Abstract
We describe a genome annotation service provided by the Entrez browser, http://www.ncbi.nlm.nih.gov/entrez. All protein products identified in fully sequenced microbial genomes have been compared with proteins with known 3-D structure by use of the BLAST sequence comparison algorithm. For the ∼20% of genome proteins in which unambiguous sequence similarity is detected, Entrez provides a link from the gene product to its predicted structure. The service uses the Cn3D molecular graphics viewer to present a 3-D view of the known structure, together with an alignment display mapping conserved residues from the genome protein onto the known structure. Using an example from Aeropyrum pernix, we illustrate how mapping to a 3-D structure can confirm predictions of biological function.
The rapid growth of sequence data from genome sequencing projects has produced a huge set of new protein sequences. These sequences are generally derived by translation of ORFs in the genomic sequence, with no experimental evidence for expression or function. Functional characterization is most often obtained by comparison with sequences already in public databases. When some of these sequence neighbors have known 3-D structure, this information may further assist assignment of biological function. For example, a biologist may check whether the residues involved in specific ligand binding in the known structure are conserved in the newly sequenced protein. Further examination of structure neighbors, as detected by structure–structure comparison, may identify additional binding sites and/or ligands.
Current efforts in structural genomics aim to provide either an experimental structure or detailed theoretical model for every protein from fully sequenced genomes (Kim 1998; Brenner et al. 1999; Burley et al. 1999; Eisenstein et al. 2000; Mallick et al. 2000; Shapiro and Harris 2000; Skolnick et al. 2000). While these projects are in progress, however, much can be learned by simply mapping residues of the newly sequenced protein onto the known structures of homologs. This analysis requires relatively straightforward computational tools, and it can be performed systematically and the results updated as additional genomes are sequenced and new structures are determined. What is needed is an efficient method for sequence comparison and molecular-graphics software for display of the corresponding sequence-structure alignments. The results may then be incorporated into a retrieval service accessible to genome researchers.
Here we describe an annotation service of this kind, on the basis of links between the Genome database of Entrez (Tatusova et al. 1999) and its 3-D-structure Database, MMDB (Wang et al. 2000a). Significant sequence similarity between genome proteins and proteins with known structure is detected by use of the BLAST algorithm (Altschul et al. 1997) and recorded as a genome-to-structure link. When a user identifies a protein of interest and follows this link, the corresponding sequence-to-structure alignment is displayed by use of Cn3D (Wang et al. 2000b), a molecular-graphics viewer that operates on a PC, MacIntosh, and other popular computers. As an example, we examine a hypothetical protein from Aeropyrum pernix K1 (Kawarabayasi et al. 1999), showing how Entrez's link to 3-D structure strongly supports identification as a phosphoenolpyruvate carboxykinase (PCK).
RESULTS AND DISCUSSION
Using Entrez to Find Links to 3-D Structure
Links from genome proteins to 3-D structures are available from the Entrez Genomes home page for each fully sequenced microbial genome. One may find a list of the available genomes by choosing http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Genome, followed by Microbial Genomes. The listing contains 28 fully sequenced genomes at the time of writing, but is updated as new genome sequence data are deposited in GenBank (Benson et al. 2000). The home page for an individual organism is reached by choosing its name, for example Aeropyrum pernix K1. Fully sequenced genomes are also recorded in Entrez's nucleic acid sequence database, and predicted proteins in Entrez's protein sequence database. Thus, one may also search for a particular protein name, author name, or other term using Entrez's sequence databases, and then follow the links to the home page of the corresponding genome (Wheeler et al. 2000). A list of proteins from all complete genomes with sequence similarity to proteins of known 3-D structure is provided at http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/PDB_bact.html.
By choosing the 3-D structure link from the genome home page, the user is presented with a tabular listing of genome proteins in which BLAST has identified significant similarity to one or more proteins with known 3-D structure. By choosing the Aeropyrum pernix protein coded by gene APE0033, for example, one sees that BLAST has identified significant similarities to 36 other proteins, 2 of which have known structure. Following the link to the ;neighbors with 3-D structure, one finds the list of structures shown in Figure 1. The home page for each genome also provides ways to search for a particular protein. The tabular listing of links-to-structure contains gene names and protein names, and one may search for a protein of interest using the find feature of a WWW browser. One may also select a region from the physical map of the genome, or use the listing of BLAST sequence neighbors grouped by taxonomic superkingdom. These routes also lead to a listing of any available links to 3-D structure.
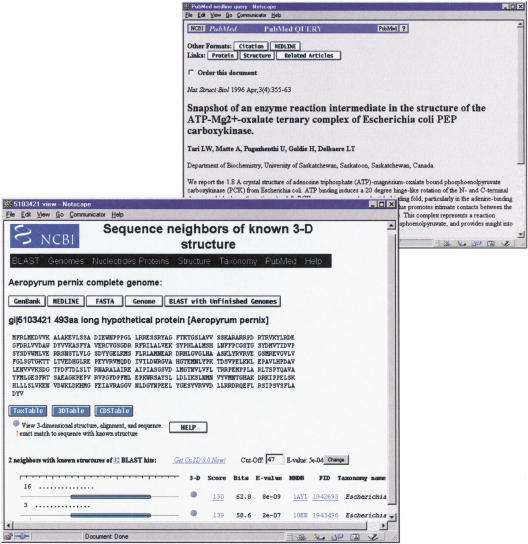
Figure 1.
The web page showing sequence-structure alignments for the protein product of the Aeropyrum pernix gene APE0033. Links from 1AYL, a homolog with known structure, lead to the MEDLINE abstract describing the enzymatic mechanism of this PCK.
Because BLAST may detect significant similarity to several proteins of known structure, genome-to-structure links are presented as rows in a table, as shown in Figure 1. Links in the column labeled 3-D launch the Cn3D molecular graphics viewer to display the genome protein mapped onto any one of the known 3-D structures. The Cn3D program can be downloaded freely by use of the get Cn3D link provided. The graphical representation of the sequence-structure alignment shows the region of the genome protein that has been aligned with each known structure. The column labeled MMDB brings one to the home page of each structure, where further links describe that structure. These include links to MEDLINE citations and links to the structure neighbors of that protein, as calculated by structure-structure comparison. Also shown are BLAST similarity scores, with links that display conventional alignments.
Using 3-D Structure Links in Genome Annotation
The product of the Aeropyrum pernix gene we consider as an example, APE0033, was originally annotated only as a hypothetical protein (Kawarabayasi et al. 1999). BLAST comparison with other protein sequences suggests a possible function, as many (although not all) of these sequence neighbors are annotated as phosphoenolpyruvate carboxykinases (PCKs). Sequence similarity is not very high, however, with only 25% identical residues in the best-scoring BLAST alignments, and it is far from certain that PCK function is conserved. It is in this situation that the availability of a 3-D-structure link can be particularly valuable. From the 3-D structure one can often identify residues required for molecular function, such as those required for PCK activity. Using the sequence-structure alignment, one may then ask whether these specific residues are conserved in the genome protein. This computational analysis cannot prove that function is conserved, or that APE0033 is a PCK, but it can provide stronger evidence than sequence similarity alone.
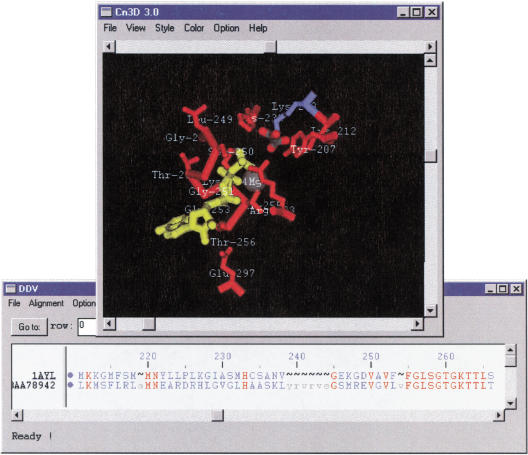
Figure 2 shows the sequence-structure alignment of the APE0033 protein with one of its sequence neighbors with known 3-D structure, 1AYL, a PCK. In Cn3D's structure window, one may identify the sites in which substrates adenosine-triphosphate (ATP), magnesium (Mg2+), and oxalate are bound. In Cn3D's sequence window, one sees the BLAST alignment of the APE0033 protein onto 1AYL. PCK is an important gluconeogenic enzyme, catalyzing the diversion of tricarboxylic acid cycle intermediates toward gluconeogenesis. Its catalytic mechanism has been studied well, and the crystallographers reporting the structure identified several active site residues (Tari et al. 1996). By selecting these residues in the structure window, one sees the corresponding residues highlighted in the sequence window. One may also see that these residues, which include the ATP-binding motifs (GX4GKTT) and (X4D), are highly conserved in the aligned sequence of the APE0033 protein.
Figure 2.
Cn3D display of 1AYL's structure and the sequence-structure alignment between 1AYL and the protein product of APE0033. Aligned residues are shown in uppercase in the sequence window, and identical ones are colored in red. Selected functional residues in 1AYL are shown in the structure window, along with substrates and cofactors. It can be seen from the alignment, that the majority of these residues are conserved in 1AYL and APE0033.
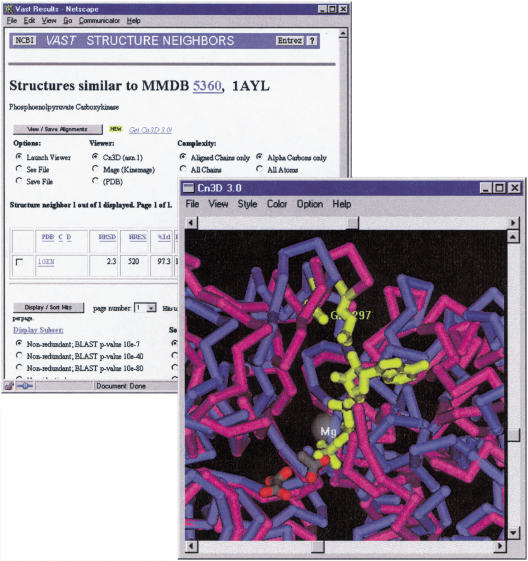
Further insight into structure-function relationships can sometimes be gained by examining other structures related to the genome protein. Structure 1OEN (Matte et al. 1996), for example, appears as both a sequence neighbor of the genome protein APE0033 and as a structure neighbor of 1AYL. This is a PCK almost identical to 1AYL, but crystallized in the unliganded state. By selecting 1OEN from among the structure neighbors of 1AYL, one may view the 3-D superposition of these two proteins, as shown in Figure 3. By using the Cn3D viewer to display an alternating animation, one may see that the protein undergoes a significant hinge motion to close the active site cleft when the substrate is bound. One may also identify residues that are brought into proximity to one another and/or the substrate by this motion, for example, 1AYL residue E297. By examining the sequence-structure alignment of APE0033 onto 1AYL, one may see that this residue is also conserved in APE0033, further suggesting that this protein possesses the same enzymatic mechanism as known PCK.
Figure 3.
Three-dimensional superposition of two PCK structures, in the presence and absence of substrates. The liganded structure 1AYL is rendered in magenta and the substrate-free structure 1OEN in blue. It can be seen that the active site cleft is closed in upon binding and that residue E297 moves into close proximity to the substrate. E297 is conserved in the protein product of APE0033 (not shown).
METHODS
Protein sequences from microbial genomes are extracted from the Genome division of Entrez (Tatusova et al. 1999) and compared with the NCBI non-redundant sequence database. Similarity searches are performed with the BLAST 2.0 engine (Altschul et al. 1997) with default parameters (substitution matrix BLOSUM-62, gap penalties: existence: 11, extension: 1). Low-complexity regions on the query sequence are masked out with SEG (Wootton and Federhen 1993). Due to the large number of the resulting alignments, the output is stored in a binary index file containing sequence identifiers, taxonomy index, raw score, and the positions of the matching regions. The results are presented in a table as shown in Figure 1. For each pair of protein sequences, raw score, calculated normalized score (bits), and expectation value (E-value) are presented, allowing evaluation of the statistical significance of the similarity. The user can specify the score cutoff (in bits) to select the most significant alignments. With the current database size of ∼150,000,000, residues for a typical query protein of 250 amino acids to achieve a marginally significant E-value of 10e-4, a normalized score of ∼47 bits is necessary. Sequences from Entrez's 3-D structure database are selected from all the BLAST neighbors and linked to a visualization system.
Entrez's 3-D structure database, MMDB (Molecular Modeling Database) (Wang et al. 2000a), contains all experimentally determined structures obtained from the Protein Data Bank (Berman et al. 2000). It provides pre-calculated structure neighbors on the basis of systematic structure-structure comparisons by use of the VAST algorithm (Gibrat et al. 1996), as well as links to MEDLINE and other resources. Entrez's 3-D viewer, Cn3D (Wang et al. 2000b), is a visualization tool for structure, sequence, and alignment, which functions as a helper application for WWW browsers. The highlighting function is a useful way for mapping residues between the sequence and structure view. Further information is available from http://www.ncbi.nlm.nih.gov/Structure/CN3D.
Acknowledgments
We thank Tom Madden for technical advice and Jim Ostell for useful discussions. We thank the NIH intramural research program for support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tatiana@ncbi.nlm.nih.gov; FAX (301) 480-9241.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.143200.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Barken D, Levitt M. The PRESAGE database for structural genomics. Nucleic Acids Res. 1999;27:151–153. doi: 10.1093/nar/27.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Almo SC, Bonanno JB, Capel M, Chance MR, Gaasterland T, Lin D, Sali A, Studier FW, Swaminathan S. Structural genomics: Beyond the human genome project. Nat Genet. 1999;23:151–157. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- Eisenstein E, Gilliland GL, Herzberg O, Moult J, Orban J, Poljak RJ, Banerjei L, Richardson D, Howard AJ. Biological function made crystal clear – annotation of hypothetical proteins via structural genomics. Curr Opin Biotechnol. 2000;11:25–30. doi: 10.1016/s0958-1669(99)00063-4. [DOI] [PubMed] [Google Scholar]
- Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- Kim SH. Shining a light on structural genomics. Nature Struct Biol. 1998;(Suppl):643–645. doi: 10.1038/1334. [DOI] [PubMed] [Google Scholar]
- Mallick P, Goodwill E, Fitz-Gibbon S, Miller JH, Eisenberg D. Selecting protein targets for structural genomics of Pyrobaculum aerophilum: Validating automated fold assignment methods by using binary hypothesis testing. Proc Natl Acad Sci. 2000;97:2450–2455. doi: 10.1073/pnas.050589297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matte A, Goldie H, Sweet RM, Delbaere LT. Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: A new structural family with the P-loop nucleoside triphosphate hydrolase fold. J Mol Biol. 1996;256:126–143. doi: 10.1006/jmbi.1996.0072. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Harris T. Finding function through structural genomics. Curr Opin Biotechnol. 2000;11:31–35. doi: 10.1016/s0958-1669(99)00064-6. [DOI] [PubMed] [Google Scholar]
- Skolnick J, Fetrow JS, Kolinski A. Structural genomics and its importance for gene function analysis. Nat Biotechnol. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- Tari LW, Matte A, Pugazhenthi U, Goldie H, Delbaere LT. Snapshot of an enzyme reaction intermediate in the structure of the ATP-Mg2+-oxalate ternary complex of Escherichia coli PEP carboxykinase. Nat Struct Biol. 1996;3:355–363. doi: 10.1038/nsb0496-355. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Madden T. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Karsch-Mizrachi I, Ostell J. Complete genomes in WWW Entrez: Data representation and analysis. Bioinformatics. 1999;15:536–543. doi: 10.1093/bioinformatics/15.7.536. [DOI] [PubMed] [Google Scholar]
- Wang Y, Addess K, Geer L, Madej T, Marchler-Bauer A, Zimmerman A, Bryant S. MMDB: 3D structure data in Entrez. Nucleic Acids Res. 2000a;28:243–245. doi: 10.1093/nar/28.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lewis G, Chappey C, Kans J, Bryant SH. Cn3D: Sequence and structure views for Entrez. Trends Biochem Sci. 2000b;6:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Chappey C, Lash AE, Leipe DD, Madden TL, Schuler GD, Tatusova TA, Rapp BA. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2000;28:10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Computers & Chemistry. 1993;17:149–163. [Google Scholar]