Abstract
Misfolded, luminal endoplasmic reticulum (ER) proteins must be recognized before being degraded by a process called ERAD-L. Using site-specific photo-crosslinking in S. cerevisiae, we tested luminal interactions of a glycosylated ERAD-L substrate with potential recognition components. Major interactions were observed with Hrd3p. These are independent of the glycan and of other ERAD components, and can occur throughout the length of the unfolded substrate. The lectin Yos9p only interacts with a polypeptide segment distant from the degradation signal. Hrd3p may thus be the first substrate-recognizing component. Der1p appears to have a role in a pathway that is parallel to that involving Hrd3p.
Keywords: ER-associated degradation, misfolded proteins, Hrd3p, Yos9p, Der1p, endoplasmic reticulum
1. Introduction
Proteins that misfold in the lumen of endoplasmic reticulum (ER) are retro-translocated across the ER membrane into the cytosol, poly-ubiquitinated, and ultimately degraded by the proteasome [1, 2]. The process is known as luminal ER-associated protein degradation (ERAD-L) [3–5]. In S. cerevisiae, ERAD-L requires a hetero-tetrameric membrane protein complex, the Hrd1 complex, which consists of Hrd1p, Hrd3p, Usa1p, and Der1p [3–5]. Hrd1p is a ubiquitin ligase that spans the ER membrane six times and has a cytosolic RING finger domain [6–8]. Hrd3p is a single-spanning protein with a luminal domain that recruits the lectin Yos9p. Usa1p spans the membrane twice with N- and C-termini in the cytosol. Finally, Der1p spans the membrane four times. Recent evidence suggests that Hrd1p is the central component of the complex, required for moving misfolded proteins across the membrane [6–9]. To do so, Hrd1p needs to oligomerize, which is facilitated by Usa1p [9–11].
How misfolded proteins are recognized and distinguished from folding intermediates is only poorly understood. One mechanism by which misfolded glycoproteins are recognized is through a terminal α1,6-mannose residue in a carbohydrate chain [2, 12–14], which is generated by the glycosidase Htm1p when a misfolded protein spends excessive time in the ER. The α1,6-mannose residue is specifically recognized by the lectin Yos9p [12, 15]. However, this cannot be the only recognition mechanism, as the same α1,6-mannose signal does not trigger degradation when placed at other locations in the polypeptide chain [2]. It has therefore been proposed that the unstructured nature of the polypeptide segment around the carbohydrate attachment site serves as an additional signal [16]. Both Yos9p and the luminal domain of Hrd3p have been implicated in recognizing these unfolded polypeptide segments [17–19], but evidence for direct interactions is lacking. In addition, it is uncertain whether Yos9p and Hrd3p are the only components of the Hrd1p complex involved in substrate recognition. Transfer of substrate to Hrd1p is only completely abolished when both Yos9p and Der1p are absent, suggesting that there is an additional pathway of substrate delivery involving Der1p [9, 20].
Here we use an in vivo site-specific photocrosslinking method to analyze interactions of an ERAD-L substrate with the potential recognition proteins Hrd3p, Yos9p, and Der1p.
2. Materials and Methods
Yeast Strains and Plasmids
Standard PCR-based recombination methods were used to make gene deletions and to tag proteins [21]. Strains with multiple gene deletions and genomically encoded protein tags were made either by PCR-based homologous recombination or by crossing haploid cells of opposite mating types, followed by sporulation and tetrad dissection [22]. The strains used were isogenic to FY251 (Mata ura3-52 his3Δ200 leu2Δ1 trp1Δ63). Supplementary Tables 1 and 2 list the yeast strains and plasmids used, respectively.
Site-specific in vivo photocrosslinking
Yeast cells were transformed with two plasmids, one encoding a CPY*-derived construct with an amber codon at a selected position, and another encoding both a tRNA that suppresses the amber stop codon and a modified tRNA-synthetase that charges this tRNA with photoreactive benzoyl amino acid [23, 24]. The protocol for photocrosslinking and analysis by immunoblotting has been described [9].
3. Results
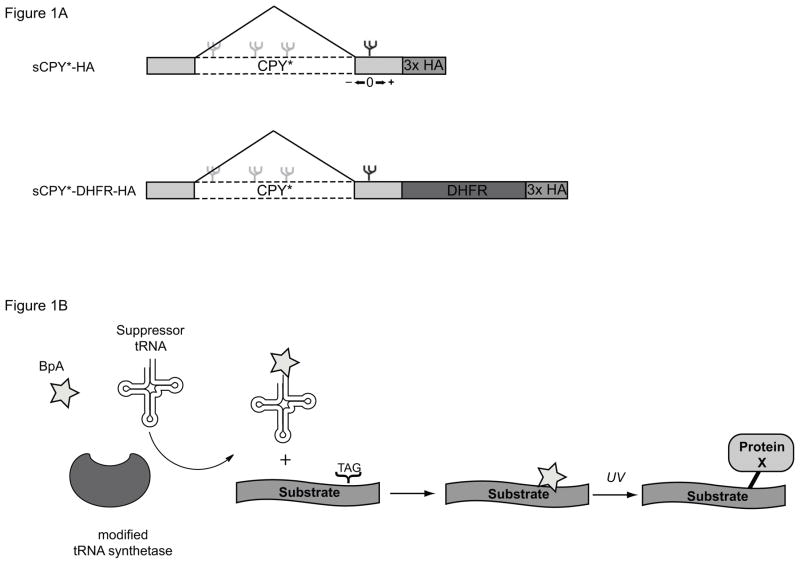
To analyze interactions of a luminal misfolded protein, we employed a shortened version of the well-characterized ERAD-L substrate CPY* (sCPY*). This protein contains the last 180 amino acids, including the N-glycosylation site that is part of the degradation signal. This protein is degraded more rapidly than the original CPY* but requires the same ERAD-L components [9]. We also used a version of sCPY* in which dihydrofolate reductase (DHFR) is fused to the C-terminus (sCPY*-DHFR), which slows the degradation of the protein [9, 17]. Both sCPY* and sCPY*-DHFR also contain three hemagglutinin (HA)-tags at the C-terminus for detection with HA antibodies.
To analyze direct interactions of sCPY*-HA or sCPY*-DHFR-HA with potential recognition components, we employed a site-specific photocrosslinking method in intact yeast cells [23, 24]. Single amber stop codons were introduced at various positions of the polypeptide chain (Fig. 1A; positions upstream and downstream of the glycosylation site are given negative and positive numbers, respectively). These constructs were expressed together with a suppressor tRNA and a modified tRNA synthetase that charges the tRNA with a phenylalanine derivative containing a photoreactive benzophenone (Bpa). The photoreactive amino acid is then incorporated at the position specified by the amber codon. Irradiation of intact cells leads to crosslinks between the ERAD-L substrate and neighboring proteins (see scheme in Fig. 1B).
Figure 1. Experimental design of in vivo site-specific photocrosslinking experiments.
(A) Schematics of the ERAD-L substrates used. (B) Explanation of the in vivo photocrosslinking method. BpA, phenylalanine derivative containing a benzophenone.
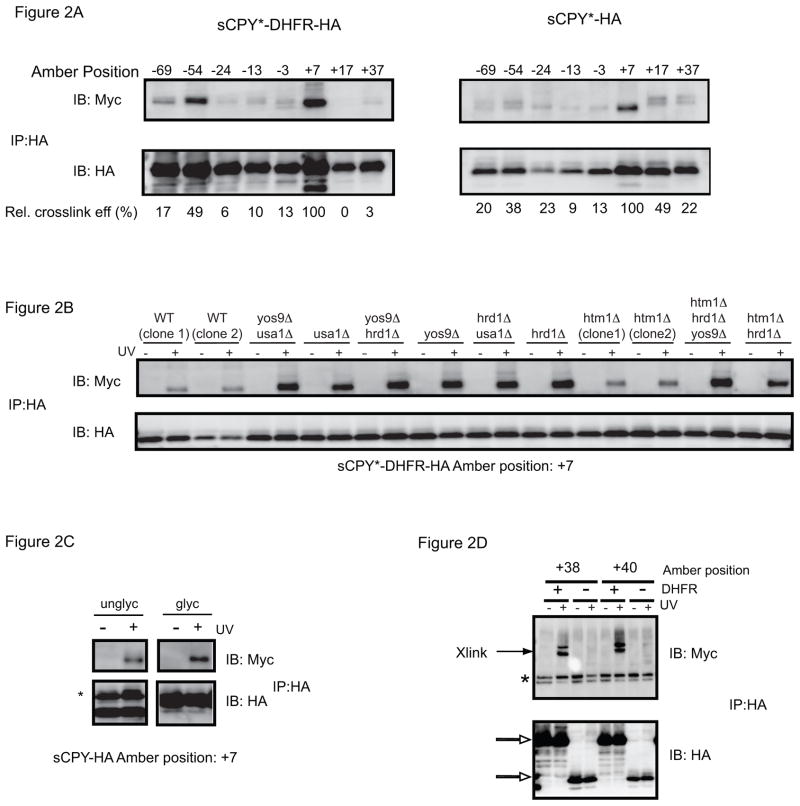
We first tested interactions of sCPY*-HA and sCPY*-DHFR-HA with the soluble luminal domain of Hrd3p (1-767) fused to 13 Myc-tags, a construct that fully complements a HRD3 deletion [9]. After irradiation, cell extracts were subjected to immunoprecipitation with HA antibodies, followed by SDS-PAGE and immunoblotting with HA- or Myc- antibodies. The HA blot demonstrates that sCPY*-DHFR-HA is degraded less efficiently than the protein lacking the DHFR moiety, resulting in elevated steady-state levels. The Myc-blot shows that the most prominent crosslinks occurred with position +7 near the N-glycosylation site, although crosslinks were also observed with other positions. As expected, without irradiation, no crosslinks occurred ([9] and Supplementary Fig. S1A). When similar experiments were performed with a Hrd1p construct containing 13 Myc tags at its C-terminus, crosslinks were only observed with sCPY*-DHFR-HA, and not with sCPY* (Fig. 2D). As reported before [9], the crosslinks with sCPY*-DHFR-HA were observed primarily at positions +38 to +42. Given that the presence of the DHFR moiety increases the substrate population contacting Hrd1p but not that contacting Hrd3p, it appears that the rate-limiting step in the degradation of this substrate occurs after Hrd3p- function.
Figure 2. Photocrosslinking to Hrd3p.
(A) sCPY*-HA or sCPY*-DHFR-HA with amber codons at the indicated positions were expressed together with Hrd3p-13myc and subjected to in vivo photocrosslinking. Membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. The numbers underneath the lanes give the relative crosslinking efficiency. For quantification, the intensity of the myc band was divided by that of the corresponding HA band, and normalized to the ratio determined for position +7. The comparison of crosslinking efficiency is only approximate, as the level of photocrosslinker incorporation may vary between positions. (B) sCPY*-HA with a photoreactive probe at position +7 was crosslinked to Hrd3p-13myc in cells lacking ERAD components. Where indicated, UV irradiation was omitted. (C) sCPY*-HA containing the N-glycosylation site (glyc) or lacking it (unglyc; mutation of N to A), both with a photoreactive probe at position +7, were crosslinked to Hrd3p-13myc. The asterisk indicates immunoglobulin light chain. (D) For comparison with (A), crosslinking to Hrd1p-13 myc was tested with sCPY*-HA or sCPY*-DHFR-HA containing a photoreactive probe at the indicated positions. The open arrows indicate the position of substrate, and the asterisk denotes non-specific bands reacting with Myc antibodies.
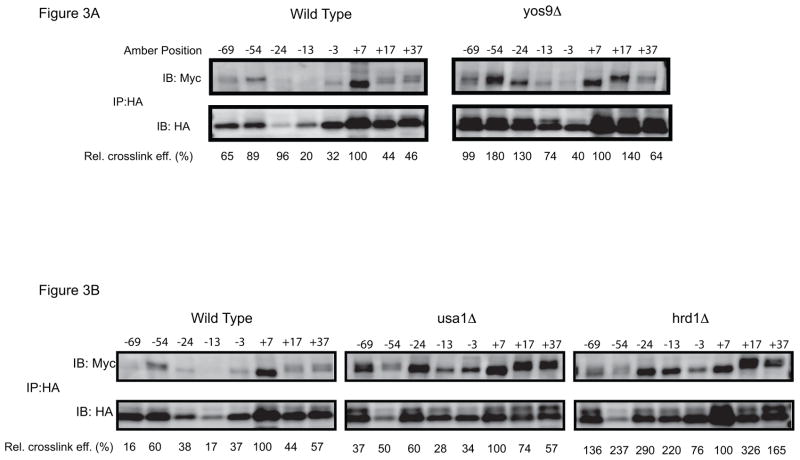
Consistent with the idea that substrate-Hrd3p interaction occurs early in ERAD-L, the crosslinking efficiency seen with sCPY* containing a probe at position +7 was not affected by deletion of ERAD components (Fig. 2B). Even the simultaneous deletion of several ERAD components had no effect (Fig. 2B). The glycosidase Htm1p was also not required, indicating that Hrd3p interaction is independent of an α1,6-mannose signal in the carbohydrate chain. Indeed, sCPY* lacking a N-glycosylation site crosslinked with the same efficiency as the glycosylated protein (Fig. 2C). Substrate interaction requires residues 1-392 of Hrd3p (Fig. S1B), consistent with previous suggestions [25]; the putative Yos9p-/Hrd1p-binding domain can be deleted without affecting crosslinking to substrate. Collectively, these data suggest that Hrd3p is involved in the early recognition of unstructured polypeptide segments. Such a chaperone-like function of Hrd3p is supported by the observation that Hrd3p interacts with many positions in sCPY* when substrate accumulates in the ER lumen. For example, positions that interacted only weakly with Hrd3p in wild type cells, crosslinked significantly stronger in ERAD-deletion mutants (Fig. 3A, B) or in a Hrd1p mutant with a defective RING finger domain (C399S) (Fig. S2). Thus, weaker binding sites for Hrd3p appear to exist throughout the unfolded polypeptide chain and become more prominent at elevated substrate levels.
Figure 3. Photocrosslinking to Hrd3p in different ERAD mutants.
(A) sCPY*-HA with amber codons at the indicated positions was expressed together with Hrd3p-13myc in wild type cells or cells lacking Yos9p. After in vivo photocrosslinking, membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. The numbers underneath the lanes give the relative crosslinking efficiency. For quantification, the intensity of the myc band was divided by that of the corresponding HA band, and normalized to the ratio determined for position +7. (B) As in (A), but crosslinking in wild type cells is compared with that in cells lacking Usa1p or Hrd1p.
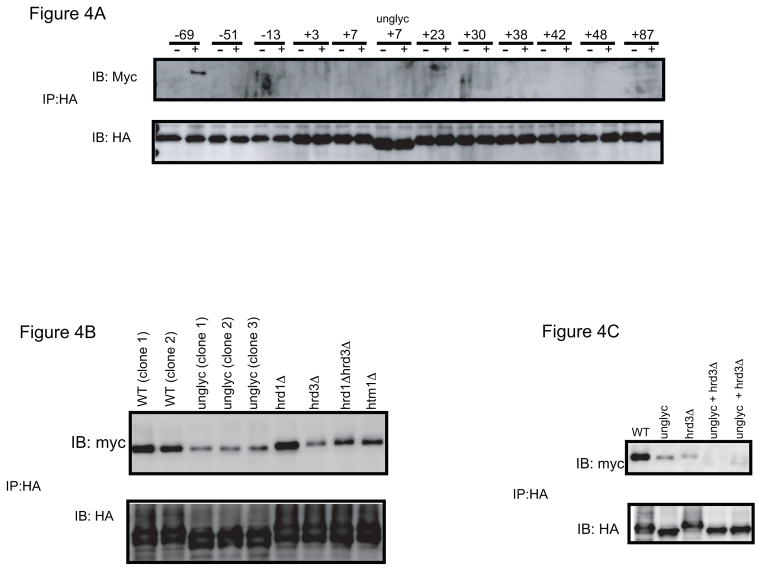
To investigate whether Yos9p contributes to polypeptide binding or is exclusively involved in sugar recognition, we performed similar crosslinking experiments in a strain expressing Yos9p containing 13 Myc-tags and an HDEL ER-retention signal at its C-terminus. As with Hrd3p, the tagged protein complements a deletion strain (data not shown). Using sCPY*-DHFR-HA, crosslinks were only observed with position −69 (Fig. 4A); no crosslinks were seen around the carbohydrate attachment site. The crosslinks at position −69 were significantly reduced when the substrate was non-glycosylated or when Hrd3p was absent (Fig. 4B). When both effects were combined, essentially no crosslinking to Yos9p was observed (Fig. 4C). Deletion of HTM1 had only a moderate effect on Yos9p crosslinking, while deletion of HRD1 did not affect it at all (Fig. 4B). The specificity for position −69 was not altered in a hrd1Δ strain (Supplementary Fig. S3). These results are consistent with the idea that Yos9p does not interact with the polypeptide segment around the carbohydrate attachment site that signals degradation. Rather, it appears that a distant segment comes in contact with Yos9p once Yos9p binds to the α1,6-signal in the carbohydrate chain. It should be noted that sCPY* without the attached DHFR domain did not give crosslinks at all (data not shown), consistent with the assumption that Yos9p acts downstream of Hrd3p.
Figure 4. Photocrosslinking of sCPY*-DHFR-HA to Yos9p.
(A) sCPY*-HA with amber codons at the indicated positions was expressed together with Yos9p-13myc and subjected to in vivo photocrosslinking. sCPY*-HA lacking an N-glycosylation site (unglyc) with a photoreactive probe at position +7 was also tested. Membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. (B) sCPY*-HA with a photoreactive probe at position −69, with or without a N-glycosylation site was crosslinked to Yos9p-13myc-HDEL in wild type cells or cells lacking different ERAD components. (C) As in (B), but crosslinking was also tested when both a carbohydrate chain in the substrate and Hrd3p were absent.
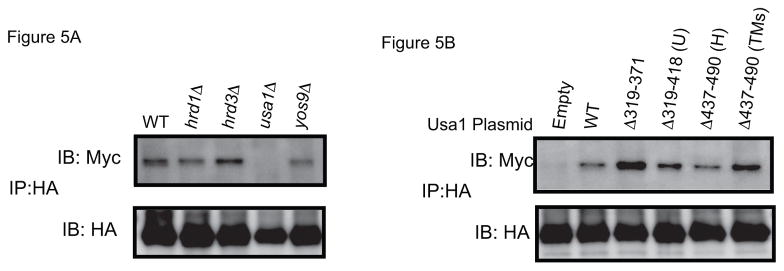
Finally, we studied the interaction of sCPY*-DHFR-HA with Der1p containing 13 myc-tags (Der1p-13myc), which retains partial activity [9]. Previous work has shown that position +24 of the substrate gives the most prominent crosslinks [9]. These crosslinks were still present in yos9Δ or hrd3Δ cells, but disappeared when USA1 was deleted (Fig. 5A). The interaction could be restored when fragments of Usa1p were expressed that are defective in either Usa1p-Hrd1p or Usa1p-Usa1p interaction (Fig. 5B). All constructs retain the C-terminal segment (residues 584-838) that has been proposed to interact with Der1p [11]. These data therefore support the idea that there are parallel pathways of substrate recruitment, one involving Hrd3p/Yos9p and another involving Der1p. In addition, it appears that Der1p does not need to be recruited to the Hrd1p complex to interact with substrate. The role of Usa1p may be in stabilizing Der1p, as the levels of Der1p are reduced in a usa1Δ strain [11].
Figure 5. Photocrosslinking to Der1p.
(A) sCPY*-HA with a photoreactive probe at position +24 was expressed together with Der1p-13myc in cells lacking different ERAD components. After in vivo photocrosslinking, membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. (B) Der1p-13myc crosslinking was examined in usa1Δ cells containing an empty vector or plasmids coding for the indicated segments of Usa1p. U and H, segments involved in Usa1p oligomerization and Hrd1p interaction, respectively [9].
4. Discussion
Our results suggest that there are two distinct pathways by which a misfolded luminal substrate is recognized for ERAD-L. The first pathway involves Hrd3p and Yos9p. Hrd3p appears to function upstream of Yos9p and interacts directly with the polypeptide substrate, including a segment around the carbohydrate attachment site. Yos9p may recognize exclusively the α1,6-mannose residue in the carbohydrate chain, as it crosslinks only far away from the degradation signal in the substrate, probably with segments that are irrelevant for actual recognition. The simultaneous interactions of Hrd3p with an unfolded polypeptide segment and of Yos9p with the α1,6-mannose signal would direct the complex of the two proteins to the region around the critical N-glycosylation site. This would explain why the degradation signal is bipartite, with neither an unfolded segment nor an α1,6-mannose signal alone being capable of directing a substrate to ERAD-L. Hrd3p may recognize many unfolded polypeptide segments, as it can crosslink throughout a polypeptide chain, particularly when the substrate is accumulated in the ER lumen. Hrd3p thus appears to be a general chaperone.
The second pathway of substrate recognition involves Der1p. Substrate delivery to Der1p does not require Yos9p or Hrd3p. It is not clear what feature in the substrate is recognized by Der1p, but it is interesting that Der1p interacts close to the degradation signal. Usa1p’s role in substrate interaction appears to be restricted to stabilizing Der1p. Both substrate delivery pathways subsequently converge at Hrd1p, which is the major component required for movement of the substrate across the membrane. Although Hrd3p is not required for substrate recognition in the Der1p pathway, it is involved in the transfer of substrate from Der1p to Hrd1p; substrate crosslinks to Hrd1p are abolished in the absence of Hrd3p or when Yos9p and Der1p are simultaneously absent [9]. Furthermore, because both Yos9p and Der1p are essential in ERAD-L, they must have non-redundant functions in addition to providing parallel pathways of substrate delivery to Hrd1p.
It is surprising that so many ERAD components are involved in substrate recognition. One reason may be the need to distinguish between ERAD substrates and folding intermediates. However, one would think that this could be achieved by the dual recognition of a processed carbohydrate signal (the terminal α1,6-mannose residue) and of an associated unfolded polypeptide segment by the Hrd3p/Yos9p complex. Perhaps, this type of recognition alone would not be specific enough, and it would not explain how non-glycosylated substrates are recognized. Der1p may serve as an additional recognition device, ensuring that only proper substrates are passed on to the central Hrd1p component.
Supplementary Material
(A) sCPY*-HA with amber codons at the indicated positions were expressed together with Hrd3p-13myc and subjected to in vivophotocrosslinking. Membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. Where indicated, UV irradiation was omitted. The arrow indicates the position of crosslinked bands. Some non-crosslinked Hrd-13myc is seen in the immunoprecipitations (asterisk). The open arrow indicates the position of substrate and of co-migrating immunoglobulin light chain. The double asterisk (**) denotes the position of immunoglobulin heavy chain. (B) sCPY*-HA with a photoreactive probe at position +7 was expressed together with Hrd3p-13myc comprising either the entire luminal domain of Hrd3 (residues 1-767) or residues 1-392.
sCPY*-HA with amber codons at the indicated positions were expressed together with Hrd3p-13myc in hrd1Δ cells containing a plasmid coding for either wild type (WT) Hrd1p or a Hrd1p mutant with the C399S mutation in the RING finger domain. Cells were UV-irradiated as indicated, membranes were isolated, and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. The prominent UV-independent band in the Myc blot is non-specifically precipitated non-crosslinked Hrd3-13myc. The prominent UV-dependent band seen with some positions in the HA blot corresponds to crosslinkedsCPY*-HA dimers.
sCPY*-HA with amber codons at the indicated positions were expressed together with Yos9p-13myc in cells lacking HRD1. After in vivophotocrosslinking, membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies.
Acknowledgments
We thank P. Schultz and J. Weissman for reagents. A.M.S. was supported by a Ruth L. Kirschstein National Research Service Award postdoctoral fellowship. P.C. is supported by a Leukemia and Lymphoma Society special fellow award. T.A.R. is supported by NIH grant GM052586 and is a Howard Hughes Medical Institute Investigator.
Abbreviations
- CPY
carboxypeptidase Y
- DHFR
dihydrofolate reductase
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- HA
hemagglutinin
- sCPY
short carboxypeptidase Y
Footnotes
Structured summary of protein interactions:
sCPY physically interacts with sCPY by cross-linking study (View interaction)
Hrd3 and sCPY physically interact by cross-linking study (View interaction)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bagola K, Mehnert M, Jarosch E, Sommer T. Protein dislocation from the ER. Biochim Biophys Acta. 2011:925–936. doi: 10.1016/j.bbamem.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 2.Xie W, Ng DTW. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. 2010:533–539. doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 6.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 7.Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a Misfolded Luminal ER Protein by the Ubiquitin-Ligase Hrd1p. Cell. 2010:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll SM, Hampton RY. Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J Biol Chem. 2010:5146–5156. doi: 10.1074/jbc.M109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn SC, Hanna J, Hirsch C, Volkwein C, Schütz A, Heinemann U, Sommer T, Jarosch E. Usa1 Functions as a Scaffold of the HRD-Ubiquitin Ligase. Mol Cell. 2009:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 14.Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spear ED, Ng DTW. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol. 2005:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie W, Kanehara K, Sayeed A, Ng DTW. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell. 2009:3317–3329. doi: 10.1091/mbc.E09-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 18.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 19.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie CR, Fink G. Guide to Yeast Genetics and Molecular Biology. Vol. 194. San Diego, CA: Academic Press; 1991. [Google Scholar]
- 23.Chen S, Schultz PG, Brock A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J Mol Biol. 2007:112–122. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 25.Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) sCPY*-HA with amber codons at the indicated positions were expressed together with Hrd3p-13myc and subjected to in vivophotocrosslinking. Membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. Where indicated, UV irradiation was omitted. The arrow indicates the position of crosslinked bands. Some non-crosslinked Hrd-13myc is seen in the immunoprecipitations (asterisk). The open arrow indicates the position of substrate and of co-migrating immunoglobulin light chain. The double asterisk (**) denotes the position of immunoglobulin heavy chain. (B) sCPY*-HA with a photoreactive probe at position +7 was expressed together with Hrd3p-13myc comprising either the entire luminal domain of Hrd3 (residues 1-767) or residues 1-392.
sCPY*-HA with amber codons at the indicated positions were expressed together with Hrd3p-13myc in hrd1Δ cells containing a plasmid coding for either wild type (WT) Hrd1p or a Hrd1p mutant with the C399S mutation in the RING finger domain. Cells were UV-irradiated as indicated, membranes were isolated, and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies. The prominent UV-independent band in the Myc blot is non-specifically precipitated non-crosslinked Hrd3-13myc. The prominent UV-dependent band seen with some positions in the HA blot corresponds to crosslinkedsCPY*-HA dimers.
sCPY*-HA with amber codons at the indicated positions were expressed together with Yos9p-13myc in cells lacking HRD1. After in vivophotocrosslinking, membranes were isolated and SDS-denatured proteins were immunoprecipitated with HA antibodies. The samples were analyzed by SDS-PAGE and immunoblotting with either HA or Myc antibodies.