Abstract
Riboswitches are cis-acting RNA fragments that regulate gene expression by sensing cellular levels of the associated small metabolites. In bacteria, the class I preQ1 riboswitch allows the fine-tuning of queuosine biosynthesis in response to the intracellular concentration of the queuosine anabolic intermediate preQ1. When binding preQ1, the aptamer domain undergoes a significant degree of secondary and tertiary structural rearrangement and folds into an H-type pseudoknot. Conformational “switching” of the riboswitch aptamer domain upon recognizing its cognate metabolite plays a key role in the regulatory mechanism of the preQ1 riboswitch. We investigate the folding mechanism of the preQ1 riboswitch aptamer domain using all-atom Gō-model simulations. The folding pathway of such a single domain is found to be cooperative and sequentially coordinated, as the folding proceeds in the 5’ → 3’ direction. This kinetically efficient folding mechanism suggests a fast ligand binding response in competition with RNA elongation.
Riboswitches represent a whole class of RNA structural motifs exerting a gene expression regulatory mechanism widely utilized among eubacteria.1,2 Riboswitches are typically found in the 5’ untranslated regions (UTRs) of mRNAs and comprise a highly specific metabolite-binding aptamer domain and an adjoining expression platform.3,4 The structural organization of the aptamer domain upon recognizing its cognate ligand directs adoption of a specific secondary structure of the expression platform, which in turn dictates the expression level of the downstream gene(s). Thus, as a metabolite sensor, a riboswitch provides a simple and direct strategy for the feed-back regulation of expression of a (set of) related gene(s).
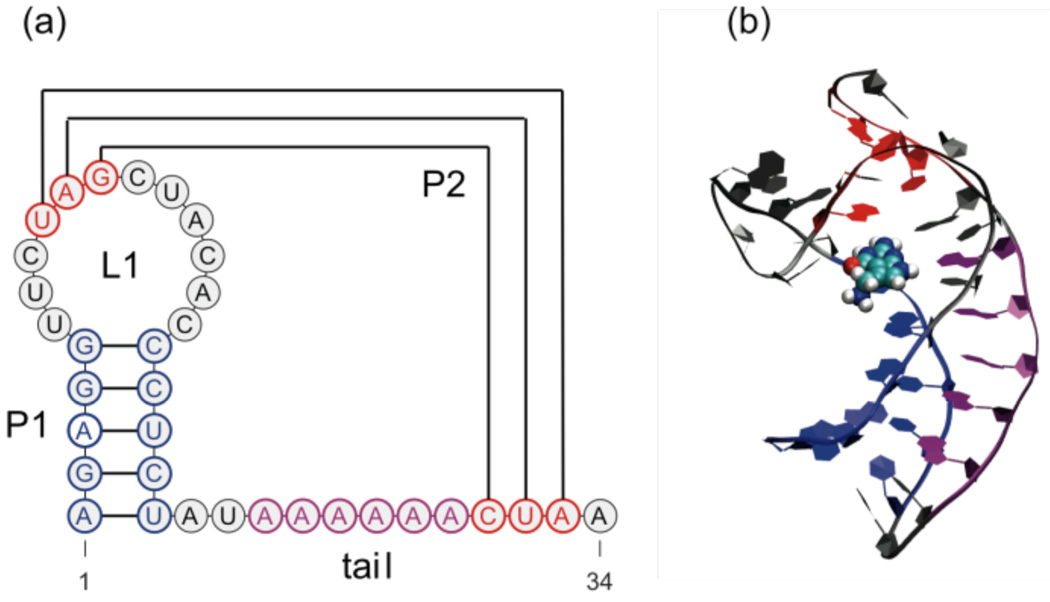
In bacteria, the class I preQ1 riboswitch modulates the gene transcription of the queuosine synthetic enzymes, in response to the intracellular concentration of preQ1 (7-aminomethyl-7-deazaguanine), a queuosine biosynthetic intermediate.5 The class I, type II, B. subtilis queC preQ1 riboswitch aptamer domain (Figure 1), which resides in the 5’ UTR of the queuosine biosynthetic operon, is the smallest naturally occurring aptamer domain known to date, comprising minimally 34 residues. The secondary structure of this modular RNA element is predicted to simply consist of a stem-loop followed by a short adenine-rich single-stranded “tail”.5 The preQ1 unbound aptamer domain appears to be largely unstructured.6 When binding its metabolite, the aptamer domain folds into a classic H-type pseudoknot,7 as evidenced by both x-ray crystallography and NMR spectroscopy.6,8 In the folded state, the 5-base pair (bp) stem P1 consolidates into the hairpin region with the 3’ end of the tail forming a second 3-bp stem P2 with its upstream loop L1. P2 coaxially stacks on P1 via the associated ligand. The adenine-tract (A-tract) of the tail forms a stretch of A-minor motif contacts in the minor groove of the P1 stem. Folding of the aptamer domain down-regulates gene transcription as it sequesters part of a downstream anti-terminator sequence and favors formation of an alternative terminator hairpin.9 The folding decision of the aptamer domain is essential to the regulatory mechanism of the riboswitch. Although high-resolution tertiary structures of the preQ1-RNA complex are available, critical information on the conformational “switch” from the ligand-free to the folded ligand-bound state is lacking and the folding pathway triggered by the ligand is yet to be investigated in any detail.
Figure 1.
Representation of the preQ1 bound aptamer domain structure. (a) Secondary structure (rendered using VARNA10). (b) Tertiary structure (rendered using VMD http://www.ks.uiuc.edu/Research/vmd/) in a ribbon cartoon. P1, P2 and the A-tract of the tail are represented in blue, red and purple, respectively, and preQ1 is rendered as a van der Waals volume.
Computer simulation can be used to provide insights into the folding mechanisms and folding landscapes of biomacromolecules. Energy landscape theory11,12 states that the free energy landscape of a protein is funnel shaped, minimally frustrated, and strongly biased toward the native state, which lies at the bottom of the funnel. Implementing this idea into a physical model, Gō-model simulations have been widely applied to study the folding landscape of proteins and nucleic acids.13–19 The energy function of the topology-based Gō model is strongly biased toward the native (or target) structure. In the non-bonded potential, attractive interactions are only assigned to contact pairs (contacts found in the native state) within a specific distance cutoff, whereas all non-native contacts are mutually repulsive. As a consequence, the biased potential models a smooth, funneled landscape that significantly reduces local thermodynamic traps and minimizes the energetic frustration on the folding pathway.
In this study, we use Gō model simulations to investigate the folding of the preQ1 riboswitch aptamer. All heavy atoms of the RNA are represented explicitly. We present the folding kinetics, folding landscape and detailed folding pathways of the aptamer domain. We employ the fraction of native contacts (Q) as a measure of folding progress to characterize secondary and tertiary structure formation.
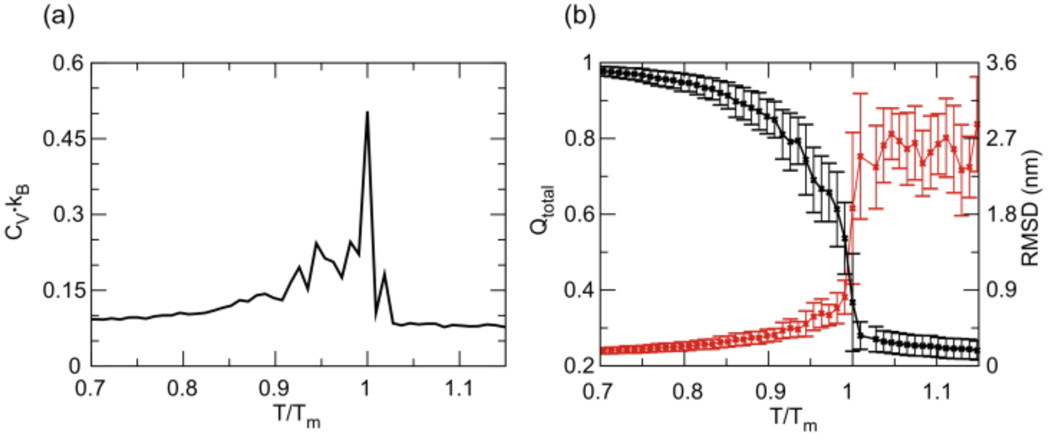
We first use thermodynamic simulations in which the RNA was simulated over a wide temperature range to capture the aptamer (un)folding transition. The heat capacity Cv is calculated to monitor aptamer folding and unfolding, following , where is the energy fluctuation at a given temperature T and kB is the Boltzmann constant. Thermal unfolding of the aptamer domain, shown in Figure 2, displays a single sharp peak in the temperature dependent heat capacity profile, indicating a two-state folding transition. As expected, structural transitions indicated by significant changes in root-mean-square displacement and loss of native contacts occur collectively about the melting temperature (Tm).
Figure 2.
Melting of the aptamer-ligand complex. (a) Heat capacity profile as a function of temperature, scaled by Tm. (b) Average fraction of native contacts (black) and RMSD (red) of RNA as a function of scaled temperature. Standard deviation of average values at each temperature are shown as error bars.
Kinetic simulations of the aptamer folding are performed at 11 temperatures under Tm, each starting with 78 representative unfolded conformations. We observe fast conformational transitions among unfolded states. We do not find any rate-limiting intermediate states that could pose significant barriers along the folding reaction coordinates. The sharp peak in the heat capacity at Tm suggests a highly cooperative folding/unfolding transition for the aptamer domain. To quantify this behavior, we use a dimensionless index Ω to measure the folding cooperativity. The index Ω is defined as , where ΔT is the width at the half-maximum of the peak of |dQ/dT|max and Tmax is the temperature at which |dQ/dT|max has a peak.20 Ω goes to infinity for a sharp two-state transition and approaches zero for a completely non-cooperative transition. The aptamer appears to fold in a highly cooperative manner as indicated by a large Ω, on the order of 106.
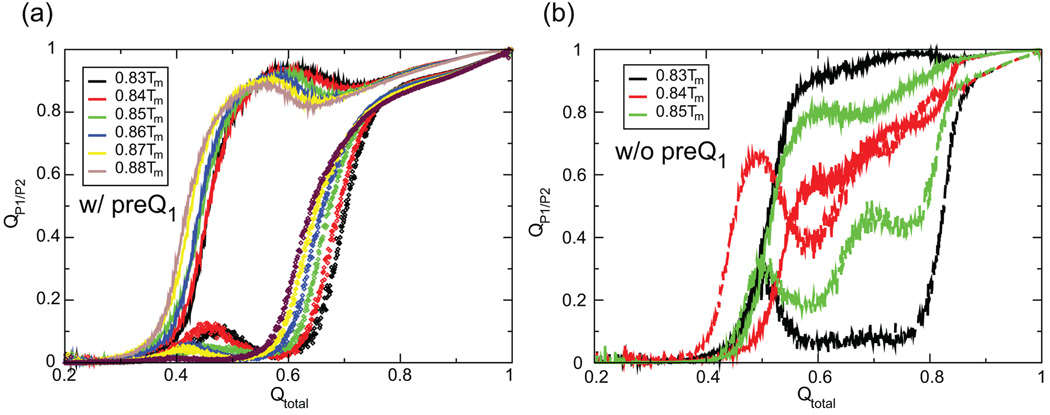
The folded aptamer domain resembles an H-type pseudoknot and is stabilized largely by the base-pairing interactions in the two helices P1 and P2, respectively. We fold the aptamer domain at temperatures below Tm in the absence and presence of the cognate ligand. Figure 3 shows the ligand influence on the folding progress variables representing the two helices. Apparently, without the assistance of preQ1 the aptamer domain encounters a topologically frustrated folding pathway. On the contrary, folding of the aptamer domain with its ligand appears to be smoothly coordinated, where folding of the P2 stem always follows the consolidation of the P1 stem. The small bumps in Figure 3a indicate that premature formation of the P2 stem can hamper the folding of the P1 stem, and that P2 has to unfold for the development of P1, which is known as “backtracking”21 as opposed to misfolding. It is noteworthy that the structural changes that occur as the aptamer domain folds are reversed in the loss of secondary structure elements in the melting simulations (Supplementary Material). However, it should be noted that in the absence of the ligand the 3-bp P2 stem is thermodynamically unstable, is not highly populated under physiological conditions,6,22 and in our simulations is simply imposed by the parameters of the Gō model.15,16 Henceforth, we will focus on folding in the presence of ligand.
Figure 3.
Fraction of native contacts in P1 (solid line) and P2 (dashed line) as a function of fraction of total contacts in the folding simulations with and without ligand. Note the premature formation of structure in P2 in the absence of preQ1. For clarity, folding at 0.86Tm− 0.88Tm in the absence of ligand can be found in the supplementary information.
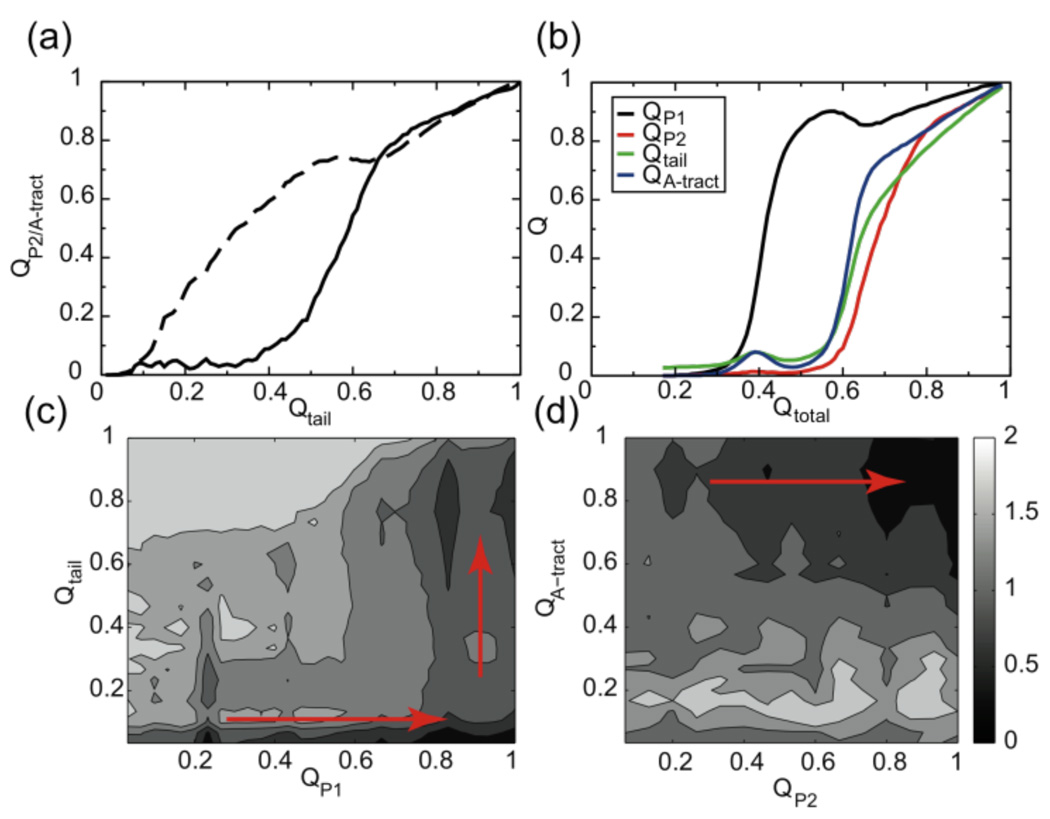
Aptamer folding in the presence of ligand is hierarchical and is monitored by the native contacts shown in Figure 4. The tail of the aptamer domain consists of 2 nucleotides at the 5’ end connecting the upstream hairpin, the 6-adenine tract and 4 bases at the 3’end (Figure 1a). This tail folds after the formation of the P1 stem-loop (QP1). The A-tract region first establishes tertiary contacts with the P1 stem (QA-tract). The bases at the 3’end then interact with their counterparts in the L1 loop to form the 3-bp P2 stem (QP2).
Figure 4.
Detailed folding pathway of the aptamer in the presence of preQ1 at 0.88Tm (Folding at other temperatures behaves similarly and thus is not shown). a. Fraction of contacts in the A-tract (dashed line) and P2 (solid line) as a function of fraction of contacts in the tail. b. Fraction of contacts in P1, P2, tail, A-tract as a function of fraction of total contacts. c. and d. Free energy profile F(QP1, Qtail) and F(QA-tract, QP2). The dominant folding pathway follows the orderly establishment in QP1, QA-tract and QP2.
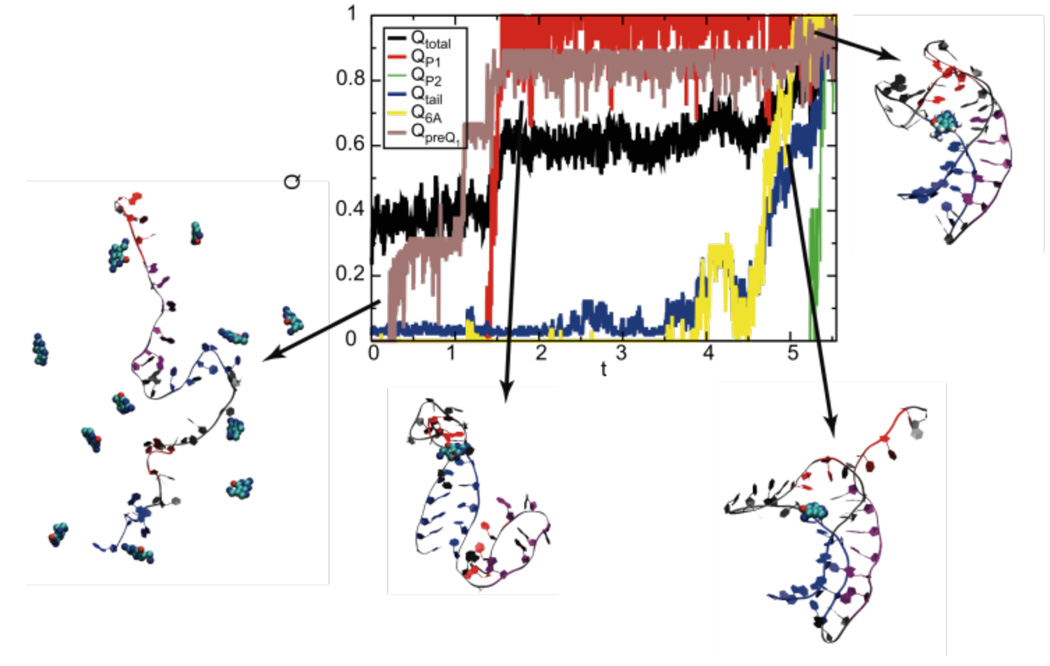
The sequential structural ordering is also evident in the free energy profile F(Qtail, QP1) and F(QA-tract, QP2), calculated using WHAM (Figure 4c,d).23 Figure 5 illustrates a representative folding pathway, which is observed in 85% of 858 independent folding simulations, regarding all structural aspects of the aptamer domain (P1, P2, tail, A-tract). The RNA structural ordering begins with the formation of the 5’ P1 stem, followed by the insertion of the A-tract segment and, finally, is completed with the consolidation of the P2 stem. The preQ1 aptamer domain folds essentially along a single route from 5’ to 3’ end, in the same direction as RNA transcription proceeds.
Figure 5.
A representative folding pathway of the aptamer with ligand. The fraction of native contacts formed in the various segments of the RNA, P1, P2, tail, A-tract, and preQ1 increases as time (arbitrary t) evolves. The surrounding structures are representative of the conformations observed during the folding process.
RNAs are known to have rugged folding landscapes.24 Folding of large and structurally complex RNAs is thought to be hierarchal, where the assembly of the thermodynamically more stable secondary structural elements precedes the collapse of the remote tertiary contacts.25 However, an RNA pseudoknot is a rather simple and compact structural motif that is a widespread building block of RNA tertiary structures. In protein folding studies, small single domain proteins have often been reported to fold according to a first-order rate law.26–28 Analogously, Gō-model folding of the preQ1 riboswitch aptamer appears to follow a simple two-state kinetics when forming the ligand-bound structure in a 5’ to 3’ direction. However, it should be noted that our Gō-like potentials are topologically biased. Gō-model simulations do not provide information related to any metastable intermediate states with non-native interactions that might be trapped in local minima, unless such interactions occur as a result of early formation of native contacts that must be lost for folding to proceed. In fact, both two-state and multi-state transitions of RNA pseudoknots have been observed in thermal29,30 and mechanical31,32 folding/unfolding experiments. Our study investigates the folding mechanism of the isolated aptamer domain, which displays minimal topological frustration along the folding pathway when its cognate ligand is present.
The current view concerning the folding pathway of an RNA pseudoknot is often based on dissecting folding into the formation of its individual constitutes, which include two helical stems (P1 and P2) and tertiary stem-loop contacts. A recent folding study33 proposed that relative stabilities of the helical stems are the main determinant of the folding mechanism, based on the observation of distinct assembly pathways of three structurally related pseudoknots. It was argued that if there is sufficient difference in the stability of the two stems, the relatively more stable one forms first. Otherwise, parallel folding pathways could exist for folding the two stems with similar stability. In the preQ1 aptamer domain study presented here, it is then perhaps not surprising that the thermodynamically more stable 5-bp P1 stem folds before the 3-bp P2 stem. But we also observe that the 6 consecutive adenines in the A-tract of the aptamer tail make unusually extensive tertiary contacts with the stem, even ahead of the secondary structure formation of P2. Thermodynamic stabilities of individual structural elements are directly coupled to the kinetics of the folding process, consistent with the prevailing hierarchical perspective of RNA folding.
Folding of the aptamer domain is key to the functional control of the preQ1 riboswitch. Putting aptamer folding in the context of mRNA transcription is instrumental for our understanding of gene regulation by alternative RNA folding. It has been appreciated that many transcriptionally responsive riboswitches rely on kinetic rather than thermodynamic control, as evidenced by the large difference between the apparent binding affinity (KD) of the ligand and the concentration (C50) of ligand needed to reach half-maximal transcriptional control in vitro.34,35 After all, once RNA polymerase passes through the riboswitch element, mRNA elongation will continue irrespective of upstream formation of a transcription terminator/anti-terminator. To ascertain its impact, the ligand binding kinetics must therefore be tightly coupled to the rate of transcription.3 The 5’ to 3’ directional folding of the preQ1 riboswitch aptamer domain observed here suggests that folding can occur concomitantly with aptamer synthesis by transcription. A picture emerges wherein, once ligand is encountered, a partially prefolded aptamer domain competes efficiently with mRNA elongation by quickly capturing its 3’ tail to signal the “off” state to the downstream gene expression platform. Interestingly, a similar 5’ to 3’ sequential folding strategy has been successfully harnessed in computational pseudoknot prediction.36 The exceedingly conserved preQ1 riboswitch aptamer domain5 ensures the preservation of this exquisite folding design throughout evolution.
In summary, we here have employed Gō-model simulations to study the folding mechanism of the preQ1 riboswitch aptamer domain and delineate the sequence of folding events. Our simulations suggest that folding of the preQ1 riboswitch aptamer domain follows a highly cooperative and nearly single-routed pathway, where the pseudoknot in the presence of its cognate ligand assembles sequentially by formation of the P1 stem, stem-loop contacts and finally the P2 stem. This folding mechanism is consistent with and in fact exploits the commonly observed hierarchical folding of RNA. The resulting directional folding of the preQ1 aptamer is proposed to provide an efficient ligand binding mechanism for conformational “switching” and riboswitch function.
Supplementary Material
Acknowledgment
We thank the center for Multiscale Modeling Tools in Structural Biology (MMTSB) for providing the computing facility. This work is supported by grants from NIH (RR012255) and NSF (PHY0216576).
Footnotes
Supporting Information Available: Materials, methods and additional figures included as supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Winkler W, Nahvi A, Breaker RR. Nature. 2002;419:952. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 2.Nudler E, Mironov AS. Trends Biochem. Sci. 2004;29:11. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Garst AD, Batey RT. Biochim. Biophys. Acta - Gene Reg. Mech. 2009;1789:584. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth A, Breaker RR. Annu. Rev. Biochem. 2009;78:305. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, Iwata-Reuyl D, Breaker RR. Nat. Struct. Mol. Biol. 2007;14:308. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 6.Kang M, Peterson R, Feigon J. Mol. Cell. 2009;33:784. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Pleij CWA. Curr. Opin. Struct. Biol. 1994;4:337. [Google Scholar]
- 8.Klein DJ, Edwards TE, Ferre-D'Amare AR. Nat. Struct. Mol. Biol. 2009;16:343. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rieder U, Kreutz C, Micura R. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10804. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darty K, Denise A, Ponty Y. Bioinformatics. 2009;25:1974. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Proteins: Struct., Funct., Genet. 1995;21:167. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 12.Brooks CL, Onuchic JN, Wales DJ. Science. 2001;293:612. doi: 10.1126/science.1062559. [DOI] [PubMed] [Google Scholar]
- 13.Ueda Y, Taketomi H, Go N. Biopolymers. 1978;17:1531. [Google Scholar]
- 14.Hills RD, Brooks CL. Int. J. Mol. Sci. 2009;10:889. doi: 10.3390/ijms10030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitford PC, Noel JK, Gosavi S, Schug A, Sanbonmatsu KY, Onuchic JN. Proteins: Struct., Funct., Bioinf. 2009;75:430. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Biophys. J. 2009;96:L7. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimada J, Shakhnovich EI. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11175. doi: 10.1073/pnas.162268099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyeon C, Thirumalai D. Proc. Natl. Acad. Sci. U.S. A. 2005;102:6789. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JC, Thirumalai D. J. Am. Chem. Soc. 2008;130:14080. doi: 10.1021/ja8063638. [DOI] [PubMed] [Google Scholar]
- 20.Klimov DK, Thirumalai D. Fold. Des. 1998;3:127. doi: 10.1016/s1359-0278(98)00018-2. [DOI] [PubMed] [Google Scholar]
- 21.Gosavi S, Chavez LL, Jennings PA, Onuchic JN. J. Mol. Biol. 2006;357:986. doi: 10.1016/j.jmb.2005.11.074. [DOI] [PubMed] [Google Scholar]
- 22.Rieder U, Lang K, Kreutz C, Polacek N, Micura R. Chembiochem. 2009;10:1141. doi: 10.1002/cbic.200900155. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. J. Comp. Chem. 1995;16:1339. [Google Scholar]
- 24.Thirumalai D, Lee N, Woodson SA, Klimov DK. Annu. Rev. Phys. Chem. 2001;52:751. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 25.Tinoco I, Bustamante C. J. Mol. Biol. 1999;293:271. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 26.Zwanzig R. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9801. doi: 10.1073/pnas.92.21.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson SE. Fold. Des. 1998;3:R81. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 28.Plaxco KW, Simons KT, Ruczinski I, David B. Biochemistry. 2000;39:11177. doi: 10.1021/bi000200n. [DOI] [PubMed] [Google Scholar]
- 29.Nixon PL, Giedroc DP. Biochemistry. 1998;37:16116. doi: 10.1021/bi981726z. [DOI] [PubMed] [Google Scholar]
- 30.Theimer CA, Giedroc DP. RNA. 2000;6:409. doi: 10.1017/s1355838200992057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, Wen JD, Tinoco I. RNA. 2007;13:2175. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green L, Kim CH, Bustamante C, Tinoco I. J. Mol. Biol. 2008;375:511. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho SS, Pincus DL, Thirumalai D. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17349. doi: 10.1073/pnas.0906625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. Biochemistry. 2005;44:13404. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 35.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. Mol. Cell. 2005;18:49. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 36.Dawson WK, Fujiwara K, Kawai G. PLos ONE. 2007:2. doi: 10.1371/journal.pone.0000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.