Abstract
A mouse locus called Lgn1 determines differences in macrophage permissiveness for the intracellular replication of Legionella pneumophila. The only regional candidate genes for this phenotype difference lie within a cluster of closely linked paralogs of the Neuronal Apoptosis Inhibitory Protein (Naip) gene. Previous genetic and physical mapping of the Lgn1 phenotype narrowed it to an interval containing only Naip2 and Naip5, suggesting that there is not complete functional overlap among the mouse Naip loci. In order to gather more information about polymorphisms among the Naip genes of the 129 mouse haplotype, we have determined the genomic sequence of a substantial portion of the 129 Naip gene array. We have constructed an evolutionary model for the expansion of the Naip gene array from a single progenitor Naip gene. This model predicts the presence of two distinct families of Naip paralogs: Naip1/2/3 and Naip4/5/6/7. Unlike the divergences among all the other Naip paralogs, the splits among Naip4, Naip5, Naip6, and Naip7 occurred relatively recently. The high degree of sequence conservation within the Naip4/5/6/7 family increases the likelihood of functional overlap among these genes.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF242431-AF242435.]
Macrophages isolated from C57BL/6J and A/J mice exhibit differences in permissiveness for intracellular replication of L. pneumophila (Yamamoto et al. 1988). This phenotype difference segregates as a single-gene trait in crosses between C57BL/6J and A/J and maps to a locus on distal chromosome 13 (Yamamoto et al. 1991; Yoshida et al. 1991; Dietrich et al. 1995; Beckers et al. 1995). Detailed physical mapping of this locus, called Lgn1, reveals that it contains a series of 50 to 80 kb highly homologous direct repeats and that a cluster of Naip gene paralogs map inside these direct repeats (Scharf et al. 1996; Growney et al. 2000).
The region of the human genome that is orthologous to the mouse Lgn1 region also contains a series of highly homologous repeated segments. The human spinal muscular atrophy (SMA) region has what appears to be an inverted duplication of some 500 kb (Lefebvre et al. 1995). This amplified genomic segment contains several transcriptionally active genes, including copies of survival motor neuron (SMN); NAIP; general transcription factor II H, polypeptide 2 (GTF2H2); and small EDRK-rich factor 1 (SERF1) (reviewed by Growney et al. 2000). However, the only gene in common between the amplified segments from the mouse and human Lgn1/SMA intervals is Naip/NAIP (Growney et al. 2000).
The fact that the mouse and human Lgn1/SMA regions both have divergently organized sets of closely linked repeats indicates that these amplified segments originated independently in the mouse and human lineages. This observation begs the question of whether the amplification of Naip/NAIP in either mouse or human has any functional significance. Although most of the mouse Naip paralogs are transcriptionally active and encode similar but not identical proteins, it is not known whether these transcripts provide redundant or diverse functions (Huang et al. 1999). These questions about the functionality of the mouse Naip loci are important to the identification of the Lgn1 mutation because the current critical interval for the Lgn1 phenotype contains two different transcriptionally active Naip genes (Naip2 and Naip5) (Growney and Dietrich 2000; Huang et al. 1999).
Mapping and sequence analysis of the mouse Lgn1 interval suggests that the Naip genes have arisen through a series of several distinct amplification events emanating from a single ancestral Naip. This model of the origins of the mouse Naip array relies heavily on the sequences (Fig. 1A) of a single exon from the clustered Naip paralogs to build a phylogenetic tree (Growney et al. 2000). A more rigorous basis for determining the relationships of the mouse Naip genes would be to compare their entire genomic sequences.
Figure 1.
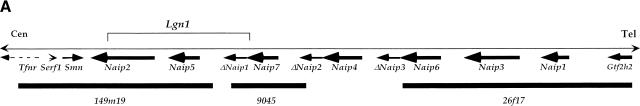
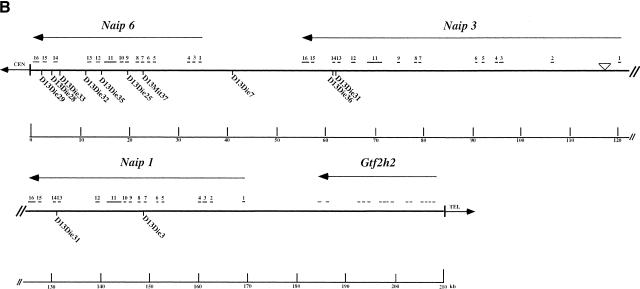
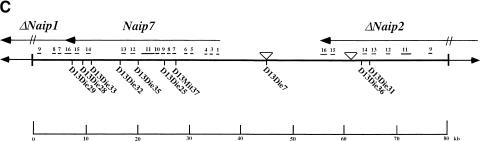
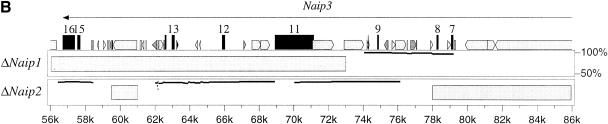
Map of the 129 mouse Naip array and annotation of the genomic sequences. (A) The map of the 129 mouse Naip array that was described previously in Growney et al. (2000) is indicated. The named arrows show the position and orientation of the Naip gene loci. The ΔNaip regions are pseudogenes that have been deleted for several of the 5′ terminal exons. The current critical interval for Lgn1 is indicated above the map (Growney and Dietrich, 2000). The positions of genomic clones with sequence reported in this work (9045 and 26f17) or elsewhere (149m19, Endrizzi et al. 1999) are indicated by bold lines beneath the gene map. The positions of other nearby genes are indicated to provide context for the map. For Fig. 1B,C, the identification and annotation of Naip gene sequences were obtained through simple alignments of known Naip cDNA sequences to the genomic fragments (see Methods). The sequences were also analyzed using Genotator/Genotator Browser (see Methods). The relative orientations of named transcription units, gene exons, and markers in each clone are shown in these figures. The scale at the bottom is in kb. The arrows represent the direction of transcription of the genes and the position and size of exons from within the genes are shown by the small numbered lines, except in the case of Gtf2h2, which has its coding exons indicated, but its 5′ and 3′ untranslated-region sequences in the mouse are unknown. (B) Annotation of 26f17 (AF242431 and AF242432). The triangle indicates the position of an approximately 7-bp gap in the sequence that cannot be determined with certainty. (C) Annotation of 9045 (AF242433-AF242435). The triangles indicate the positions of two small gaps (each are ∼500 bp) in the sequence.
In this paper, we report the complete annotated sequence of 26f17, a 220-kb bacterial artificial chromosome (BAC) clone that contains the three Naip genes on the centromere-distal side of the array in the 129 haplotype (Naip1, Naip3, and Naip6) (Fig 1A; Growney et al. 2000). In addition, we present three large annotated fragments of genomic sequence from 9045, a 75-kb P1 clone mapping to the central portion of the 129 Naip array (Fig 1A; Growney et al. 2000). Our analysis of these genomic sequences has provided additional markers to refine the map of the Lgn1 interval (Growney and Dietrich 2000) and allowed us to refine the previously reported model of the origins of the mouse Naip array.
RESULTS
Genomic Sequence Determination
The 220-kb BAC clone 26f17 was roughly mapped to the distal side of the Lgn1 region by others (Diez et al. 1997). Subsequent precise mapping of the clone identified it as an ideal template for sequencing the Lgn1 interval because it covered a large extent of the distal side of the Naip gene array (Fig 1A; Growney et al. 2000).Our prior map information about this clone suggested that it was likely to contain multiple copies of Naip gene sequences; so we used a tiered strategy for the sequence assembly (see Methods; Endrizzi et al. 1999).
The final sequence assembly of this clone consists of two contiguous sequences covering 117,791 bp and 90,650 bp (GenBank accession nos. AF242431 and AF242432). We could not complete the sequence across the remaining gap with certainty because it was composed of a 300-bp simple sequence repeat. We were able to link the two contiguous sequences using the polymerase chain reaction (PCR), and our estimate of the total sequence length (208,448 bp) suggests an extremely small gap of only 7 bp (Fig. 1B). The two consensus sequences were derived from 3960 sequencing reactions, with every base in the consensus representing data from at least one sequencing reaction on each strand. The average per-base sequencing redundancy is over fivefold. The sequence assembly was analyzed extensively for consistency with known restriction digest and PCR amplification patterns from clone and genomic DNA, indicating that the sequence represents both the clone and the genomic structure with fidelity (data not shown).
P1 clone 9045 was identified by us several years ago and subsequently mapped with precision into the center of the Naip array (Fig. 1A) in 129 (Scharf et al. 1996; Growney et al. 2000). We chose to sequence this clone because of its position in the center of the Naip array because it could reveal significant discrepancies from our model of the origin of this repeat. We used a similar strategy for sequence assembly as we did for 26f17.
The final sequence assemblies for 9045 consist of three contiguous sequences totaling 72,460 bp (GenBank Accession nos. AF242433–AF242435). The holes in the sequence represent areas that are difficult to sequence because they contain microsatellite sequences. However, we measured the size of the remaining gaps in the sequence using PCR and found them to be quite small (Fig. 1C). The three consensus sequences were derived from a total of 1355 sequencing reactions and as with 26f17, every base in the sequence represents data from each strand. The average per-base sequencing redundancy is approximately fivefold. The total size of the known sequence and our estimates of the gap sizes are in accordance with our estimates of the size of 9045 from NotI digestion and pulsed field gel analysis (data not shown).
Discovery and Annotation of Genes in 26f17 and 9045
We have used several methods to discover and annotate genes in our new genomic sequences. Because we knew that the clones were going to contain Naip gene loci, the first—and most straightforward—annotation relied on aligning known Naip cDNA sequences to the clones (Fig. 1).
Naip Loci in 26f17:
Naip1. The distal-most Naip gene in the cluster, Naip1, spans 45 kb and has 16 exons, including an exon 2 in its 5′ untranslated region (UTR), which is a sequence found only in Naip1, Naip3, and Naip2 (see below; Endrizzi et al. 1999). This gene is transcriptionally active but has been genetically excluded from the Lgn1 interval (Yaraghi et al. 1998; Huang et al. 1999; Growney et al. 2000).
Naip3. Naip3, which spans approximately 65 kb, is likely to be a nonfunctional gene sequence. We have never isolated a cDNA corresponding to transcripts from this locus, and the genomic sequence shows that the region corresponding to exon 10 of this gene is completely absent, which likely creates a frameshift if exon 9 is spliced directly to exon 11 (Huang et al. 1999). As suggested previously, Naip3 is likely to be the direct progenitor of the so-called fragmentary ΔNaip sequences (see below; Growney and Dietrich 2000).
Naip6. As has been seen with the genomic sequence of Naip5, this gene sequence, which spans approximately 35 kb, has only 15 exons and contains a number of polymorphic marker sequences that characterize members of the central Naip repeat (Endrizzi et al. 1999; Growney et al. 2000). This gene is likely to be transcriptionally active because cDNAs from close relatives of this locus have been isolated (Huang et al. 1999). The 3′ UTR of these cDNAs contain unspliced exons from an adjacent ΔNaip locus. Unfortunately, our sequence of 26f17 does not extend into the region where these ΔNaips should reside. Nevertheless, a marker called D13Die30, that specifically amplifies ΔNaips from genomic DNA, maps proximally to Naip6 (Growney et al. 2000). Furthermore, we have determined the sequences of ΔNaip loci from our assembly of 9045 (see below). The only ortholog of Naip6 contained in the C57BL/6J genome has been excluded from the Lgn1 interval (Growney and Dietrich 2000).
Naip Loci in 9045:
Naip7. Naip7, which spans approximately 30 kb, has many similarities to Naip5 and Naip6, including the number of exons and the presence of repeated microsatellite markers characteristic of the central Naip array. In addition, it is similar to Naip6 but diverges from Naip5 in that it has a ΔNaip juxtaposed at its 3′ end. As we noted for Naip6, it is possible that this gene is transcriptionally active, since cDNAs from a relative of this locus in another mouse strain have been isolated (Huang et al. 1999).
ΔNaips. We have sequenced portions of two different ΔNaip loci in 9045. From these two partial ΔNaip sequences, we discerned two important features. First, the ΔNaip loci, which span approximately 20 kb, begin with an exon 7 that is juxtaposed extremely close to the exon 16 of the adjacent Naip. Second, the marker content of the ΔNaip loci are similar to that of Naip3, as can be seen by the presence of D13Die36, the size of its intron 13, and the absence of an exon 10. All these data point strongly to the possibility that ΔNaips are recently diverged relatives of Naip3. However, one significant difference between Naip3 and the ΔNaips is seen in exon 11, which is present in only a fragmentary form in the ΔNaips.
In addition to aligning our sequences with cDNAs known to map into the interval, we subjected them to a series of homology searches and gene prediction programs using the Genotator and Genotator-Browser packages (Harris 1997). We identified only one other gene in our sequences using this method. Consistent with prior data, we found sequences from 26f17 having significant BLAST homologies to human GTF2H2 sequences (Growney et al. 2000). Because the cDNA sequence for the mouse ortholog has not been determined, we aligned the human cDNA to 26f17 and determined the intron-exon structure of the coding portion of the mouse Gtf2h2 gene, but we could not definitively identify the 5′ and 3′ UTR sequences. For that reason, we have not numbered the exons of Gtf2h2 that are depicted in Figure 1.
Alignments of Mouse Naip Sequences
Given the sequence relatedness of the mouse Naip gene loci, it is likely that they all share a single common progenitor. We have done alignments of the known mouse Naip sequences in order to shed some light about the nature of the events that have taken place since the divergence from a single Naip gene (see Methods). The data from these alignments is presented in Figure 2 and Table 1.
Figure 2.
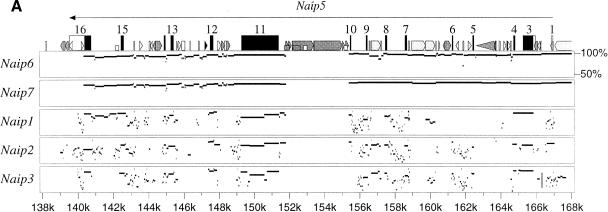
Percent Identity Plot (PIP) Analysis of Naip Genomic Sequences. The alignments have been generated and drawn as described in Methods. The figure indicates regions for which there are alignments having >50% identity. Before alignment, the genomic sequences were masked by RepeatMasker. Interspersed repeats in the mouse sequence are indicated as follows: (white pointed box) L1; (light gray box) SINE other than MIR; (black box) MIR or LINE2; (dark gray box) all others. Other elements in the sequence are indicated as follows: (arrows), positions and directions of transcription of known genes in the query sequence; (numbered black rectangles) positions of exons within the transcription units; (short gray rectangles), position of CpG islands. The figure shows several PIPs between mouse Naip genes. (A) Comparison of the Naip5 to all the other sequenced Naips, showing the existence of two distinct families of gene loci: Naip1/2/3 and Naip4/5/6/7. (B) Comparison of Naip3 with the ΔNaip loci. The elongated gray boxes inside the PIP panels indicate regions in which a comparison between the two sequences is not possible because one of the sequences ends.
Table 1.
Comparison of Alignments of Mouse Naip Paralogsa
| Naip1b | Naip2c | Naip3d | Naip5e | Naip6f | Naip7g | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| align | ident | align | ident | align | ident | align | ident | align | ident | align | ident | |
| Naip1b | — | — | 36 | 78 | 46 | 82 | 48 | 80 | 37 | 79 | 45 | 77 |
| Naip2c | 46 | 81 | — | — | 34 | 81 | 49 | 75 | 36 | 80 | 44 | 73 |
| Naip3d | 67 | 80 | 37 | 79 | — | — | 46 | 72 | 46 | 78 | 34 | 85 |
| Naip5e | 27 | 80 | 21 | 76 | 17 | 77 | — | — | 63 | 95 | 78 | 94 |
| Naip6f | 29 | 80 | 22 | 77 | 24 | 80 | 86 | 94 | — | — | 100 | 97 |
| Naip7g | 28 | 80 | 21 | 75 | 15 | 85 | 86 | 96 | 82 | 98 | — | — |
The genomic sequence for each Naip gene locus was aligned with each other Naip locus (see Methods). The similarities between these alignments are expressed in terms of the percentage of the gene named in the column head that appears in a local alignment with the gene named in the row head (column labeled “align”) and of the percentage of sequence identity within those local alignments (column labeled “ident”). Because of the differences in the overall length of the different Naip genes, it is important for the reader to confine their comparisons to looking for trends within a column. In this way, one can see the relationships among the different families of Naip genes. For example, by looking in the Naip1 column, one can see that it most resembles Naip3, because of the extensive proportion of Naip1 that aligns with Naip3. On this basis, one can also see that Naip1 is more closely related to Naip2 and Naip3 than it is to Naip5, Naip6, or Naip7. The parameters used in generating the local alignments prohibit little variation in the percent identity of the alignments. An exception to this is seen in the homologies among the Naip5/6/7 family, in which the percent identities typically exceed 90%.
Bases 6546-51581 of GenBank no. AF242432.
Bases 68968-128492 of GenBank no. AF131205.
Bases 140365-165807 of GenBank no. AF131205.
Bases 22-34589 of GenBank no. AF242431.
Bases 5565-34032 of GenBank no. AF242433.
Inspection of Figure 2A, in which the alignments of the Naip genes are represented as a Percent Identity Plot (PIP), shows that Naip5, Naip6, and Naip7 are extremely closely related to each other, confirming either that they are the result of recent gene duplications or that they are subject to homogenization via gene conversion. Similarly, Naip1, Naip2, and Naip3 share extensive alignments with each other, indicating that they are closely related (Fig. 2A; Table 1). The amount of alignment and levels of homology among the two groups of paralogs suggest an early duplication of an ancestral Naip, leading to the progenitors of what can be called the Naip1/2/3 and Naip4/5/6/7 families (Fig. 2A; Table 1). Even though we do not have genomic sequence for Naip4, we have included it in the Naip4/5/6/7 group based on prior published data demonstrating a high degree of similarity in marker content (Growney et al. 2000).
Although the amplification of the Naip5, Naip6, and Naip7 gene loci seems to be a recent event (as demonstrated by their extremely high level of sequence conservation and their virtually complete alignment that is broken only by the insertion of interspersed repeat elements), the amplification and divergence of the Naip1, Naip2, and Naip3 loci appears to have happened longer ago (as suggested by their lower level of sequence conservation and alignment). Our analysis of the overall conservation of alignments between the Naip1/2/3 sequences, suggests that Naip2 diverged from Naip1/3 before a more recent split between Naip1 and Naip3 (Fig. 2A; Table 1).
Our alignments of the Naip3 locus confirmed our suspicion that the ΔNaip loci are extremely close relatives of Naip3—No other Naip locus exhibited such extensive alignment and high level of sequence identity (Fig. 2B). This suggests that the formation of the ΔNaip loci occurred after the split between Naip1 and Naip3. Similarly, because the structure of the ΔNaip loci are identical throughout the central Naip repeat, the formation of the ΔNaip loci likely occurred before or as part of the amplifications that created Naip5, Naip6, and Naip7. We summarized our interpretation of these data in a model of expansion of the mouse Naip array in Figure 3.
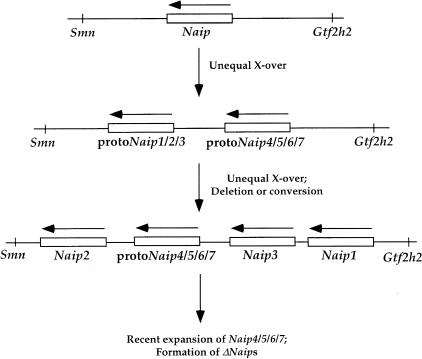
Figure 3.
Model of the Origin of the Naip Gene Array in 129. The essential features of this model are as follows. First, the single ancestral Naip gene became duplicated. This duplication may have occurred due to an unequal crossing over event between different copies of an interspersed repetitive element. This original duplication event is strongly suggested by the sequence similarity profiles between different Naip genes and represents the ancient split between the Naip1/2/3 and the Naip4/5/6/7 families. Second, the proto Naip4/5/6/7 locus becomes flanked by Naip2 on its centromere proximal side and by Naip1 and Naip3 on its centromere distal side. The mechanisms whereby this occurred are obscure, but the possibilities include additional duplications of the array via unequal crossing over and deletion or gene conversion of some of the resulting distal loci. Third, the origin of the central portion of the Naip array, including the ΔNaip loci, occurred much more recently; a model for this is described elsewhere (Growney et al. 2000).
DISCUSSION
The arrangement of highly related genes in closely linked clusters is commonly seen in mammalian genomes. Broadly speaking, these arrays are of two types: those whose members have acquired important divergent functions and those whose members are redundant in function. Examples of closely linked gene families whose members have divergences in function are seen in the cases of the color-vision genes and the beta-globins (Nathans et al. 1986; Yokoyama et al. 1993; Fritsch et al. 1980; Hardies et al. 1984). Similarly, there are examples of the occurrence of closely linked gene copies that are redundant in function, such as is seen in the observed amplification of ribosomal RNA genes in various organisms and in the cellular aquisition of resistance to chemotherapeutic agents (Nath and Bollon 1977; Raymond et al. 1990).
The mouse Naip gene cluster is interesting because we currently do not know if it represents an example of functional diversity, functional redundancy having some important phenotypic consequence or even perhaps a fixation of an amplification that has no functional impact on the organism. Furthermore, the mouse Naip cluster is interesting because one of the members of this family must play an important role in determining the permissiveness of macrophages to the intracellular replication of L. pneumophila (Growney et al. 2000). In light of these unanswered questions, we have determined the genomic sequence of substantial portions of the mouse Naip gene array from 129 in an attempt to measure the relatedness of all the Naip genes.
In our analyses of these genomic sequences, we have definitively ascertained that the mouse Naip gene cluster can be divided into two families: the Naip1/2/3 family and the Naip4/5/6/7 family. The sequence relations of the members of these two families suggests that the Naip4/5/6/7 family members have diverged from each other relatively recently and may, as a consequence, share more functional relatedness than the members of the Naip1/2/3 family. However, since the molecular functions of each of the mouse Naip paralogs have been incompletely described, the sequence data alone cannot be used to make definitive statements about potential similarities or differences in function.
Nevertheless, two lines of additional evidence indicate that the functions of the different mouse Naip paralogs can be separated from each other. First, the recent report of a knockout of the Naip1 gene illustrates a function of this gene in neuronal survival during physiological insult (Holcik et al. 2000). It is unclear whether the inability of the other Naip gene paralogs to compensate for the loss of Naip1 function has to do with differences in the molecular activity of the Naip proteins, with an overall diminishment of Naip function or with some tissue specificity in expression of the Naip paralogs.
The second line of evidence in favor of divergent functions of the mouse Naip genes comes from our knowledge of the genetic map position of the mouse Legionella susceptibility locus (Lgn1). Lgn1 has been mapped to an interval that only includes Naip2 and Naip5, suggesting that the other Naip paralogs cannot compensate for a mutation in one of these genes (Growney and Dietrich 2000). Unfortunately, based on the current information, it is impossible to tell which of the two remaining candidates is responsible for the Lgn1 phenotype.
Remaining unanswered is the broader question of whether the differences in Naip/NAIP gene content in the mouse and human genomes indicate differences in gene function between the two species. Based on previously published data, it seems that there is only a single human NAIP locus that produces an intact, translationally competent transcript (Roy et al. 1995). Unfortunately, critical pieces of information about the human region are missing or unclear.
For example, while it is well documented that differences in the structure of the SMA region exist among human individuals, only a few haplotypes have been mapped in detail (Lefebvre et al. 1995; Roy et al. 1995). The situation is further complicated by the fact that human genomic libraries consist of clones from at least two different haplotypes. Given that assembling a sensible map of the mouse Lgn1 region was extremely difficult in a situation where only one haplotype was being assembled, the complexity of making a consistent human map from mixed haplotype libraries presents even more of a challenge (Growney et al. 2000; Growney and Dietrich 2000). Indeed, it remains possible that there is more variation in the number of functional NAIP sequences among human individuals than had been previously believed because of the technical difficulties involved in mapping the region. In addition, the extent of human variation in permissiveness to Legionella replication is currently unknown, making any cross-species structure-function comparisons impossible.
Because of the complexities of mapping and studying the human interval, it seems likely that the mouse will serve as a springboard for progress into understanding the origins and functional diversity of the Naip array. Not only can the structures of the Naip array be well described in inbred mouse strains, but we and others are making significant progress in elucidating the functional roles of these genes in a variety of processes. With regard to identifying the Lgn1 gene, it is most likely that further comparative sequencing in search of causative mutations in Naip2 or Naip5 and/or attempts to complement the phenotype will resolve the matter. These experiments are currently underway in our laboratory.
METHODS
Sequencing
The strategy used for determining the sequence of clones that contain multiple copies of highly related regions was described extensively elsewhere (Endrizzi et al. 1999). Here, we briefly describe the technical aspects to the sequencing.
BAC DNA Isolation
We isolated BAC (26f17) DNA from 100 ml overnight cultures (LB with 12.5 μg/ml chloramphenicol) following Research Genetics' BAC miniprep protocol. We isolated P1 (9045) DNA from 500 ml overnight cultures (LB with 50 μg/ml kanamycin) using Qiagen's Large Construct Kit.
Library Construction
We sheared 10 μg of BAC DNA in 50 μl of 1X Mung Bean buffer (New England Biolabs) using a sonicator and made the fragment ends blunt by incubating 0.5 μl of Mung Bean nuclease with the sheared DNA for 30 min at 30° C. We ran total DNA through a 1% low-melt agarose gel (FMC) in 1X TAE buffer at 1.5 V/cm for 16 hr alongside a 1 kb DNA ladder (GIBCO). We excised DNA fragments in the range of 3.5 to 4.5 kb, extracted with buffer-saturated phenol and after ethanol precipitation, resuspended in 20 μl dH2O. We quantified the size-selected DNA against a low mass ladder (GIBCO) using an agarose gel. We ligated 150 ng of blunt-end murine DNA to 50 ng of dephosphorylated, SmaI blunt-cut pUC18 vector (Pharmacia) at 14° C for 16 hr and used 2 μl of the ligation reaction for transforming DH5α ultracompetent Escherichia coli cells (GIBCO).
Sequencing Template Preparation
We picked colonies by hand and inoculated in 96 deep-well plates containing 1.25 ml of TB plus ampicillin (50 μg/ml final). Cultures grew at 37° C for 20 hr while shaking at 225 rpm. We isolated plasmids using a 96-well alkali lysis protocol (Edge Biosystems) and resuspended in 30 μl of 1 mM Tris-Cl.
Sequencing Reactions
We sequenced 500 ng of template using ABI Big Dye terminator chemistry (Perkin Elmer) according to the manufacturer's specifications. We performed the reaction in an MJ Research thermal cycler (PTC-225). We purified reactions with 96-well filter plates (Edge), dried samples in a Speedvac evaporator, and stored the samples at −20° C until resuspending in loading buffer. We used both an ABI 377 and an ABI 3700 for detection. We extracted DNA sequences using Bass, Grace, and Trout (Whitehead/MIT) for ABI 377 data and ABI Data Collection software (Perkin Elmer) for ABI 3700 data.
Assembly
We imported approximately 4X coverage for each genomic clone in sequence reads from both ends of 4-kb subclones into a Gap4 database (Staden 1996). We used an initial threshold of 5% mismatch for automated assembly. We then manually removed and reassembled misaligned reads based on our observations of consistent sequence polymorphisms with the consensus. Ultimately, this low-level sequence coverage of the clones yielded a manageable number of contiguous sequences that were ordered and oriented by linking subclones (Chen et al. 1993). We isolated the inserts of these subclones and sequenced sheared, cloned 500-bp fragments to obtain sequence coverage of the gaps.
Long PCR to Obtain Gap-spanning Fragments
We chose primers for long PCR using Primer 0.5 on consensus sequence from the ends of assembled contiguous sequences for which we had no linking subclones (Lincoln et al. 1991). We designed long PCR reactions to cover all possible orders and orientations of contiguous sequences. We repeated three reactions for each positive PCR product to eliminate early-round mutations introduced in any one reaction and pooled products together for either direct sequencing or library construction.
Confirmation of Sequence
To check the sequence assembly for errors, we compared the restriction digest pattern of each clone to a virtual digest of the consensus sequence. In all cases, the predictions were consistent with the digest pattern (data not shown).
Analysis and Annotation of the Sequence
Alignment with Known cDNA Sequences
We assembled sequences of Naip cDNAs (Huang et al. 1999) to genomic consensus sequence using Sequencher 3.0.
Genotator
After the assembly was complete, we utilized Genotator/Genotator Browser (Harris 1997) to annotate the final sequence with BLAST homologies to the expressed-sequence-tag and GENPEPT databases, open reading frames, and exons predicted by the programs Genie, GENSCAN, GRAIL, and GeneFinder (Kulp et al. 1996; Burge and Karlin 1997; Uberbacher and Mural 1991; Solovyev et al. 1994). See the paper by Endrizzi et al. (1999) for more details.
Alignments with Mouse Paralogous Sequences
Sequences were aligned using a program called Blastz (Schwartz et al. 2000), which can be run on user-supplied data at http://bio.cse.psu.edu/. We aligned unmasked sequences using the default alignment scores (match, 1; mismatch, −1; gap of length k, −6–0.2k) and the Chaining option, which forces aligned regions to have the same order and orientation in the two sequences.
Display of Alignments
For overviews of the alignment results, we used a visual representation called the percent identity plot (PIP) (Oeltjen et al. 1997; Hardison et al. 1997; Ansari-Lari et al. 1998). The PIPs, unlike the traditional representation of these alignments as dot-plots, lose some of the spatial relationships with one of the compared sequences but accurately depict the level of identity at each position in the alignment.
Acknowledgments
We thank Victor Boyartchuk, James Watters, and Rebecca Mosher for critical evaluation of the manuscript and Jeremiah Scharf and Lou Kunkel for helpful discussions. This work was supported by a grant from the Muscular Dystrophy Association to W.F.D., who is an assistant investigator of the Howard Hughes Medical Institute. W.M. was supported by grant LM05110 from the National Library of Medicine.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dietrich@rascal.med.harvard.edu; FAX (617) 432-3993.
REFERENCES
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny OM, Lu J, Gorrell JH, Chinault AC, Belmont JW, Miller W, Gibbs RA. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Beckers MC, Yoshida S, Morgan K, Skamene E, Gros P. Natural resistance to infection with Legionella pneumophila: Chromosomal localization of the Lgn1 susceptibility gene. Mamm Gen. 1995;6:540–545. doi: 10.1007/BF00356173. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chen EY, Schlessinger D, Kere J. Ordered shotgun sequencing, a strategy for integrated mapping and sequencing of YAC clones. Genomics. 1993;17:651–656. doi: 10.1006/geno.1993.1385. [DOI] [PubMed] [Google Scholar]
- Cheng S, Fockler C, Barnes W, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Damron DM, Isberg RR, Lander ES, Swanson MS. Lgn1, a gene that determines susceptibility to Legionella pneumophila, maps to mouse chromosome 13. Genomics. 1995;26:443–450. doi: 10.1016/0888-7543(95)80161-e. [DOI] [PubMed] [Google Scholar]
- Diez E, Beckers MC, Ernst E, DiDonato CJ, Simard LR, Morissette C, Gervais F, Yoshida SI, Gros P. Genetic and physical mapping of the mouse host resistance locus Lgn1. Mamm Gen. 1997;8:682–685. doi: 10.1007/s003359900536. [DOI] [PubMed] [Google Scholar]
- Diez E, Yaraghi Z, MacKenzie A, Gros P. The Neuronal Apoptosis Inhibitory Protein (Naip) is expressed in macrophages and is modulated after phagocytosis and during intracellular infection with Legionella pneumophila. J Immunol. 2000;164:1470–1477. doi: 10.4049/jimmunol.164.3.1470. [DOI] [PubMed] [Google Scholar]
- Endrizzi M, Huang S, Scharf JM, Kelter A-R, Wirth B, Kunkel LM, Miller W, Dietrich WF. Comparative Sequence Analysis of the Mouse and Human Lgn1/SMA interval. Genomics. 1999;60:137–151. doi: 10.1006/geno.1999.5910. [DOI] [PubMed] [Google Scholar]
- Fritsch EF, Lawn RM, Maniatis T. Molecular cloning and characterization of the human beta-like globin gene cluster. Cell. 1980;19:959–972. doi: 10.1016/0092-8674(80)90087-2. [DOI] [PubMed] [Google Scholar]
- Growney, J.D., and Dietrich, W.F. 2000. High resolution genetic and physical map of the Lgn1 interval in C57BL/6J inplicates Naip2 or Naip5 in Legionella pneumophila pathogenesis. Genome Res. This issue. [DOI] [PMC free article] [PubMed]
- Growney JD, Scharf JM, Kunkel LM, Dietrich WF. Evolutionary divergence of the mouse and human Lgn1/SMA repeat structures. Genomics. 2000;64:62–81. doi: 10.1006/geno.1999.6111. [DOI] [PubMed] [Google Scholar]
- Hardies SC, Edgell MH, Hutchison CA. Evolution of the mammalian beta-globin gene cluster. J Biol Chem. 1984;259:3748–3756. [PubMed] [Google Scholar]
- Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Harris NL. Genotator: A workbench for sequence annotation. Genome Res. 1997;7:754–762. doi: 10.1101/gr.7.7.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Scharf JM, Growney JD, Endrizzi MG, Dietrich WF. The mouse Naip gene cluster on Chromosome 13 encodes several distinct functional transcripts. Mamm Gen. 1999;10:1032–1035. doi: 10.1007/s003359901155. [DOI] [PubMed] [Google Scholar]
- Holcik M, Thompson CS, Yaraghi Z, Lefebvre CA, MacKenzie AE, Korneluk RG. The hippocampal neurons of neuronal apoptosis inhibitory protein 1 (NAIP1)-deleted mice display increased vulnerability to kainic acid-induced injury. Proc Natl Acad Sci. 2000;97:2286–2290. doi: 10.1073/pnas.040469797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. A generalized hidden marker mode for the recogniton of human genes in DNA. Proc Intelligent Syst Mol Biol. 1996;4:134–142. [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lincoln, S., Daly, M., and Lander, E.S. 1991. Whitehead Institute for Biomedical Research http://www.genome.wi.mit.edu.
- Nath K, Bollon AP. Organization of the yeast ribosomal RNA gene cluster via cloning and restriction analysis. J Biol Chem. 1977;252:6562–6571. [PubMed] [Google Scholar]
- Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- Oeltjen JC, Malley TM, Muzny DM, Miller W, Gibbs RA, Belmont JW. Large-scale comparative sequence analysis of the human and murine Bruton's typosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rose E, Housman DE, Gros P. Physical mapping, amplification, and overexpression of the mouse mdr gene family in multidrug-resistant cells. Mol Cell Biol. 1990;10:1642–1651. doi: 10.1128/mcb.10.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- Scharf JM, Damron D, Frisella A, Bruno S, Beggs AH, Kunkel LM, Dietrich WF. The mouse region syntenic for human spinal muscular atrophy lies within the Lgn1 critical interval and contains multiple copies of Naip exon 5. Genomics. 1996;38:405–417. doi: 10.1006/geno.1996.0644. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Fraser KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker–A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev VV, Salamov AA, Lawrence CB. Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res. 1994;22:5156–5163. doi: 10.1093/nar/22.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Uberbacher EC, Mural RJ. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci USA. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Klein TW, Newton CA, Widen R, Friedman H. Growth of Legionella pneumophila in thioglycollate-elicited peritoneal macrophages from A/J mice. Infect Immun. 1988;56:370–375. doi: 10.1128/iai.56.2.370-375.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Klein TW, Friedman H. Legionella pneumophila growth in macrophages from susceptible mice is genetically controlled. Proc Exp Biol Med. 1991;196:405–409. doi: 10.3181/00379727-196-43207. [DOI] [PubMed] [Google Scholar]
- Yaraghi Z, Korneluk RG, MacKenzie A. Cloning and characterization of the multiple murine homologs of NAIP (neuronal apoptosis inhibitory protein) Genomics. 1998;51:107–113. doi: 10.1006/geno.1998.5378. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Starmer WT, Yokoyama R. Paralogous origin of the red- and green-sensitive visual pigment genes in vertebrates. Mol Biol Evol. 1993;10:527–538. doi: 10.1093/oxfordjournals.molbev.a040024. [DOI] [PubMed] [Google Scholar]
- Yoshida SI, Goto Y, Mizuguchi Y, Nomoto K, Skamene E. Genetic control of natural resistance in mouse macrophages regulating intracellular Legionella pneumophila multiplication in vitro. Infec and Imm. 1991;59:428–432. doi: 10.1128/iai.59.1.428-432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]