Abstract
Obesity, a major risk factor for type II diabetes, is becoming more prevalent in Western populations consuming high calorie diets while expending less energy both at the workplace and at home. Most human obesity, and probably most type II diabetes as well, reflects polygenic rather than monogenic inheritance. We have genetically dissected a polygenic mouse model of obesity-driven type II diabetes by outcrossing the obese, diabetes-prone, NZO (New Zealand Obese)/HlLt strain to the relatively lean NON (Nonobese Nondiabetic)/Lt strain, and then reciprocally backcrossing obese F1 mice to the lean NON/Lt parental strain. A continuous distribution of body weights was observed in a population of 203 first backcross males. The 22% of first backcross males developing overt diabetes showed highest peripubertal weight gains and earliest development of hyperinsulinemia. We report a complex diabetes-predisposing (“diabesity”) QTL (Quantitative Trait Loci) on chromosome 1 contributing significant main effects to increases in body weight, plasma insulin, and plasma glucose. NZO contributed QTL with significant main effects on adiposity parameters on chromosomes 12 and 5. A NON QTL on chromosome 14 interacted epistatically with the NZO obesity QTL on chromosome 12 to increase adiposity. Although the main effect of the diabetogenic QTL on chromosome 1 was on rapid growth rather than adiposity, it interacted epistatically with the obesity QTL on chromosome 12 to increase plasma glucose levels. Additional complex epistatic interactions eliciting significant increases in body weight and/or plasma glucose were found between the NZO-contributed QTL on chromosome 1 and other NZO-contributed QTL on chromosomes 15 and 17, as well as with an NON-contributed QTL on chromosome 2. We further show that certain of these intergenic interactions are predicated on, or enhanced by, the maternal postparturitional environment. We show by cross-fostering experiments that the maternal environmental influence in part is because of the presence of early obesity-inducing factors in the milk of obese F1 dams. We also discuss a strategy for using recombinant congenic strains to separate and reassemble interacting QTL for future study.
Simpson's paradox (Yule 1903; Simpson 1951) is a statistical phenomenon in which marginal effects, e.g., effects associated with a single genetic locus, can be masked, enhanced, or even reversed in the presence of interactions that are not detected and accounted for. The implications of Simpson's paradox for the study of complex diseases such as type II (non-insulin dependent) diabetes are that, in some cases, it may not be possible to predict a phenotype from a given genotype if the interactions among many components of the system cannot be fully characterized (Clark 2000). It is becoming increasingly clear that understanding a complex multifactorial disease such as type II diabetes requires methodology to identify interactions between multiple susceptibility genes expressing within certain environmental limits. To date, genome-wide scans of relatively isolated human populations with a high frequency of obesity-associated type II diabetes have uncovered only a few susceptibility genes with major effects (Hanis et al. 1996; Mahtani et al. 1996; Stern et al. 1996). Statistical methods to uncover intergenic interactions recently have been applied to genome-wide scan data that established the presence of NIDDM1, a major locus on human chromosome 2, and an unlinked susceptibility locus on chromosome 15 (Cox et al. 1999). Similar analysis of intergenic interaction uncovered another human susceptibility QTL (Quantitative Trait Loci) on chromosome 2 that interacted with NIDDM2, a diabetes locus on human chromosome 12, in a relatively isolated Finnish population (Mahtani et al. 1996). The genome-wide scan analysis for main effects had indicated only weak evidence for linkage of this QTL (F. Collins, pers. comm.).
The ability to control both genotype and environment in inbred populations of rodents greatly simplifies analysis of these complex gene–gene and gene–environment interactions. When compared with human populations, the genetics of an inbred line cross are relatively simple. Allelic variation is restricted to a small or moderate number of loci segregating at most two alleles and IBD (Identity by Descent) status can be inferred unambiguously from marker data. The ability to produce large populations of mice in a controlled environment provides an optimal situation for the statistical detection of genetic effects. Even with these advantages, we show in this report that type II diabetes in mice presents a challenge for statistical analysis because of interactions among multiple genetic and environmental factors.
Visceral obesity, a phenotype with high heritability, is the major risk factor for type II diabetes development in humans (Bjorntorp 1993). In mice, diabesity genes are defined as obesity-predisposing genes capable of interacting deleteriously with other susceptibility loci, as well as with environmental factors, to elicit a state of impaired glucose tolerance (IGT) and insulin resistance sufficiently severe to precipitate development of overt type II diabetes (Leiter and Herberg 1997). We previously have identified a collection of QTL from two unrelated inbred strains, the relatively lean NON (Nonobese Nondiabetic)/Lt strain and the markedly obese NZO (New Zealand Obese)/HlLt strain. NZO mice represent a model of polygenic, juvenile-onset obesity (Proietto and Larkins 1993). Obese NZO/HlLt mice of both genders show IGT, as well as insulin and leptin resistance (Halaas et al. 1997). However, only approximately half of NZO/HlLt males transit from IGT to overt diabetes by 24 weeks of age. NON males do not develop marked obesity, but show IGT, low insulin secretory responses (Leiter and Herberg 1997), and a more normal level of circulating leptin (data not shown). Crossing these two mouse strains showed that both parental genomes contributed to increase diabetes frequency to nearly 100% in F1 males (Leiter et al. 1998). Intercrossing these F1 hybrids to produce a segregating F2 generation permitted mapping of QTL from both strains contributing to diabetes subphenotypes (PG [Plasma Glucose] and PI [Plasma Insulin]). However, we could not identify diabesity genes because all F2 males became obese (Leiter et al. 1998). In the present report, we show that backcrossing the obese F1 hybrids to the NON/Lt parental strain allowed identification of epistatic diabesity QTL from NZO/HlLt. Furthermore, we show that penetrance of these QTL is postnatally regulated by factors in milk from obese dams.
RESULTS
Segregation of Diabesity in BC1 Males as a Threshold Phenomenon
In contrast to our previous inability to segregate the obesity phenotype in an F2 population, the reciprocal first backcross (BC1) of obese F1 hybrids of both genders to the relatively lean NON/Lt strain resulted in a continuous distribution of BW (Body Weight) between the two parental extremes (Fig. 1A). Of the 203 BC1 males that were monitored for type II diabetes development, 38% had an abnormal PG level >225 mg/dL before termination at 24 weeks of age. Diabetes (conservatively defined as chronic PG ≥300 mg/dL) was diagnosed in 22% of the cross, with an additional 16% classified as borderline diabetics because their PG oscillated between 225 and 300 mg/dL. Analysis of the phenotypic data showed that a BW threshold must be crossed for the mice to become at risk for diabetes (Fig. 1B). The threshold was progressive; at the 16-week time point, all mice that were to develop diabetes weighed >40 g, and by the 24-week time point, nearly all of the overtly diabetic males weighed in excess of 50 g. Crossing the threshold, however, was not a guarantee of diabetes, because fully 41% of those higher than the 24-week BW threshold remained normoglycemic. Hyperinsulinemia more extreme than observed in the NZO/HlLt parental males was the additional phenotype distinguishing obese BC1 males that eventually developed diabetes versus those that did not. NON/Lt parental males have a PI in the low normal range (1–2 ng/mL), whereas PI values in NZO/HlLt males are moderately elevated, ranging between 3 and 6 ng/mL. Of the BC1 males higher than the BW threshold at 24 weeks, those with an extremely elevated PI level (>24 ng/mL) had a mean PG in the diabetic range. Those mice showing hyperinsulinemia of intermediate severity (6–24 ng/mL) or had a normal insulin level (<6 ng/mL) maintained a mean PG in the normal range (Fig. 1C). Early hyperinsulinemia was also a predictor of eventual diabetes. Those mice already showing elevated PI level at 16 weeks (the earliest time point taken) also showed the highest rate of weight gain and the highest percentage of diabetes compared with mice developing later hyperinsulinemia at 20 weeks, 24 weeks, or not at all (Fig. 1D). Presumably, the early development of hyperinsulinemia contributed to an earlier development of insulin resistance that became progressively more severe as the males aged. In fact, the phenotypic cluster of highest BW gain during the peripubertal period and earliest development of marked hyperinsulinemia accounted for most of the diabetics in the cross. Continuous distribution of multiple phenotypes (BW, logPG, PI, BMI [Body Mass Index], AI [adiposity index], individual fat depot weights, and serum leptin) in these BC1 males permitted genome-wide scan for QTL with main effects to elucidate the genetic determinants of disease.
Figure 1.
Phenotypic relationships between body weight (BW), plasma insulin, and plasma glucose (PG). (A) Markedly different BW distributions distinguish the NZO/HlLt and NON/Lt parentals, whereas the BC1 males show a continuous BW distribution between these two extremes. (B) Relationship between BW and PG in this BC1 male population at 24 weeks showing that a BW threshold exists for diabesity development, with virtually all of the diabetic males having attained a BW ≥50 g. (C) Diabetes development in mice higher than the BW threshold is associated with extreme hyperinsulinemia. Males with BW >50 g but with a normal insulin level (<6 ng/mL) or even a high insulin level (6–24 ng/m) had normal mean PGs (216 + 33, 216 + 11mg/dl respectively) whereas those with an extremely high insulin level (>24 ng/mL) had a high mean PG (360 + 16mg/dl) (P < 0.0001). (D) Relationship between early development of hyperinsulinemia with rapid prematurational rate of weight gain. Males showing hyperinsulinemia by 16 weeks of age also showed the highest rate of weight gain and the highest rate of diabetes, accounting for the majority of the diabetics in the cross by termination. In Figure 1D, 12 mice were left out of the correlation because of oscillating phenotype and 16 because of missing data at one or more time points. Standard errors in D were so small that they could not be depicted in the figure.
Dominant NZO/HlLt QTL Contributing toward Attainment of Diabetogenic Thresholds
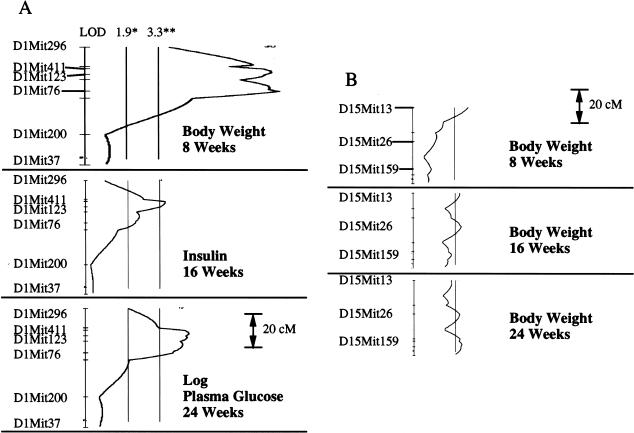
Genome-wide scan identified a region on proximal chromosome 1 that met the criterion for a diabesity QTL in affecting not only BW at 8, 16, and 24 weeks, but also in contributing significantly to the diabetes subphenotypes of increased PG at 20 and 24 weeks and early hyperinsulinemia at 16 weeks (Table 1). This NZO-derived QTL spans a large segment of chromosome 1 (delimited by the D1Mit411-D1Mit123-D1Mit76 interval, ∼14 cM) with different markers within this span affecting different phenotypes in an age-dependent fashion. Hence, it is probable that there may be more than one gene contributing to diabesity-related traits in this region (Fig. 2A). Interestingly, although the chromosome 1 effect gave a significant QTL for BMI, and suggestive linkage for three of the four fat pads weighed (inguinal, gonadal, and mesenteric; data not shown), it did not contribute significantly to AI or to serum leptin levels. This suggests that the primary effect of this QTL was on early growth parameters distinct from adipogenesis. The percentage of the variance contributed by this QTL was only 16% for BW and only 8% and 9% for PG and PI, respectively, clearly indicating that additional contributions were required for diabesity development. Indeed, our genome-wide scan also indicated significant evidence for a NZO-derived QTL on chromosome 15 affecting PG, a NZO-derived QTL on chromosome 12 affecting AI, BMI, and leptin, and a NZO-derived QTL on chromosome 5 affecting AI and leptin. Additionally, suggestive linkage by NZO-derived QTLs was indicated on chromosome 18 affecting AI and BMI, a locus on chromosome 3 affecting PI level, a locus on chromosome 13 affecting BMI, a second separate locus on chromosome 15 contributing to BW, BMI, and leptin, and a third separate locus on chromosome 15 contributing to early BW. Suggestive linkage from a homozygous NON-derived QTL was found on chromosome 6 contributing to BW and PI (Table 1). Many of these loci gave significant or suggestive QTL for one or more of the four fat pads weighed, with an additional locus, D4Mit166, that gave a significant LOD score for only gonadal fat pad weight and for no other parameter measured (data not shown). The complexity of the multiple QTL on chromosome 15 is depicted in Figure 2B and Table 1 by the demonstration of variable effects on diabesity subphenotypes over time. The most proximal locus, marked by D15Mit13, was associated with early BW increases, whereas the next distal locus, marked by D15Mit26 (20 cM away), was associated with later BW increase. The most distal locus, marked by D15Mit159 (another 20 cM away), was associated significantly with elevated PG and suggestively with BW.
Table 1.
Significant and Suggestive QTL Associations Detected by the Genome Scans for Main Effects
| Chr. | cM | Marker | BW8 | BW16 | BW24 | PI16 | LOG PG24 | BMI | AI | Leptin |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 8.3 | D1Mit296 | 4.12a | 2.69 | 2.89 | 2.18 | 1.98 | |||
| 1 | 15.0 | D1Mit211 | 7.03a | 5.36a | 6.13a | 3.01 | 3.48a | 3.42a | ||
| 1 | 18.5 | D1Mit411 | 9.28a | 7.23a | 7.54a | 4.57a | 4.85a | 4.25a | ||
| 1 | 21.0 | D1Mit123 | 9.36a | 6.69a | 7.00a | 4.37a | 4.92a | 4.07a | ||
| 1 | 25.7 | D1Mit213 | 8.32a | 5.66a | 5.75a | 4.10a | 4.63a | 3.18 | ||
| 1 | 32.8 | D1Mit76 | 8.49a | 4.91a | 4.99a | 3.82a | 4.00a | 2.00 | ||
| 1 | 43.1 | D1Mit46 | 4.61a | 2.93 | 2.77 | 2.55 | 2.09 | |||
| 3 | 33.7 | D3Mit22 | 2.53 | |||||||
| 5 | 28.0 | D5Mit81 | 2.81 | |||||||
| 5 | 45.0 | D5Mit7 | 3.86a | 3.18 | ||||||
| 6 | 25.5 | D6Mit275 | 2.16 | 2.48 | 1.92 | |||||
| 6 | 29.0 | D6Mit123 | ||||||||
| 12 | 45.0 | D12Mit259 | 2.14 | 2.04 | 2.45 | 3.42a | 2.34 | |||
| 12 | 48.0 | D12Mit231 | 2.00 | 2.45 | 3.07 | 3.96a | 4.88a | 3.34a | ||
| 12 | 52.0 | D12Mit132 | 2.50 | 3.29a | 4.47a | 3.22a | ||||
| 12 | 53.0 | D12Mit262 | 2.85 | 4.28a | 3.5a | |||||
| 12 | 59.0 | D12Mit150 | 2.21 | 2.85 | 2.46 | |||||
| 13 | 62.0 | D13Mit53 | 2.03 | |||||||
| 15 | 6.7 | D15Mit13 | 2.89 | 2.39 | ||||||
| 15 | 15.4 | D15Mit22 | 2.20 | 2.21 | 2.06 | |||||
| 15 | 29.0 | D15Mit26 | 2.67 | 2.48 | 2.61 | 2.30 | 3.02 | |||
| 15 | 49.0 | D15Mit72 | 2.34 | 2.88 | 2.20 | 2.58 | ||||
| 15 | 49.6 | D15Mit159 | 2.01 | 2.29 | 3.55a | 2.30 | ||||
| 15 | 57.8 | D15Mit217 | 2.17 | 2.99 | 2.15 | |||||
| 15 | 59.2 | D15Mit42 | 2.11 | |||||||
| 18 | 16.0 | D18Mit60 | 1.90 | 2.47 |
(Chr) chromosome; (BW) body weight; (PG) plasma glucose; (BMI) body mass index; (QTL) quantitative trait loci; (AI) adiposity index.
BW is shown at three different time points, 8, 16, and 24 weeks of age. Plasma Insulin (PI) is shown at the 16 wk time point. LOG PG, BMI, AI, and serum leptin are shown at the 24-week time point.
Exceeds the LOD 3.3 criterion for significance proposed by Lander and Kruglyak (1995). Centimorgan position from the Mouse Genome Database maintained by The Jackson Laboratory (www.informatics.jax.org).
Figure 2.
Complexity of QTL (Quantitative Trait Loci) on chromosomes 1 and 15. (A) The peak of the QTL for BW (Body Weight) and PG (Plasma Glucose) on chromosome 1 spans a 14-cM region that probably consists of two separate but equal peaks marked by D1Mit411 and D1Mit76. For plasma insulin, the peak is much stronger for D1Mit411, although there is a distinct shoulder marked by D1Mit76. (B) Three separate loci on chromosome 15 show a suggestive effect on BW at progressive time points. The locus marked by D15Mit13 is only associated with early BW and thus probably a growth modifier. The locus marked by D15Mit26, 20 cM distal, is associated with later BW, gain, body mass index, and leptin, suggesting that it could be an adipocyte growth modifier. The third locus, another 20 cM distal, and marked by D15Mit159, is suggestively associated with late BW gain and significantly associated with PG. (LOD) logarithm of odds; (*) threshold for suggestive QTL; (**) threshold for significant QTL.
Maternal Lactational Environment as an Interactive Factor Regulating Penetrance of Diabesity QTL
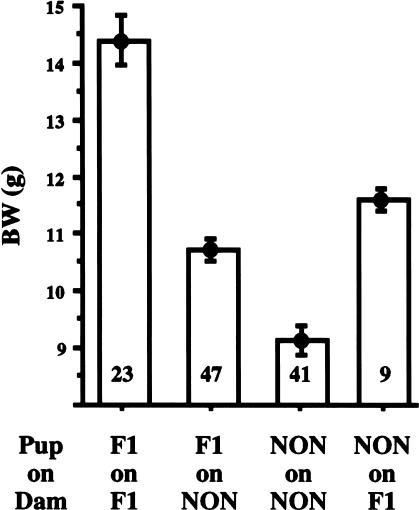
Maternal environment also contributed to the attainment of the BW-related diabetogenic threshold. Because (NZO × NON)F1 and (NON × NZO)F1 males and females were mated to NON/Lt males and females to generate the BC1 progeny, roughly half of the males had an obese F1 mother whereas the rest had a lean NON mother. The frequency of diabetes development was greater than twice as high in progeny from obese F1 mothers (32%) than from lean NON/Lt mothers (14%). This maternal influence significantly increased mean BW at 4 weeks of age, the earliest time point measured, and the increases persisted to the termination point at 24 weeks, suggesting that factors in milk from obese dams fundamentally altered early growth parameters. In addition, mean PG, PI, BMI, AI, and leptin levels were significantly higher in males with an obese F1 mother (Table 2). Contributions of X- and Y-linked genes could be excluded because reciprocal F1 hybrid combinations were used to backcross both males and females to NON mice. Although neither NZO/HlLt nor F1 virgin females develop overt diabetes, they do show IGT when challenged with a bolus of glucose administered intraperitoneally. Glucose tolerance tests performed on a pair of pregnant F1 females confirmed presence of IGT, but not fasting hyperglycemia (data not shown). That the early postnatal lactational environment was an essential covariable for diabesity gene–gene interactions to achieve diabetogenic thresholds was shown by cross-fostering experiments (Fig. 3). Litters of newborn F1 pups (in which the genetic variance is fixed) were randomized and fostered onto either lactating obese F1 or nonobese NON females. Postnatal day 18 BW of F1 pups suckled by obese F1 dams averaged 3.7 g more than F1 pups fostered onto lean NON lactating females (14.4 ± 0.4 vs. 10.7 ± 0.2 g). Similarly, NON pups fostered on F1 dams were 2.4 g larger than NON pups fostered on lactating NON dams (11.6 ± 0.2 g vs. 9.1 ± 0.3 g). Preliminary analysis indicated the lipid content of milk from obese F1 dams to be almost twice as high as that of lean NON females (40% vs. 22% v/v) and to contain four times the amount of leptin (4.3 ng/mL vs. 1.0 ng/mL). Thus, although we cannot assess the contribution of maternal obesity during gestation, it is clear that milk from obese F1 dams contains increased levels of diabesity-promoting factors compared with milk from lean NON/Lt dams.
Table 2.
Obese Maternal Environment Contributes to Diabetogenic Thresholds
| Dam | n | BW (g) 4 wk | BW (g) 24 wk | PI (ng/mL) 16 wk | PG (mg/dL) 24 wk | BMI 24 wk | AI 24 wk | Leptin (ng/mL) 24 wk |
|---|---|---|---|---|---|---|---|---|
| F1 (obese) | 89 | 23.4 ± 0.4 | 53.7 ± 0.7 | 10.3 ± 1.0 | 282 ± 14 | .422 ± .004 | 13.4 ± .25 | 51.1 ± 2.0 |
| NON (lean) | 114 | 21.5 ± 0.3 | 48.8 ± 0.6 | 6.1 ± 0.7 | 208 ± 7 | .393 ± .004 | 12.4 ± .20 | 41.8 ± 2.0 |
(BW) body weight; (PI) plasma insulin; (PG) plasma glucose; (BMI) body mass index; (AI) adiposity index.
Values are mean ± SEM. Differences all significant at P ≤ 0.0003 except P = 0.0035 for AI and P = 0.0015 for leptin.
Figure 3.
Maternal lactational effects on early weight gain. F1 and NON pups fostered on different lactational environments (obese F1 dam vs. lean NON dam) are significantly larger at postnatal day 18 if suckled on the obese F1 dam (P < 0.0001). Genotypic difference between F1 pups and NON pups is indicated by the fact that F1 pups will become significantly larger on the same lean NON lactational environment (P < 0.0001). Litter size was standardized to five to eight pups (mixed genders). Numbers of pups tested are shown in the bars. (BW) Body weight; (NON) nonobese nondiabetic.
Intergenic and Gene–Maternal Environment Interactions
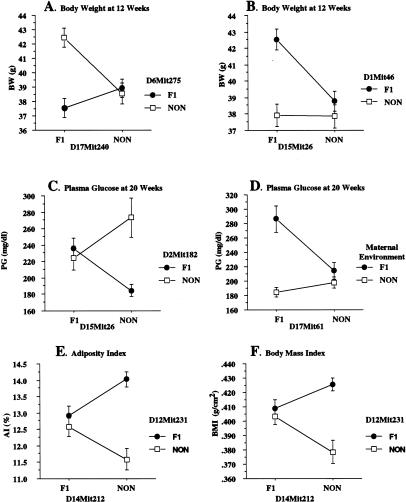
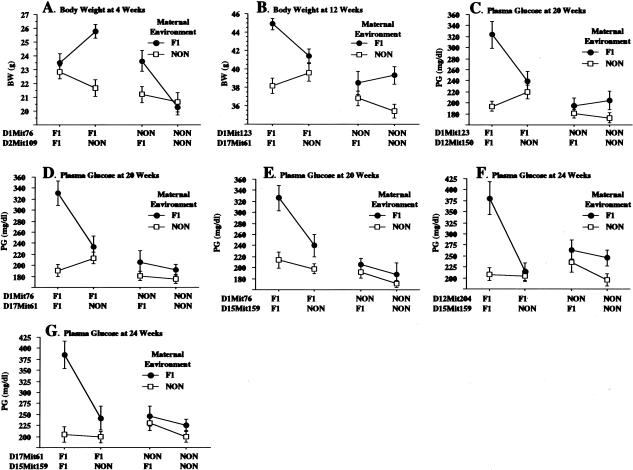
When considered individually, the diabesity QTL on chromosome 1 and the maternal effect both make significant contributions to the diabesity phenotypes. However, a general linear model analysis (ANOVA) suggested that these effects were not strictly additive. A genome-wide scan was performed to search for pairwise interactions among all pairs of loci and/or the maternal environment. Four interacting pairs were identified (Table 3 and Fig. 4), all contingent on maternal environment. These included the QTLs with significant or suggestive main effects already noted on chromosomes 1, 6, 12, and 15 as well as additional QTL on chromosomes 2, 14, and 17 (NON-derived modifier at D2Mit182, NON-derived modifier at D14Mit212, and NZO-derived modifier at D17Mit240, respectively). Note that both alleles of the QTL marked by the D15Mit26 polymorphism interact with separate loci to enhance separate phenotypes (Fig. 4B,C). Thus, heterozygosity for the NZO allele at D15Mit26 interacts epistatically with the NZO allele on chromosome 1 marked by D1Mit46 to increase the 8-week BW phenotype (Fig. 4B), whereas NON homozygosity at D15Mit26 interacts epistatically with D2Mi182 on chromosome 2 to elevate the 20-week PG phenotype (Fig. 4C). A second all-pairs genome-wide scan was conducted to search for three-way interactions. The relatively small sample size of ∼200 mice and the magnitude of the multiple testing implicit in a three-way scan reduce the power to detect three-way interactions. Thus, we restricted our search to three-way interactions between marker pairs and maternal environment. This search identified four significant interacting pairs (Table 4 and Fig. 5), each of which includes a chromosome 1 locus in combination with loci from chromosome 2, 12, or 17 (Fig. 5A–D). All diabetogenic alleles were contributed by the NZO/HlLt genome with the exception of the chromosome 2 contribution (D2Mit109, interestingly, a second separate locus 44 cM distal to D2Mit182 cited above), which represented a homozygous contribution from NON/Lt. The many interactions between NZO and NON-derived loci help explain the synergism of the two parental genomes that leads to nearly 100% diabetes in the F1 males. In addition to the significant interaction effects shown in Tables 3 and 4, several interactions achieved near-significant levels in our analysis (Table 4 and Fig. 5E–G). These involved the maternal environment interacting with the QTL on chromosomes 1 and 15 and with pairs of loci (12 × 15, 15 × 17) separate from the chromosome 1 diabesity complex. Although the main effects of some of these loci individually were on obesity phenotypes, as pairs they become diabesity loci by interacting to elevate PG.
Table 3.
Significant Pairwise Interactions between Marker Pairs or between a Marker and the Maternal Environment Are Summarized
| Trait | Marker 1 | Marker 2 | F overall | F interaction |
|---|---|---|---|---|
| BW | D6Mit275 | D17Mit240 | 10.83 (1.3 × 10−6) | 18.96 (2.1 × 10−5) |
| BW | D1Mit46 | D15Mit26 | 12.71 (1.2 × 10−7) | 9.08 (2.9 × 10−3) |
| PG | D2Mit182 | D15Mit26 | 9.72 (5.1 × 10−6) | 13.10 (3.7 × 10−4) |
| PG | D17Mit61 | F1 Dam | 12.97 (9.0 × 10−8) | 11.44 (8.7 × 10−4) |
| AI | D12Mit231 | D14Mit212 | 12.62 (1.4 × 10−7) | 13.96 (2.4 × 10−4) |
| BMI | D12Mit231 | D14Mit212 | 11.03 (9.9 × 10−7) | 12.92 (4.1 × 10−4) |
(BW) body weight; (PG) plasma glucose; (AI) adiposity index; (BMI) body mass index.
The marker names, the overall F statistic (with 3 and 199 df), and the interaction F statistic (with 1 and 199 df) are shown. Nominal p values are shown in parentheses. Marker pairs exceeding the critical value (Foverall > 8.30 and Finteraction > 8.06) were deemed to be significant.
Figure 4.
Pairwise interactions between unlinked QTL. (A) The BW-increasing interaction between the homozygous NON allele marked by D6Mit275 with the distal NZO-derived locus identified on chromosome 17 and marked by D17Mit240. Both alleles failed to show a main effect QTL because their effect is dependent on the presence of the other locus which, in a backcross, would only happen in ∼25% of the population. (B) The BW-increasing interaction between the “diabesity” QTL from NZO on chromosome 1 (here marked by D1Mit46) and the NZO-derived allele marked by D15Mit26. (C) The NON-derived effect at D2Mit182 elevating PG is only apparent when a NON-derived modifier at D15Mit26 is present in homozygous state. (D) The elevation in PG attributable to the interaction between the F1 maternal environment with the NZO-derived allele marked by D17Mit61. (E,F) Homozygosity for the NON-derived allele on chromosome 14 marked by D14Mit212 epistatically enhances the ability of the strong NZO-derived QTL on chromosome 12 marked by D12Mit231 to increase AI and BMI. (BW) body weight, (NON) nonobese nondiabetic, (PG) plasma glucose, (AI) adiposity index, (BMI) body mass index.
Table 4.
Significant Three-Way Interactions Involving a Marker Pair and Maternal Environment Are Summarized
| Trait | Marker 1 | Marker 2 | F overall | F interaction |
|---|---|---|---|---|
| BW | D1Mit76 | D2Mit109 | 8.90 (1.4 × 10−8) | 10.70 (1.3 × 10−3) |
| BW | D1Mit123 | D17Mit61 | 9.00 (1.1 × 10−8) | 9.22 (2.7 × 10−3) |
| PG | D1Mit123 | D12Mit150 | 8.40 (4.0 × 10−8) | 14.17 (2.2 × 10−4) |
| PG | D1Mit76 | D17Mit61 | 10.37 (5.4 × 10−10) | 15.24 (1.3 × 10−4) |
| PGa | D1Mit76 | D15Mit159 | 10.17 (8.3 × 10−10) | 4.07 (4.5 × 10−2) |
| PGa | D12Mit204 | D15Mit159 | 5.57 (2.3 × 10−5) | 10.92 (1.1 × 10−3) |
| PGa | D17Mit61 | D15Mit159 | 6.89 (1.2 × 10−6) | 12.15 (6.0 × 10−4) |
(BW) body weight; (PG) plasma glucose.
The marker names, the overall F statistic (with 6 and 195 df), and the interaction F statistic (with 1 and 195 df) are shown. Nominal p values are shown in parentheses.
Not significant by the screening criteria but significant for either Foverall or Finteraction and borderline for the other.
Figure 5.
Three-way interactions with maternal environment. (A) The interaction between the F1 maternal environment, the NZO-derived allele marked by D1Mit76, and the homozygous NON-derived allele marked by D2Mit109. The interaction is only discernible at the very earliest time point (4 weeks) suggesting that it is particularly age dependent. (B) The increase in BW because of the interaction of the NZO-derived alleles marked by D1Mit123 and D17Mit61 with the F1 maternal environment. In the presence of the NON maternal environment, there is no significant effect of either locus. (C–G) The effects on PG of interactions between the F1 maternal environment and five separate pairs of NZO-derived alleles on chromosomes 1 × 12, 1 × 17, 1 × 15, 12 × 15, and 17 × 15. All five show a similar pattern of dramatically increased mean PG into the diabetic range only when all three diabetogenic parameters are present. (B) body weight; (NON) nonobese nondiabetic; (PG) plasma glucose.
DISCUSSION
We have identified a major NZO/HlLt-contributed diabesity QTL (probably comprising two or more separable genes) on chromosome 1 affecting early rapid somatic growth, development of early insulin resistance, and maturity-onset diabetes. Another QTL contributing significantly to hyperglycemia also appears to be complex. We previously identified non-insulin-dependent diabetes (Nidd) QTL in a segregating F2 population of males generated from reciprocal F1 hybrids between NZO/HlLt and NON/Lt (Leiter et al. 1998). Because polygenic obesity was present in all segregants, the Nidd QTL identified were associated with the diabetes subphenotypes of PG and/or PI but were actually negatively correlated with long-term weight gain. Two of these segregated as dominant contributions from the NON/Lt genome and were provisionally designated Nidd1 (chromosome 4) and Nidd3 (chromosome 18). Another, a recessive QTL derived from the NZO/HlLt genome, was designated Nidd2 (chromosome 11). More recently, others have used Nidd nomenclature for QTL identified in various mouse models of either polygenic obesity, IGT, or both (Hirayama et al. 1999; Ueda et al. 1999). However, note that the phenotype described in these later reports was not nonfasting hyperglycemia, the benchmark clinical phenotype defining diabetes in humans, but rather, obesity and IGT. For the new QTL identified in the present analysis, we have chosen not to use the provisional Nidd nomenclature because of the complexity of the gene–gene and gene–environment interactions required for diabetogenesis. For example, the NZO/HlLt-contributed QTL on chromosome 12 identified by genome scan exerted significant main effects on obesity subphenotypes (AI, leptin, BMI), but not PG or PI, suggesting that it should be designated as an obesity QTL, or Obq (Taylor and Phillips 1996), but not necessarily a Nidd QTL. However, interaction analysis showed that chromosome 12 QTL interacted epistatically with the chromosome 1 QTL complex to elevate PG, and that detection of this interaction was absolutely contingent on an obese F1 dam providing the postparturitional environment. A recent congenic analysis of the contribution of a mouse adiposity QTL required feeding a very high fat diet (York et al. 1999). Our model system of juvenile-onset obesity and maturity-onset type II diabetes in mice has provided an excellent illustration of how Simpson's paradox relates to a complex disease wherein gene–environment interactions are essential factors in development of a clinical phenotype. The system of interacting genes, like the composition of breast milk, is quite complex but certain regularities are apparent that suggest testable hypotheses. There appear to be separate QTL controlling early somatic growth and subsequent fat deposition that, in combination, will drive obesity and additional QTL that will drive subsequent diabetes onset through exacerbation of insulin resistance (reflected by extreme hyperinsulinemia). The chromosome 1 locus and the maternal environment are the major factors that drive early weight gain. Certain of these QTL may control components of the growth hormone/insulin-like growth factor system, because elevations in serum IGF-1 is the earliest endocrine anomaly we have observed in NZO/HlLt mice (Flurkey et al. 1998). The chromosome 1 locus contributes to transition from juvenile obesity and IGT into overt diabetes, presumably by catalyzing development of severe hyperinsulinemia whereas the maternal environmental contribution, entailing factors in the milk, enhances penetrance of the combined genetic susceptibilities. The activation of these major effects depends on the presence of one or more modifier loci. Among the modifiers, the NZO/HlLt-derived chromosome 17 locus, marked by D17Mit61, has the largest effect for BW and PG when combined with the QTL on chromosomes 1 and 15. A second chromosome 17 interaction with an NON/Lt-derived locus on chromosome 6 affecting BW also appears to be a major contributor. This interaction is with a distal locus, marked by D17Mit240, suggesting that multiple genes on chromosome 17 may be contributing to diabetes development. Of course, these interactions are not occurring on their own, but simultaneously with the other pairwise and three-way interactions. Unfortunately, the sample size is not large enough to look for four-way interactions, but ANOVA analysis suggests that these three-way interactions marked by the maternal environment, chromosome 1 and a second locus (chromosome 12, 15, or 17) are dependent on a third diabetogenic allele also being present from that group. Thus, it is quite likely that a congenic strain with just a single locus will not be diabetes prone, especially in view of our experience and that of others, and that a QTL on a given chromosome entails contributions from multiple loci (Legare et al. 2000). This certainly has been true for fine mapping of the genes predisposing the NOD/Lt strain to insulin-dependent diabetes (Podolin et al. 1998; Serreze et al. 1998).
Under these circumstances, a recombinant congenic strain (made by inbreeding a second backcross to NON/Lt) would be required to fix numbers of interactive NZO/HlLt and NON/Lt QTL necessary to exceed a diabetogenic threshold. Indeed, we have completed preliminary analysis of a matched pair of incipient recombinant congenic strains. Both (sub)strains derived from a common stem mating and thus share the chromosome 1 diabesity QTL from NZO/HlLt as well as numerous other known NZO and NON diabesity-associated QTL. They differ only by a recombination event that allowed one line to be selected for a truncated segment of the NZO-derived diabesity region on chromosome 15 marked by D15Mit159. The substrain carrying NON/Lt genome at this marker is diabesity resistant, whereas the substrain with NZO genome spanning this marker develops diabetes. Although it is premature to select candidate genes on chromosome 1, the finding of a diabesity requirement for NZO genome at D15Mit159 marker suggests two interesting candidates, peroxisome proliferator activated receptor alpha (Ppara) and carnitine palmitoyltransferase (Cpt1b). Either of these two genes may be defective in their ability to interact with loci controlling fatty acid oxidation on chromosome 1. Such a potential interaction is suggested by the finding of diabetogenic accelerants in the milk of obese F1 dams, which appears to be lipid enriched. The biochemical properties of milk are quite complex and involve a plethora of nutrients and bioactive factors, many of which become active on digestion. Milk contains vitamins, minerals, sugars, and fats along with enzymes, growth hormones, and immune cells to provide the infant/pup with complete nourishment until weaning (Kunz et al. 1999). Although milk constituents have received considerable attention as potential triggers for autoimmune (type 1) insulin-dependent diabetes (Norris and Pietropaolo 1999), our results in an animal model clearly indicate the potential for milk factors from obese mothers to serve as early triggers for type II diabetes development. The mice in this study model for juvenile obesity in humans. Pima Indians represent a human population in which juvenile obesity is prevalent, and in which mothers are frequently obese. Studies analyzing the obesity status of the Pima mother have shown that children born either to obese females or to underweight females, but not normal weight females, are at higher risk to develop type II diabetes (Pratley 1998). Although a recent study in normal weight human females indicates that breast feeding is highly beneficial when compared with feeding cow's milk–based infant formulas in preventing juvenile obesity (von Kries et al. 1999), the study did not evaluate whether breast milk from overweight females might be quantitatively different. Gestational obesity in the mother has been correlated with increased obesity in offspring (Levin and Govek 1998). The present study in mice raises the clear possibility that certain genetic combinations might create an excess or dearth of one or more nutrients and/or factors in breast milk that would have a profound effect on metabolically “imprinting” the postnate for later obesity and its sequelae.
Recognizing the extent and nature of the genetic complexity that underlies the many phenotypes of interest will be a key step in unraveling the complexity of type II diabetes in humans. It is becoming increasingly apparent that many phenotypes of biomedical importance in humans are influenced by multiple gene–gene and gene–environment interactions. Thus, it is essential that we begin to see the whole of a complex phenotype as more than the sum of independent, additive effects. Identification of disease modifying QTL deriving from both sides of a pedigree is becoming increasingly more common (Ranheim et al. 1997; Kido et al. 2000; Kuida and Beier 2000). New statistical approaches for the detection and characterization of interactions are needed. Mouse models of human disease can reduce the complexity of multiple genetic and environmental interactions that confound the detection of genes in human populations. As such, they are essential tools for the eventual discovery of the relevant genes.
METHODS
Mice and Phenotype Characterization
NZO/HlLt males and females were mated to NON/Lt males and females to generate reciprocal F1 hybrids that then were backcrossed to NON/Lt males and females. Mice were fed NIH-31 (4% fat) grain and housed in double pen Plexiglass boxes with free access to food and water. All mice shared the same mouseroom with controlled temperature and humidity and a 12-h light/dark cycle. Two hundred three male progeny were aged to 24 weeks with BW (in grams) measured every week, PG (mg/dL) measured every 4 weeks (glucose analyzer; Beckman Instruments), and PI (ng/mL) measured at weeks 16, 20, and 24 by rat insulin radioimmune assay kit (Linco). On termination at 24 weeks, four fat pads (inguinal, retroperitoneal, gonadal, and mesenteric) were removed and weighed to calculate total fat (sum of four fat pad weights taken), AI (weight of four fat pads [g]/body weight [g] × 100], and BMI (body weight [g]/body length [cm2]).
Genotyping
DNA was initially isolated from 5-mm tail clips and later from frozen tissue (liver or kidney). A genome scan was conducted by polymerase chain reaction using 83 microsatellite markers under conditions recommended by the supplier (Research Genetics) using various cyclers (MJ Research and Perkin-Elmer), and the products were separated on 4% Metaphor/LE (3:1) agarose gels (BMA). Markers were distributed ∼20 cM apart with higher concentrations of markers in areas around suggestive QTL linkage.
Statistical Analysis
We performed three genome scans: first to look for QTL with main effects; second to look for pairwise interactions, including interaction with maternal environment; and third to look for three-way interactions involving pairs of loci interacting with maternal environment. Genome scans for main effects were performed by one-way ANOVA of the phenotype across the genotype classes at each marker locus. Significance of the ANOVA F statistic (with 1 and 201 degrees of freedom) was assessed by permutation analysis (Churchill and Doerge 1994). Based on 1000 permutations, the 0.05 critical value was estimated to be 11.76, corresponding to a LOD (Logarithm of Odds) score of 2.6. Pairwise interactions were assessed by a two-stage procedure. In the first stage, significant marker pairs among all possible pairs were identified by computing the overall F statistic in a two-way ANOVA. This statistic compares the likelihood under the full model (with two main effects and an interaction) with the likelihood under a null model of no genetic effects. It has 3 and 199 d.f. Significance of marker pairs was assessed by permutation analysis of the overall F statistic to account for multiple testing across markers and traits. Marker pairs with an overall F statistic exceeding 8.3 were deemed to be significant at a genome-wide 0.05 level. Once a significant marker pair was identified, a second stage test to assess the significance of the interaction term was conducted. An F statistic (with 1 and 199 d.f.) is used to compare the full model likelihood to the likelihood of a model with two main effects but no interaction term. A nominal significance level is appropriate here because the marker pairs already have been screened for genome-wide significance. We chose a conservative 0.005 level (F > 8.06) for this test to ensure that only the most significant interactions would be selected. Maternal environment was included as a “marker” in this genome scan, which allowed us to detect pairwise interaction of a marker locus with the maternal environment. For the three-way interaction search, which is analogous to the pairwise search, maternal environment is included in the model, and we then search through all marker pairs. The initial screen is based on an overall F statistic (with 6 and 195 d.f.) that compares the full model (with three main effects, three pairwise interactions, and a three-way interaction) with a null model that includes only the maternal effect. Genome-wide significance was determined by permutation analysis of the overall F statistics. The genome-wide 0.05 critical value was found to be 7.7. A second F test (with 1 and 195 d.f.) that compares the full model to a model with no three-way interaction was performed at a nominal 0.005 level (F > 8.06) and was used to identify those triplets (two markers plus maternal environment) that show significant three-way interactions. All linkages detected by genome scans were calculated by marker regression using MATLAB software (Mathworks, Inc.). Source code and analysis scripts are available at http://jax.org/research/churchill. MapManager QTb28ppc (www.mcbio.med.buffalo.edu/mapmgr.html) (Manley and Olson 1999) was used to identify QTL during data acquisition and for generating the graphics in Figure 2. ANOVA graphics in Figures 4 and 5 were generated by Stat View (Abacus Concepts).
Acknowledgments
We thank Jenn Kintner and Bruce Regimbal for excellent technical assistance. This research was supported by National Institutes of Health-National Center for Research Resources 88911. Institutional shared services were supported by National Cancer Institute Center Support Grant CA34196.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ehl@jax.org; FAX (207) 288-6079.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.147000.
REFERENCES
- Bjorntorp P. Androgens, the metabolic syndrome, and non-insulin dependent diabetes mellitus. Ann NY Acad Sci. 1993;676:242–252. doi: 10.1111/j.1749-6632.1993.tb38738.x. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold value for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG. Limits to prediction of phenotype from knowledge of genotypes. In: Clegg MT, Hecht M, MacIntyre RJ, editors. Limits to knowledge in evolutionary genetics. New York: Kluwer Academic/Plenum Publishers; 2000. pp. 205–224. [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A. Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet. 1999;21:213–215. doi: 10.1038/6002. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Rosen C, Partke H-J, Herberg L, Leiter EH. Rapid maturational growth as a predictor of NIDDM in NZO male mice. Diabetes. 1998;47: (Suppl.):A318. doi: 10.2337/diab.47.8.1287. [DOI] [PubMed] [Google Scholar]
- Halaas JL, Boozer C, Denton DA, Blairwes J, Friedman JM, Fidahuse N. Physiological response to long-term peripheral and central leptin infusion in lean and obese mice. Proc Natl Acad Sci. 1997;94:8878–8883. doi: 10.1073/pnas.94.16.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanis C, Boerwinkle E, Chakraborty R, Ellsworth D, Concannon P, Stirling B, Morrison V, Wapelhorst B, Spielman R, Gogolin-Ewens K, et al. A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet. 1996;13:161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- Hirayama I, Yi Z, Izumi S, Arai I, Suzuki W, Nagamachi Y, Kuwano H, Takeuchi T, Izumi T. Genetic analysis of obese diabetes in the TSOD mouse. Diabetes. 1999;48:1183–1191. doi: 10.2337/diabetes.48.5.1183. [DOI] [PubMed] [Google Scholar]
- Kido Y, Burks DJ, Withers D, Bruning JC, Kahn CR, White MF, Accili D. Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J Clin Invest. 2000;105:199–205. doi: 10.1172/JCI7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida S, Beier DR. Genetic localization of interacting modifiers affecting severity in a murine model of polycystic kidney disease. Genome Res. 2000;10:49–54. [PMC free article] [PubMed] [Google Scholar]
- Kunz C, Rodriguez-Palmero M, Koletzko B, Jensen R. Nutritional and biochemical properties of human milk. Part I: General aspects, proteins, and carbohydrates. Clin Perinatol. 1999;26:307–333. [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Legare ME, Bartlett FS, II, Frankel WN. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- Leiter EH, Herberg L. The polygenetics of diabesity in mice. Diabetes Rev. 1997;5:131–148. [Google Scholar]
- Leiter EH, Herberg L, Reifsnyder PC, Flurkey K, Partke H-J, Junger E, Herberg L. Non-insulin dependent diabetes genes in mice: Deleterious synergism by both parental genomes contributes to diabetogenic thresholds. Diabetes. 1998;47:1287–1295. doi: 10.2337/diab.47.8.1287. [DOI] [PubMed] [Google Scholar]
- Levin B, Govek E. Gestational obesity accentuates obesity in obesity-prone progeny. Am J Physiol. 1998;275:R1375–1379. doi: 10.1152/ajpregu.1998.275.4.R1374. [DOI] [PubMed] [Google Scholar]
- Mahtani M, Widen E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, et al. Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet. 1996;14:90–94. doi: 10.1038/ng0996-90. [DOI] [PubMed] [Google Scholar]
- Manley K, Olson JM. Overview of QTL mapping software and introduction to MapManager QT. Mamm Genome. 1999;10:327–334. doi: 10.1007/s003359900997. [DOI] [PubMed] [Google Scholar]
- Norris JM, Pietropaolo M. Controversial topics series: Milk proteins and diabetes. J Endocrinol Invest. 1999;22:568–580. doi: 10.1007/BF03343611. [DOI] [PubMed] [Google Scholar]
- Podolin PL, Denny P, Armitage N, Lord CJ, Hill NJ, Levy ER, Peterson LB, Todd JA, Wicker LS, Lyons PA. Localization of two insulin-dependent diabetes (Idd) genes to the Idd10 region on mouse Chromosome 3. Mamm Genome. 1998;9:283–286. doi: 10.1007/s003359900749. [DOI] [PubMed] [Google Scholar]
- Pratley RE. Gene-environment interactions in the pathogenesis of type 2 diabetes mellitus: Lessons learned from the Pima Indians. Proc Nutr Soc. 1998;57:175–181. doi: 10.1079/pns19980029. [DOI] [PubMed] [Google Scholar]
- Proietto J, Larkins RG. A perspective on the New Zealand obese mouse. In: Shafrir E, editor. Lessons from animal models of diabetes IV. London: Smith-Gordon; 1993. pp. 65–73. [Google Scholar]
- Ranheim T, Dumke C, Schueler KL, Cartee GD, Attie AD. Interaction between BTBR and C57BL/6J genomes produces an insulin resistance syndrome in (BTBR x C57BL/6J) F1 mice. Arterioscler Thromb Vasc Biol. 1997;17:3286–3293. doi: 10.1161/01.atv.17.11.3286. [DOI] [PubMed] [Google Scholar]
- Serreze DV, Bridgett MB, Chapman HD, Chen E, Richard SB, Leiter EH. Subcongenic analysis of the Idd13 locus in NOD/Lt mice: Evidence for several susceptibility genes including a possible diabetogenic role for β2-microglobulin. J Immunol. 1998;160:1472–1478. [PubMed] [Google Scholar]
- Simpson EH. The interpretation of interaction in contingency tables. J Roy Stat Soc. 1951;13:238–241. [Google Scholar]
- Stern M, Mitchell B, Blangero J, Reinhart L, Kammerer C, Harrison C, Shipman P, O'Connell P, Frazier M, MacCluer J. Evidence for a major gene for type II diabetes and linkage analyses with selected candidate genes in Mexican-Americans. Diabetes. 1996;45:563–568. doi: 10.2337/diab.45.5.563. [DOI] [PubMed] [Google Scholar]
- Taylor B, Phillips S. Detection of obesity QTLs on mouse chromosome 1 and 7 by selective DNA pooling. Genomics. 1996;34:389–398. doi: 10.1006/geno.1996.0302. [DOI] [PubMed] [Google Scholar]
- Ueda H, Ikegami H, Kawaguchi Y, Fujisawa T, Yamato E, Shibata M, Ogihara T. Genetic analysis of late-onset type 2 diabetes in a mouse model of human complex trait. Diabetes. 1999;48:1168–1174. doi: 10.2337/diabetes.48.5.1168. [DOI] [PubMed] [Google Scholar]
- von Kries R, Koletzko B, Sauerwald T, von Mutius E, Barnert D, Grunert V, von Voss H. Breast feeding and obesity: Cross sectional study. BMJ. 1999;319:147–150. doi: 10.1136/bmj.319.7203.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, Truett AA, Monteiro MP, Barry SJ, Warden CH, Naggert JK, Maddatu TP, West DB. Gene-environment interaction: A significant diet-dependent obesity locus demonstrated in a congenic segment on mouse chromosome 7. Mamm Genome. 1999;10:457–462. doi: 10.1007/s003359901023. [DOI] [PubMed] [Google Scholar]
- Yule GU. Notes on the theory of association of attributes in statistics. Biometrika. 1903;2:121–134. [Google Scholar]