A. (1983). Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41, 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J. P., Kaper J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor S. W., Low J. C., Besser T. E., Mahajan A., Gunn G. J., Pearce M. C., McKendrick I. J., Smith D. G., Gally D. L. (2003). Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71, 1505–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor S. W., Nart P., Sales J., Flockhart A., Gally D. L., Low J. C. (2007). Impact of the direct application of therapeutic agents to the terminal recta of experimentally colonized calves on Escherichia coli O157:H7 shedding. Appl. Environ. Microbiol. 73, 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor S. W., Roe A. J., Nart P., Spears K., Smith D. G. E., Low J. C., Gally D. L. (2005). Escherichia coli O157:H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 151, 2773–2781 10.1099/mic.0.28060-0 [DOI] [PubMed] [Google Scholar]
- Oelschlaeger T. A., Barrett T. J., Kopecko D. J. (1994). Some structures and processes of human epithelial cells involved in uptake of enterohemorrhagic Escherichia coli O157:H7 strains. Infect. Immun. 62, 5142–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton J. C., Paton A. W. (1998). Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11, 450–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna N. T., Mayhew G. F., Posfai G., Elliott S., Donnenberg M. S., Kaper J. B., Blattner F. R. (1998). Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66, 3810–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D. H., Sheng H. Q., Wynia S. A., Hovde C. J. (2003). Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J. Clin. Microbiol. 41, 4924–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas L., Coffey B., McAuliffe O., McDonnell M. J., Burgess C. M., Coffey A., Ross P. R., Duffy G. (2010). The in vivo and ex vivo evaluation of bacteriophage e11/2 and e4/1c for the control of Escherichia coli O157:H7. Appl. Environ. Microbiol. 76, 7210–7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenshine I., Donnenberg M. S., Kaper J. B., Finlay B. B. (1992a). Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 11, 3551–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenshine I., Duronio V., Finlay B. B. (1992b). Tyrosine protein kinase inhibitors block invasin-promoted bacterial uptake by epithelial cells. Infect. Immun. 60, 2211–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P. J., Ryter A., Clerc P., Maurelli A. T., Mounier J. (1986). Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 51, 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H., Davis M. A., Knecht H. J., Hovde C. J. (2004). Rectal administration of Escherichia coli O157:H7: novel model for colonization of ruminants. Appl. Environ. Microbiol. 70, 4588–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H., Knecht H. J., Kudva I. T., Hovde C. J. (2006a). Application of bacteriophages to control intestinal Escherichia coli O157:H7 levels in ruminants. Appl. Environ. Microbiol. 72, 5359–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H., Lim J. Y., Knecht H. J., Li J., Hovde C. J. (2006b). Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74, 4685–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H., Lim J. Y., Waktins M. k., Minnich A. S., Hovde C. J. (2008). Characterization of an Escherichia coli O157:H7 O-antigen deletion mutant and effect of the deletion on bacterial persistence in the mouse intestine and colonization at the bovine terminal rectal mucosa. Appl. Environ. Microbiol. 74, 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small P. L., Isberg R. R., Falkow S. (1987). Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocolitica to enter and replicate within HEp-2 cells. Infect. Immun. 55, 1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. R. (1998). Escherichia coli O157 infections: the Scottish experience. Hosp. Med. 59, 164. [PubMed] [Google Scholar]
- Stamm I., Mohr M., Bridger P. S., Schropfer E., Konig M., Stoffregen W. C., Dean-Nystrom E. A., Baljer G., Menge C. (2008). Epithelial and mesenchymal cells in the bovine colonic mucosa differ in their responsiveness to Escherichia coli Shiga toxin 1. Infect. Immun. 76, 5381–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuno I., Horie M., Abe H., Miki T., Makino K., Shinagawa H., Taguchi H., Kamiya S., Hayashi T., Sasakawa C. (2001). toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69, 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlich G. A., Gunther N. W., IV, Bayles D. O., Mosier D. A. (2009). The CsgA and Lpp proteins of an Escherichia coli O157:H7 strain affect HEp-2 cell invasion, motility, and biofilm formation. Infect. Immun. 77, 1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlich G. A., Keen J. E., Elder R. O. (2002). Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect. Immun. 70, 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlisidou I., Dziva F., La Ragione R. M., Best A., Garmendia J., Hawes P., Monaghan P., Cawthraw S. A., Frankel G., Woodward M. J., Stevens M. P. (2006). Role of intimin-tir interactions and the tir-cytoskeleton coupling protein in the colonization of calves and lambs by Escherichia coli O157:H7. Infect. Immun. 74, 758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis B. C., Liebler J. M., Luby-Phelps K., Nicholson A. G., Crandall E. D., du Bois R. M., Borok Z. (2005). Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am. J. Pathol. 166, 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Doyle M. P., Shere J., Garber L. (1995). Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61, 1290–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]