Abstract
The crystal structure of Escherichia coli 3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase in complex with E. coli thioredoxin 1 (Trx1) has been determined to 3.0 Å resolution. The two proteins are covalently linked via a mixed disulfide that forms during nucleophilic attack of Trx’s N-terminal cysteine on the Sγ atom of the PAPS reductase S-sulfocysteine (E-Cys-Sγ-SO3−), a central intermediate in the catalytic cycle. For the first time in a crystal structure, residues 235–244 in the PAPS reductase C-terminus are observed, depicting an array of interprotein salt bridges between Trx and the strictly conserved glutathione-like sequence, Glu238Cys239Gly240Leu241His242. The structure also reveals a Trx-binding surface adjacent to the active site cleft and regions of PAPS reductase associated with conformational change. Interaction at this site strategically positions Trx to bind the S-sulfated C-terminus and addresses the mechanism for requisite structural rearrangement of this domain. An apparent sulfite-binding pocket at the protein–protein interface explicitly orients the S-sulfocysteine Sγ atom for nucleophilic attack in a subsequent step. Taken together, the structure of PAPS reductase in complex with Trx highlights the large structural rearrangement required to accomplish sulfonucleotide reduction and suggests a role for Trx in catalysis beyond the paradigm of disulfide reduction.
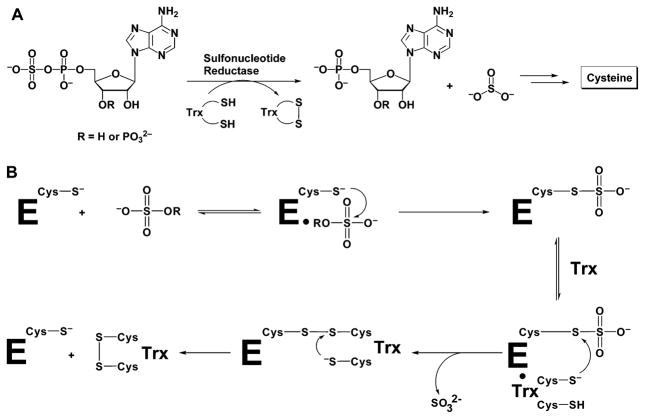
Sulfonucleotide reductases (SRs)1 catalyze the reduction of adenylated sulfate to sulfite, the first committed step in the production of reduced sulfur that is required for de novo cysteine biosynthesis in plants, fungi, and many bacteria (Figure 1A) (1, 2). The two classes of enzymes in this family are distinguished by substrate specificity and by the presence or absence of an iron–sulfur cluster (3, 4). Adenosine-5′-phosphosulfate (APS) is reduced by the [4Fe-4S]-containing enzyme APS reductase and is found in archae, Gram-positive bacteria, and plants (Figure 1A) (4, 5). In contrast, 3′-phosphoadenosine-5′-phosphosulfate (PAPS) is reduced by PAPS reductase, an enzyme that lacks the cluster and is present in enteric bacteria and yeast (Figure 1A) (6, 7). The sequences of APS and PAPS reductases are approximately 25–30% identical, and with the determination of the crystal structure of Pseudomonas aeruginosa APS reductase, it is established that these enzymes are structurally homologous (8, 9).
Figure 1.
(A) Depending on the organism, APS or PAPS is reduced to sulfite by APS reductase or PAPS reductase, respectively. The sulfite product formed in this reaction is further reduced to sulfide, and the sulfur is incorporated into cysteine. This amino acid is then converted into numerous metabolites, including methionine and cofactors, such as coenzyme A. (B) Mechanism proposed for sulfonucleotide reduction.
SRs are found in many human pathogens, such as Mycobacterium tuberculosis, which accounts for 3 million deaths per year worldwide (10). In addition to lethal synergy with HIV, this epidemic is also fueled by the increasing number of individuals infected with multidrug resistant strains of M. tuberculosis, presently estimated at 50 million (11). Identification of new targets and antibiotics is paramount to addressing this growing problem (12, 13). In M. tuberculosis, the SR belongs to the class of APS reducing enzymes (3, 14) and the cysteine product of this metabolic pathway can be transformed into methionine, essential cofactors such as coenzyme A, biotin, thiamine, and the antioxidant mycothiol (the mycobacterial equivalent of glutathione) (15). APS reductase was identified in a screen for essential genes in Mycobacterium bovis BCG (16), and recently, it has also been demonstrated that APS reductase activity is required for the survival of M. tuberculosis during the persistence phase of infection in a murine model of tuberculosis (17). Parallel studies of APS and PAPS reductase enzymes address the general mechanism of sulfonucleotide reduction (3) (see details below), consistent with their structural homology (8, 9). In particular, PAPS reductase is an attractive model for investigating general features of sulfonucleotide reduction due to the absence of an oxygen sensitive iron–sulfur cluster. Additionally, PAPS reductase and sulfate-dependent pathways are present in pathogens such Escherichia coli, Salmonella typhimurium, and Yersinia pestis, providing further motivation for study of this enzyme. Notably, SRs are not found in humans, further increasing their interest as potential targets for therapeutic intervention.
The input of two electrons is required for sulfonucleotide reduction, and these can be supplied by a protein cofactor with a redox-active disulfide bond, known as thioredoxin (Trx) (18–20). Biochemical, spectroscopic, and mass spectrometry investigations of sulfonucleotide reductases in both APS- and PAPS-dependent enzymes are consistent with a two-step mechanism (Figure 1B), in which the sulfonucleotide undergoes nucleophilic attack by an absolutely conserved cysteine to form an enzyme S-sulfocysteine intermediate, E-Cys-Sγ-SO3− (previously termed “thiosulfonate intermediate”) (3, 8, 21, 22). In a subsequent step, sulfite is released in a Trx-dependent reaction (3, 8, 21, 22). Hence, APS and PAPS reductases perform the same overall chemistry to catalyze sulfonucleotide reduction, despite their differences in substrate specificity (3). In a manner analogous to Trx reduction of protein disulfide bonds, we have postulated that nucleophilic attack by Trx takes place at the Sγ atom of the S-sulfocysteine intermediate (Figure 1B) (3). This reaction would result in the formation of a mixed disulfide between Trx and the SR, concomitantly releasing sulfite. Nonetheless, nucleophilic attack at the more highly oxidized sulfur atom is also chemically plausible, and distinction between these models has awaited further experimental examination.
In addition to the chemical transformations that are taking place, biochemical and structural data are consistent with significant structural rearrangement during the SR catalytic cycle (3, 8). In particular, the 25 C-terminal residues of SRs (including the essential cysteine nucleophile) are highly mobile (8, 9). Defining the molecular interactions of Trx with the S-sulfated C-terminus and associated conformational rearrangements is, therefore, essential to elucidating the mechanism of sulfonucleotide reduction. Molecular interactions distal from the C-terminal residues and the scissile S-sulfocysteine bond could also play a role in Trx–enzyme recognition, and in addition to SRs (sulfur assimilation), Trx is involved in the reduction of numerous other enzymes such as ribonucleotide reductase (DNA synthesis) (23) and methionine sulfoxide reductase (protein repair) (18, 24). Gaining insight into the molecular determinants that govern Trx–target recognition is therefore a question of fundamental importance. Three-dimensional structural information obtained from Trx–protein complexes, “caught” in the act of reduction, is a necessary step in addressing this question. However, with the exception of barley Trx h isoform 2 (HvTrxh2) in complex with α-amylase/subtilisin inhibitor (BASI) determined to 2.3 Å (25), no other structures of a mixed disulfide intermediate with a full-length target protein have been described, and in particular, structural insight into the complex that forms between Trx and an enzyme target during its catalytic cycle has yet to be reported.
Here we describe the structure of E. coli PAPS reductase in complex with E. coli Trx1, a key intermediate present during the catalytic cycle, at a resolution of 3.0 Å. The two proteins are covalently linked via a mixed disulfide that forms during nucleophilic attack of Trx’s N-terminal cysteine on the Sγ atom of the PAPS reductase S-sulfocysteine intermediate. For the first time in a crystal structure, residues 235–244 of the C-terminus of PAPS reductase are ordered, permitting analysis of the interactions between Trx and the strictly conserved glutathione-like sequence, Glu238Cys239-Gly240Leu241His242. A second site of protein–protein interaction is also observed near the PAPS reductase active site and adjacent to the flexible C-terminal tail. Taken together, the structure of PAPS reductase in a mixed disulfide complex with Trx provides a molecular rationale for interaction of these proteins, illuminates the large conformational rearrangements required for sulfonucleotide reduction, and reveals a unique protein–protein interface as a potential candidate for disruption by small molecule or peptide inhibitors.
EXPERIMENTAL PROCEDURES
Cloning
Construction of the E. coli PAPS reductase pET24b (Novagen; contains a plasmid-encoded C-terminal His tag) protein expression vector has been previously described (3). Trx1 was amplified from E. coli genomic DNA ATC 700926 (ATCC) using the forward primer (5′-gggcatatgatgagcgataaaat-3′) and the reverse primer (5′-ggggggatccttacgccaggttag-3′). PCR mixtures contained each primer at 0.25 μM, 10 ng of genomic DNA template, and 2.5 units of Pfu DNA polymerase in a reaction buffer supplied by the manufacturer. The PCR product was ligated into a Zero Blunt Topo cloning vector (Invitrogen), digested with NdeI and BamHI, and ligated to similarly cut pET14b (Novagen; contains a plasmid-encoded N-terminal His tag). Construction of the P. aeruginosa APS reductase pET24b protein expression vector has been described previously (3). The gene encoding P. aeruginosa Trx was amplified from P. aeruginosa genomic DNA ATCC 47085D (ATCC) using the forward primer (5′-ggggcatatgagcgaacatatcgtcaacg-3′) and the reverse primer (5′-ggggcatatgagcgaacatatcgtcaacg-3′). The E. coli Trx1 and P. aeruginosa Trx Cys35Ala variants were constructed using the Quikchange PCR mutagenesis kit (Stratagene) according to the manufacturer’s specifications. Successful incorporation of the desired mutation was confirmed by sequencing, and once it was verified, the plasmid was transformed into BL21(DE3) (Novagen) for protein expression as described below.
Protein Purification
Plasmids containing E. coli PAPS reductase, P. aeruginosa APS reductase, or Trx were transformed into BL21(DE3) cells, grown to an OD of 0.6 at 37 °C, and induced with 0.4 mM IPTG. Cells were harvested after growth for 4 h at 30 °C (or 37 °C for Trx). His-tagged PAPS reductase and Trx were purified using a previously established procedure via affinity and size-exclusion chromatography (3). Briefly, after affinity chromatography, proteins were incubated with 10 mM DTT at 37 °C for 15 min and concentrated, and DTT-free protein was prepared by size-exclusion chromatography on a 10/30 Superdex 200 column (GE Healthcare) equilibrated in gel filtration buffer [50 mM Tris-HCl (pH 8.0) with an ionic strength of 150 mM with NaCl]. Desired fractions were collected, frozen in liquid nitrogen, and stored at −80 °C.
Formation of the Sulfonucleotide Reductase–Trx Complex
For protein crystallography, PAPS reductase S-sulfocysteine intermediate was formed by incubating PAPS reductase with 2-fold molar excess of PAPS substrate for 5–10 min at 37 °C. Quantitative formation of the PAPS reductase S-sulfocysteine intermediate was verified by the expected shift in the molecular mass of PAPS reductase by 80 Da (the molecular mass of a covalently bound sulfite) via electrospray mass spectrometry (3, 21). Subsequently, the PAPS reductase S-sulfocysteine intermediate was incubated with a 2-fold molar excess of Trx Cys35Ala for 5–10 min at 37 °C. For protein crystallography, size-exclusion chromatography was employed to purify the resulting complex from excess Trx Cys35Ala. For reversed phase HPLC and nonreducing SDS–PAGE analysis, PAPS reductase and a 2-fold molar excess of PAPS substrate (or buffer only) were incubated as described above. The final concentration of the PAPS reductase S-sulfocysteine intermediate or PAPS reductase alone was varied from 0 to 25 μM, as reported in the figure legends. Subsequently, Trx Cys35Ala was added to reaction mixtures at final concentrations of 1–10 μM. Complex formation was allowed to proceed for 20 s to 10 min at room temperature. The reverse set of experiments was carried out by employing a constant amount of PAPS reductase (1–10 μM) with increasing concentrations of Trx1 Cys35Ala (0–25 μM). Reactions were quenched, and the extent of complex formation was quantified as described below. Identical experiments were carried out for APS reductase and Trx Cys35Ala from P. aeruginosa.
Reversed Phase HPLC
Reactions between the PAPS reductase S-sulfocysteine intermediate or PAPS reductase alone and Trx1 Cys35Ala were quenched by the addition of 30 μL of 0.1% trifluoroacetic acid (TFA). The low pH of the quench buffer protonates free thiols and inhibits formation of the SR–Trx complex by preventing formation of the requisite thiolate. Reaction mixtures were separated on a protein–peptide 218TP54 C18 column (Vydac) employing a gradient of 40 to 55% acetonitrile and 0.1% TFA over a period of 20 min. Protein peaks were collected and their molecular masses determined by electrospray mass spectrometry.
Nonreducing SDS–PAGE
Reactions were quenched by the addition of iodoacetamide (25 mM), as described previously (3). Addition of this reagent covalently modifies remaining unreacted free thiols and, thus, prevents further disulfide formation during reactions and in subsequent sample preparation steps (e.g., unfolding in nonreducing SDS–PAGE load dye and heating samples prior to loading). Samples were separated on 12% Bis-Tris Criterion XT Precast Gels, and protein was visualized by staining with Coomassie Blue or Sypro Orange (Invitrogen). For Sypro Orange, gels were stained in the dark for 40 min and destained with 7.5% acetic acid in the dark for 1 min. Reaction products were visualized on a Typhoon gel imaging system (GE Healthcare) and quantified using Image Quant. The rate of formation of the PAPS reductase–Trx Cys35Ala complex was plotted as a function of increasing Trx Cys35Ala or PAPS reductase S-sulfocysteine intermediate concentration. Data were fit to the equation for active site-directed irreversible inhibition (26) using Kaleidagraph (Synergy Software). The reaction of the inhibitor (Trx Cys35Ala) with the PAPS reductase S-sulfocysteine intermediate involves the initial formation of a reversibly bound PAPS reductase S-sulfocysteine–Trx complex followed by covalent modification (e.g., mixed disulfide formation) and, thus, irreversible inhibition:
The scheme is analogous to the Michaelis–Menten mechanism, and the reaction exhibits saturation kinetics with an increasing inhibitor concentration (26):
Kinetic data were obtained in at least two independent experiments, and the standard error was less than 15%. Due to the limit of detection, the minimum practical protein concentration used in these experiments was 0.5 μM, and therefore, the reported apparent Ki represents an upper limit on the affinity between the PAPS reductase S-sulfocysteine intermediate and Trx.
Crystallographic Analysis
Crystals were grown by vapor diffusion in sitting drops containing 2 μL of the E. coli PAPS reductase–Trx Cys35Ala complex prepared as described above at 1.5 mg/mL in 50 mM Tris-HCl (pH 8.0) with NaCl added to an ionic strength of 150 mM, and 2 μL of a reservoir solution consisting of 10% (w/v) PEG3400 in 0.2 M KH2-PO4 (pH 4.6). Orthorhombic crystals with dimensions of 200 μm × 30 μm × 20 μm grew at 18 °C over 3–4 weeks. Crystals were briefly soaked in a cryoprotectant solution of 7% (w/v) PEG3400, 0.2 M KH2PO4 (pH 4.6), and 20% (v/v) glycerol and flash-frozen in liquid N2. Forty-seven crystals were evaluated at SSRL BL 1-5 using automated screening Blu-ice hutch control software and subjected to in situ annealing between 100 K and room temperature for 4–10 s intervals. One crystal annealed twice for 4 s displayed significant unit cell shrinkage and anisotropic diffraction to 2.8 Å resolution; a complete data set was collected to 3.0 Å (Table 1).
Table 1.
Crystallographic Statistics
| Unit Cell and Data Collection | |
| space group | C222(1) |
| a, b, c (Å) | 40.93, 111.15, 153.43 |
| SSRL beamline | BL 1-5 |
| wavelength (Å) | 0.9794 |
| resolution (Å) | 3.00 |
| no. of observations | 33575 |
| no. of reflections | 7334 |
| redundancy | 4.6 |
| completenessa (%) | 99.7 (99.6) |
| 〈I〉/〈σI〉a | 9.7 (2.1) |
| Rsymm(I)a | 0.126 (0.340) |
| Refinement | |
| resolution range (Å) | 50.0–3.00 |
| no. of reflections >0.0σF | 7319 |
| R-factora | 0.297 (0.381) |
| Rfreea (5.0% of data) | 0.308 (0.454) |
| rmsd for bonds (Å) | 0.004 |
| rmsd for angles (deg)b | 1.24 |
| Model
| ||
|---|---|---|
| protein | residues | 〈B-factor〉 (Å2) |
| PAPS reductase | 2–220, 235–244 | 56.4 |
| Trx1 | 1–108 | 58.5 |
Values for the highest-resolution shell (3.11–3.00 Å) are in parentheses.
Ramachandran plot: 79.6% of residues in most favored regions, 18.4% in allowed regions, and 2.0% in generously allowed regions.
The structure was determined by molecular replacement using Phaser (27) to locate the PAPS reductase molecule and, subsequently, Molrep (28) to locate Trx. The search models comprised residues 2–216 of E. coli PAPS reductase from an independently determined structure (data not shown) homologous with PDB entry 1SUR (9) and E. coli Trx (PDB entry 2TRX). Following rigid body refinement using Refmac (29), σA-weighted 2|Fo| − |Fc| maps exhibited clear density for residues 235–244 of PAPS reductase in association with Trx and for the disulfide bond between Cys239 and Cys32. The arrangement of the interacting protein molecules in the asymmetric unit is physically constrained by the lattice symmetry and length of the C-terminal peptide. Five cycles of refinement with Buster (30) were used to complete the initial model (31), which was refined with CNS (32) using tight restraints on geometry and group B-factors (Table 1). Coordinates and structure factors have been deposited with the PDB (entry 2O8V). Figures of protein structures and surface area calculations were generated using PyMOL (33). The Ramachandran plot was computed with PROCHECK in CCP4 (28).
The model of the sulfite moiety was derived from the refined structure of P. aeruginosa APS reductase (8), using the sulfite group of the substrate APS to attach it 2.0 Å from the Sγ atom of Cys239 in the PAPS reductase structure (for the Sγ–SO3− bond), while maintaining tetrahedral bond angles. The energy was not minimized, but the geometry was idealized. It was unnecessary to adjust any atoms of PAPS reductase or Trx.
RESULTS
Trapping the Covalent Trx–PAPS Reductase Intermediate
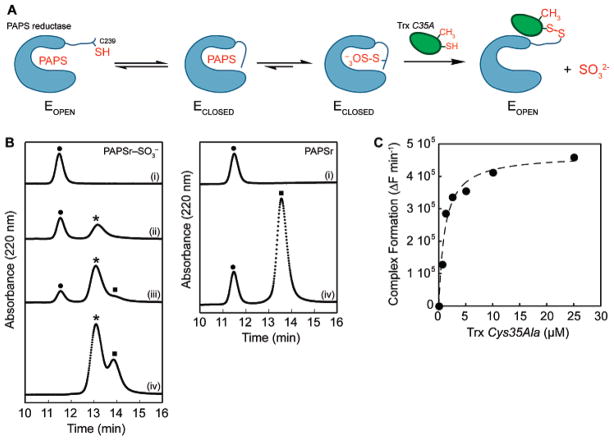
Our first experimental objective was to devise a strategy that would stabilize the complex between PAPS reductase and Trx1. Under normal conditions, the predicted protein–protein intermediate is too fleeting for direct interrogation by biochemical or X-ray crystallography methods (3). To facilitate its capture, the resolving cysteine at the active center of Trx, Cys35 (Trx residues italicized), was changed to alanine, which would selectively affect the breakdown, but not the ability to form the protein–protein intermediate (34) (Figure 2A). This variant of Trx was added either to the PAPS reductase S-sulfocysteine intermediate (formed via incubation with PAPS; see Experimental Procedures) or to PAPS reductase only. Formation of the Trx–PAPS reductase complex was monitored by reversed phase HPLC (Figure 2B), nonreducing SDS–PAGE (Figure S1 of the Supporting Information), and size-exclusion chromatography (data not shown). Mass spectrometry analysis of proteins isolated by reversed phase HPLC confirmed the identities of observed peaks (Figure 2B). The native molecular mass of the PAPS reductase–Trx complex estimated using size-exclusion chromatography was ~90 kDa, consistent with homodimeric PAPS reductase in association with two Trxs. Denaturing analysis of the Trx–PAPS reductase complex using reversed phase HPLC or SDS–PAGE disrupted the noncovalent interactions between monomers in the PAPS reductase homodimer. The Trx–PAPS reductase complex, however, remained covalently associated via the mixed disulfide bond (Figure 2 and Figure S1); the molecular mass of this species as determined by electrospray mass spectrometry and mobility via SDS–PAGE was ~45 kDa. Incubation of the protein complex with a chemical reducing agent, such as DTT, reduced the disulfide bond between Trx and PAPS reductase, as expected (data not shown).
Figure 2.
Formation of the PAPS reductase–Trx1 protein complex. (A) Strategy for trapping the E. coli PAPS reductase–Trx1 protein complex. (B) Complex formation was monitored at 220 nm by reversed phase HPLC analysis and plotted as a function of time. E. coli Trx1 Cys35Ala (10 μM) was incubated with increasing amounts PAPS reductase S-sulfocysteine intermediate [0 (i), 4 (ii), 9 (iii), and 20 μM (iv) at the left] or PAPS reductase alone [0 (i) and 20 μM (iv) at the right]. Mass spectrometry analysis of the observed peaks indicated masses of 13 937.1 Da for Trx1 [(●) 13 937.8 Da, theoretical], 30 007.3 Da for PAPS reductase [(■) 30 007.5 Da, theoretical], and 43 941.8 Da for the PAPS reductase–Trx1 complex [(*) 43 942.4 Da, theoretical]. (C) Rate of formation of the PAPS reductase–Trx1 complex from the PAPS reductase S-sulfocysteine intermediate (1 μM) plotted as a function of Trx Cys35Ala concentration, monitored by SYPRO ruby-stained nonreducing SDS–PAGE gel analysis. The dashed line represents a fit of the data to the equation derived for active site-directed irreversible inhibitors as described in Experimental Procedures and gave an apparent Ki value of 1.1 μM for the PAPS reductase S-sulfocysteine intermediate and Trx1 Cys35Ala.
The PAPS reductase–Trx1 complex formed readily from the S-sulfocysteine intermediate (Figure 2 and Figure S1). To estimate the apparent dissociation constant (Ki) of Trx Cys35Ala for the PAPS reductase S-sulfocysteine intermediate (see Experimental Procedures), the rate of complex formation could be quantified by measuring the change in peak area for free Trx using HPLC or fluorescence of SYPRO ruby-stained nonreducing SDS–PAGE gels. The later method exhibited a more sensitive limit of detection and was therefore used to determine an apparent Ki of 1.1 μM for these species (Figure 2C). Formation of a covalent complex between P. aeruginosa APS reductase and E. coli Trx Cys35Ala has been reported in previous biochemical work (35). A high affinity for the S-sulfated enzyme was observed when P. aeruginosa APS reductase was reacted with the P. aeruginosa Trx Cys35Ala variant (Figure S2 of the Supporting Information), consistent with the general mechanism proposed for this family of enzymes. The PAPS reductase–Trx complex, formed via reaction of the PAPS reductase S-sulfocysteine intermediate with Trx1, represents a stabilized mixed disulfide intermediate along the reaction pathway and was the focus of subsequent structural investigation.
Protein–Protein Interaction
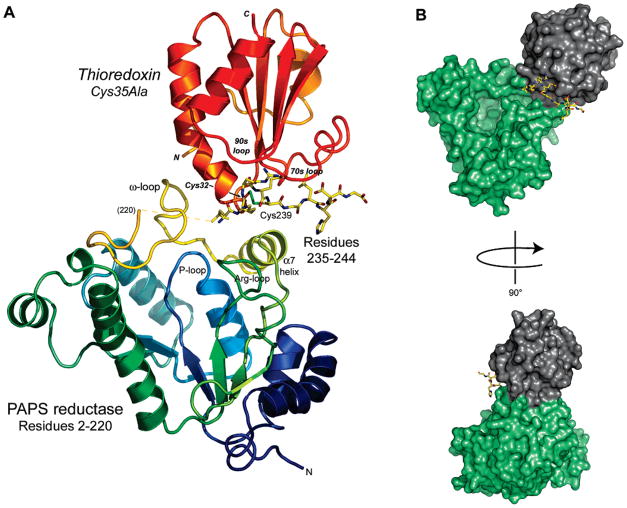
The structure of the PAPS reductase–Trx1 complex was determined to 3.0 Å (Table 1). Trx binds to PAPS reductase at the enzyme surface adjacent to the active site cleft (Figure 3A). The association of residues 2–220 and 235–244 within the same PAPS reductase molecule is uniquely defined; all other possible connections between residues 2–220 and symmetry-related residues 235′–244′ are physically impossible. With the connection of residues 2–220 and 235–244 established, the linkage of residues 235–244 to Trx is established by the continuous electron density of the mixed disulfide. Consequently, the unique association of PAPS reductase and Trx1 is defined by the crystal structure. The protein surfaces are complementary in shape, and 800 Å2 of solvent accessible surface is buried at the PAPS reductase–Trx interface; an additional 783 Å2 is buried between Trx and PAPS reductase C-terminal residues 235–244 (Figure 3B). This segment includes Cys239 in the mixed disulfide with Cys32. The preceding tail residues (221–234) span above the active site adjacent to the PAPS reductase–Trx interface but are disordered (Lys220–Leu235 distance of 13.5 Å). An increase in Trp fluorescence occurs when E. coli PAPS reductase is incubated with Trx (6). Consistent with these data, Trp221 would experience different solvent exposure as the C-terminal residues rearrange to bind Trx.
Figure 3.
Overall view of the PAPS reductase–Trx1 complex in the cocrystal structure. (A) The flexible C-terminal tail of the reductase (residues 235–244; all atoms shown) fits into a groove on Trx1 comprised of the 30s, 70s, and 90s loops (Trx1 residues 28–33, 71–77, 91–96, respectively; Trx1 residues in italics); the mixed disulfide is formed between Cys239 of the reductase and Cys32 of Trx. Trx in turn is bound to PAPS reductase among an ω-loop (residues 202–212, colored yellow), helix α7, and the C-terminal Cys239-peptide (residues 235–244). Residues 221–234 of PAPS reductase are disordered in the cocrystal structure (indicated by a dashed yellow line in front of the ω-loop). (B) Solvent accessible surface depiction of the PAPS reductase–Trx1 complex. In panel B and in Figures 4 and 5, PAPS reductase residues 1–220 are colored green, C-terminal peptide residues 235–244 are colored yellow, and Trx is colored gray.
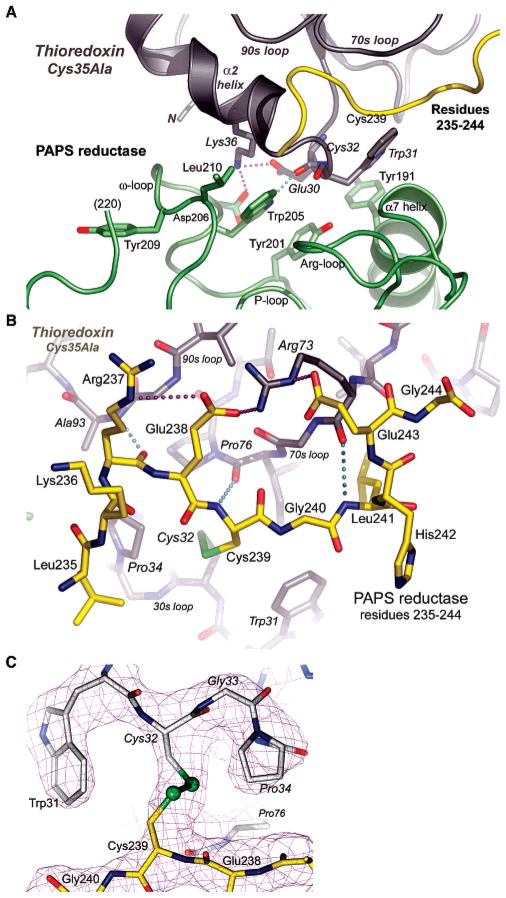
Trx’s α-helix 2 and active site Cys32Gly33Pro34Cys35 loop fit into a depression on PAPS reductase formed on three sides by α-helix 7 (following strand 6 of the β-sheet, residues 186–198), by residues 199–220 wrapping around the phosphate-binding loop (P-loop) at the back of the active site, and by PAPS reductase residues 235–244 (hereafter, identified as the “Cys239 peptide”) (Figure 4A). The segment of residues 199–220 contains an ω-loop (36) at residues 202–212 involved in specific interactions with Trx. Trp205 hydrogen bonds with the carbonyl of Glu30, and Asp206 participates in the Lys36–Glu30 salt bridge. PAPS reductase residues Asp206 and Glu207 are also proximal to Ser2 at the N-terminus of Trx. Aromatic stacking interactions occur for Tyr201 with Trp205 and for Tyr191 with Trp31. Hydrophobic contacts include Leu210 with Met37 and Gly33 in the Cys32Gly33Pro34Cys35 loop, and with Leu235. Hence, the protein–protein interface includes specific hydrogen bonds, and electrostatic, aromatic, and hydrophobic interactions.
Figure 4.
(A) Specific recognition of PAPS reductase by Trx1 occurs via residues in an ω-loop of the reductase and the 30s loop of Trx1 and involves hydrogen bonds, aromatic stacking, and hydrophobic interactions. The view is the same as in Figure 3A. (B) Specific recognition of the C-terminal peptide of PAPS reductase (yellow) by Trx1 (gray) involves three hydrogen bonds between main chain atoms, hydrophobic contacts with Leu235 in a pocket on TrxA, and interdigitation of Arg237, Glu238, Arg73, and Glu243 in a network of three salt bridges. The view is the same as in Figure 3A. (C) Electron density for the mixed disulfide formed between PAPS reductase Cys239 and Trx1 Cys32 (σA-weighted 2|Fo| − |Fc| map contoured at 1.7σ). Density for adjacent residues in the PAPS reductase C-terminal peptide (yellow) and in the TrxA 30s loop (gray) is also shown. The view is from behind the complex as shown in Figure 3A (i.e., the Glu238Cys239Gly240 peptide orientation is reversed).
Trx–PAPS Reductase Complex Cys239–Peptide Interaction
The PAPS reductase Cys239–peptide interaction clasps the Cys32Gly33Pro34Cys35 loop from the side opposite the ω-loop (Figure 4A). Reciprocally, the peptide lies in a shallow groove formed between the 30s loop (Trx residues 28–33) at the protein–protein interface (Trp31Cys32Gly33Pro34-Cys35; Ala35 in the cocrystal structure) and 70s and 90s loops (Trx residues 71–77 and 91–96, respectively) (Figure 4B). The chain directions of the 30s, 70s, and 90s loops are locally antiparallel to the Cys239–peptide. The conformation of the Cys239–Cys32 bond is (−,+,+,+,−), placing it in the commonly occurring –RHspiral class, consistent with a 5.93 Å separation between Cα atoms (Figure 4C) (37). The disulfide is surrounded by hydrophobic residues, including Trp31, Pro34, Ala35 (Cys35), Ile75, and cis-Pro76, but exposed to solvent in the direction of Cys239. The Cαi–Cβi–Sγi–Sγj, Cαj–Cβj–Sγj–Sγi, and Cβi–Sγi–Sγj–Cβj dihedral angles (i = Cys32Trx1 and j = Cys239) are 57°, 117°, and 92°, respectively. The parameters observed for the PAPS reductase–Trx intermolecular disulfide bond are quite similar to those observed in the BASI–Trx complex (25). In particular, both disulfide bonds exhibit right-handed geometry and similar dihedral angles and Cα atom distances.
Cys239–peptide residues 237–242 adopt an extended β-conformation and bridge the 70s and 90s loops to form main chain hydrogen bonds between the Arg237 carbonyl and Ala93 amide, the Cys239 amide and Ile75 carbonyl, and the Leu241 amide and Arg73 carbonyl (Figure 4B). In this conformation, Leu241 fits into a hydrophobic pocket comprised of Trx residues Trp31, Ile60, Ala67, Ile72, and Ile75. On its solvent-exposed side, the Cys239-peptide participates in an interdigitated network of salt bridges that incorporates Arg237, Glu238, Arg73, and Glu243 (Figure 4B); His242 is within 4.5 Å of Asp186 on the reductase. Consequently, the Cys239-peptide is fully recognized, with hydrophobic pockets for the mixed disulfide and Leu241, three main chain hydrogen bonds with Trx, and an interprotein array of three salt bridges. In accord with this recognition, the glutathione-like sequence, Glu238Cys239Gly240Leu241His242, is strictly conserved in PAPS reductases, residues Lys236, Arg237, and Asp186 are strongly conserved, and Trx residue Arg73 is absolutely conserved (8).
Quaternary Structure
Both E. coli PAPS reductase and the PAPS reductase–Trx1 complex crystallize as dimers in space group C2221 with local 2-fold axes superimposed on crystallographic 2-fold axes, consistent with each being a homodimer in solution (3, 9). The identity of the solution dimer cannot be deduced from symmetry considerations, because the asymmetric unit contains just one PAPS reductase–Trx1 complex and because all 2-fold axes are equivalent by the space group operators. Hence, the most likely dimer can be inferred only by homology. Because the rmsd between Cα atoms of PAPS reductase subunits is 1.09 Å in the two structures, we expect that the PAPSr–Trx1 solution dimer resembles that proposed for the PAPS reductase dimer (9) (Figure S3 of the Supporting Information). In this homodimer, the active sites face away from each other and the distance between Lys220 residues is ~36 Å (i.e., the C-terminal tail cannot interact with the opposite subunit), and a total of 790 Å2 of accessible surface area is buried at the interface. P. aeruginosa APS reductase is also structurally homologous to E. coli PAPS reductase (8). In this case, the solution homotetramer crystallizes in a triclinic unit cell with local 222-fold symmetry only. Interestingly, the proposed PAPS reductase and PAPS reductase–Trx1 homodimers correspond to the AC pair in the P. aeruginosa APS reductase crystal structure. Thus, a common homodimer interface is observed in three crystal structures, irrespective of lattice contacts or space group.
DISCUSSION
The protein complex used for these studies was formed via reaction of Trx Cys35Ala with PAPS reductase S-sulfocysteine intermediate, E-Cys-Sγ-SO3−; the S-sulfocysteine intermediate has been characterized in previous biochemical and mass spectrometry studies (3, 8, 21). Since (1) a stable enzyme–S-sulfocysteine intermediate forms upon addition of sulfonucleotide substrate, (2) reduction of the intermediate, and concomitant production of sulfite, is Trx-dependent, and (3) biochemical and structural data are not consistent with disulfide formation between Cys239s in the PAPS reductase homodimer, we have proposed the mechanism for sulfonucleotide reduction illustrated in Figure 1B (3). Structural observation of the PAPS reductase C-terminal tail outside of the active site, and in highly specific interaction with Trx, provides strong support for the central feature of the proposed mechanism, that formation of S-sulfocysteine is followed in subsequent steps by nucleophilic attack of Trx Cys32 on the Sγ atom of this intermediate. These structural findings are further supported by the high-affinity interaction between Trx and S-sulfated SRs (Figure 2 and Figure S2).
In the cocrystal structure, Trx interacts with the last 10 residues of the PAPS reductase C-terminal tail, spanning ~20 Å (Figure 4B). Recognition of this segment consists of hydrophobic contacts, main chain hydrogen bonds, and a network of interprotein salt bridge interactions. As a consequence of these extensive contacts, Cys32 is exquisitely positioned for nucleophilic attack at the Cys239 Sγ atom of the S-sulfocysteine, as modeled in Figure 5 from the cocrystal structure. The Cys35 Sγ atom would, in turn, be suitably positioned to resolve the mixed protein disulfide (distance to the Cys32 Sγ atom of <3 Å based on the position Ala35). In terms of S-sulfocysteine recognition prior to nucleophilic attack by Trx, it is interesting to note the formation of a putative sulfite-binding pocket at the protein–protein interface (Figure 5) by a constellation of strongly conserved residues (8). The pocket contains a potential hydrogen bond with the Gly33 amide, while the PAPS reductase residue Asn187 (positioned by Arg157 and Glu159) can hydrogen bond to the carbonyl of Trp31 and the sulfite moiety. Pro34 and Leu235 define the other side of the pocket. In this model, recognition of S-sulfate can entail hydrogen bonds with two amides, while sulfite release is accommodated by access to the solvent. Consistent with the conservation of mechanism, an analogous sulfite-binding pocket forms at the interface of P. aeruginosa APS reductase when it is superimposed on the PAPS reductase–Trx structure (8).
Figure 5.
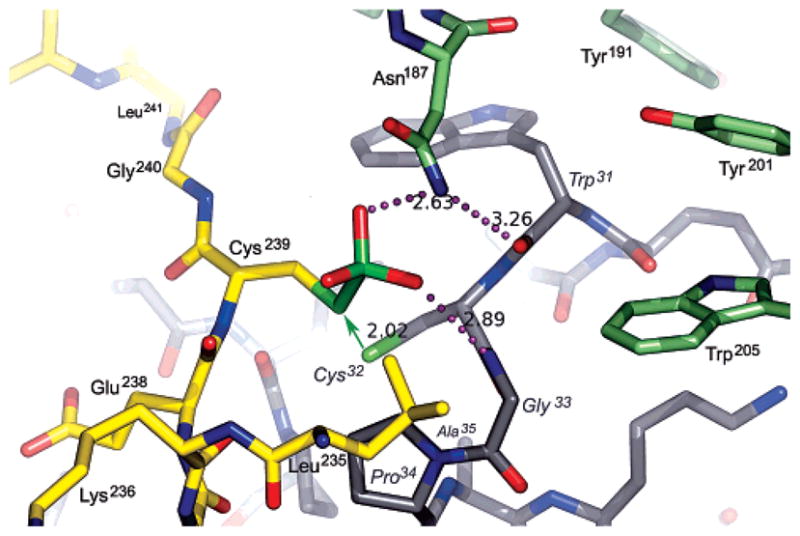
Model for the S-sulfocysteine intermediate of PAPS reductase bound to Trx1, based on the crystal structure of the mixed disulfide complex. The sulfite moiety fits into a pocket at the PAPS reductase–Trx1 interface. PAPS reductase residues in the C-terminal tail are colored yellow and other residues green; Trx1 residues are colored gray with labels in italics. Potential hydrogen bonds (dotted lines; distances in angstroms) include those from Asn187 to the sulfite and the carbonyl of Trp31 and from the amide of Gly33 to the sulfite. The green arrow represents the nucleophilic attack that must occur for formation of the mixed disulfide and displacement of sulfite; the distance between the Sγ atoms of Cys239 and Cys32 in the cocrystal structure is 2.02 Å. The resolving cysteine of Trx, Cys35, is mutated to Ala in the complex (visible below Pro34 in the 30s loop). With respect to Figure 3A, the view is approximately from the N-terminus of Trx1 looking along the axis of the C-terminal peptide.
In both NFκB and Ref-1 structures, at least nine of 13 residues adopt an extended β-conformation across the 30s, 70s, and 90s loops of Trx, analogous to the extensive interactions observed for PAPS reductase. These loop segments in Trx, aptly termed the “substrate recognition loop motif”, also play a primary role in molecular recognition of BASI residues in the BASI–Trx complex (25). The chain direction of PAPS reductase and BASI peptide residues adjacent to the mixed disulfide are antiparallel to Trx substrate binding loops. However, loop segments in HvTrxh2 interact with only three BASI residues, Asp146Trp147Cys148 (with Cys148 forming part of the mixed disulfide). One possible explanation proposed by Maeda et al. for the larger number of residues in Trx–peptide interactions is that the synthetic NFκB and Ref-1 substrates possess more conformational freedom, relative to folded BASI protein, and are therefore able to make more contacts. Consistent with this hypothesis and the mobility of the 25 C-terminal residues of PAPS reductase, more extensive interactions with Trx are observed in the structure presented here.
Glutaredoxin (Grx) can also act as a protein reductant for E. coli PAPS reductase (38) and M. tuberculosis APS reductase (K. S. Carroll, unpublished observations). Grxs belong to the Trx superfamily and share a common fold, including the position of several loop segments and the active site cysteine residues. In this context, related structures are E. coli Grx-1 in complex with a peptide fragment derived from ribonucleotide reductase (39) and Grx-1 and Grx-3 in complex with the tripeptide glutathione, GluCysGly (40, 41). The latter comparisons are particularly relevant since the C-terminal residues in PAPS reductase contain an absolutely conserved glutathione-like motif, Glu238Cys239Gly240Leu241-His242. Recognition of the tripeptide by Grx is largely electrostatic, and in accord with these observations, hydrogen bonding and salt bridge interactions dominate recognition of C-terminal peptide residues in PAPS reductase by Trx.
Chemical reductants with redox potentials similar to that of Trx, such as DTT or GSH, do not efficiently cleave the S-sulfocysteine in the folded enzyme (3, 8). The simplest interpretation of this observation is that access to the S-sulfate intermediate is restricted, as C-terminal residues remain closed over the active site. Hence, the S-sulfocysteine moiety remains sequestered in the active site until the C-terminal residues are displaced through protein–protein interaction with Trx, the state evidenced in the structure presented here. Interestingly, conformational rearrangement involving a labile intermediate is also observed in the cysteine desulfurylase, SufS, a component of the SUF iron–sulfur cluster biosynthetic system (42, 43). These pyridoxal phosphate-dependent proteins catalyze the conversion of L-cysteine to L-alanine and sulfane sulfur via a covalent persulfide intermediate (44). The catalytic cysteine nucleophile resides on a highly mobile loop that transfers a sulfide group to another protein, such as SufE, without releasing it into solution (45, 46). Hence, a general strategy for protecting labile, reactive intermediates in sulfur metabolism appears to be to incorporate mobile structural elements in the enzymes mediating transfer.
With regard to residues and protein–protein contacts that could be critical for mediating conformational changes in the PAPS reductase C-terminal domain, we consider both the cocrystal structure presented here and the structure of the E. coli PAPS reductase (PDB entry 1SUR), crystallized in the absence of substrate and Trx (9) (Figure S3). The final 30 residues in the C-terminus of 1SUR are disordered, and thus, a direct comparison of this mobile element is not possible. Nonetheless, conformational rearrangements are observed in two regions adjacent to the active site (and are not involved in crystal contacts). One of the mobile elements is the Arg loop (Arg157Arg158Glu159Gln160Ser161Gly162Ser163-Arg164) between β4 and β5. On the basis of structural similarities of substrate-bound APS reductase, Arg164 (one of only six absolutely conserved residues in PAPS reductase) is predicted to contact the oxygen that bridges the substrate phosphorus and sulfur atoms (8). In 1SUR, the Arg loop is folded over the active site, and in the structure presented here, the Arg loop is displaced onto the enzyme surface. Additional conformational differences are also observed in the ω-loop, with Tyr209 and other residues shifting relative to 1SUR. Mutation of Tyr209 to Phe alters the Michaelis constant (Km) of E. coli PAPS reductase with Trx (6), in accord with its location proximal to the protein–protein interface and to Lys220 in the C-terminal tail. Taken together, these observations suggest that conformational changes associated with Trx binding involving residues in the C-terminal tail, the flexible Arg loop (e.g., Leu235 and Ser161, respectively), and the ω-loop could collectively facilitate displacement of the S-sulfated C-terminal residues from the active site.
Integrating the cocrystal structure with prior structural and biochemical studies, we can formulate a model for conformational rearrangement during the SR catalytic cycle (Figure 2A). We hypothesize that binding of the sulfonucleotide substrate favors closure of the C-terminal residues over the active site. This is a logical step since the C-terminal residues contain the essential cysteine nucleophile and must come into the proximity of the substrate. Also consistent with the proposed ordering of these residues, formation of the S-sulfocysteine intermediate confers proteolytic resistance to the C-terminal tail and Arg loop (8). Subsequent association of Trx with PAPS reductase adjacent to the active site, and accompanying conformational rearrangements, displaces the S-sulfated C-terminus from the active site and is consistent with the state observed in our cocrystal structure. The observed protein–protein interaction strategically positions Trx to bind the displaced residues and reduce the S-sulfocysteine intermediate in subsequent steps. Consequently, key steps in the catalytic cycle, S-sulfocysteine formation and reduction, are delineated by considerable structural rearrangement in PAPS reductase and take place at distinct sites on the enzyme.
Our structure of Trx with a full-length protein target affords the opportunity to identify additional sites involved in protein–protein recognition. In this context, our structure reveals that Trx interacts with PAPS reductase at a second site adjacent to the ω-loop and involves contacts between Lys36, Glu30, Trp31, and Trp205 and Asp206, all highly conserved residues (Figure 4A). This observation (and the additional 800 Å2 of buried surface area) is an important feature that distinguishes the PAPS reductase–Trx complex from previously reported synthetic peptide fragment- or BASI–Trx complexes. In addition to substrate reduction, the functional consequences of this interaction (e.g., mediating conformational rearrangements) are significant for the mechanism of sulfonucleotide reduction and may represent a common theme among enzymes that require Trx. In addition to the BASI–Trx complex, only one other structure of Trx in a mixed disulfide with a full-length protein has been reported. This landmark structure, the complex between Trx and its in vivo reductant (Trx reductase), was determined to 3.0 Å resolution by Lugwig and co-workers (47). In this structure, Trx makes multiple contacts with Trx reductase NADPH and FAD nucleotide-binding domains to promote a large-scale domain rotation that is essential for the transfer of electrons in this reaction. Trx has also been crystallized in a noncovalent complex with T7 DNA polymerase, an interaction that stabilizes a processive conformation but does not involve redox chemistry (48). A notable feature of the cocrystal structures in which multiple Trx–protein interacting sites are observed (e.g., Trx reductase and T7 DNA polymerase) is the association of Trx with regions of mobility and conformational change, which is analogous to the situation in the PAPS reductase–Trx complex.
The structure reported here reveals unique interactions between Trx and PAPS reductase with significant implications for the chemical and conformational aspects of sulfo-nucleotide reduction. Visualization of the PAPS reductase C-terminal tail outside the active site, and in association with Trx, supports the essential feature of the mechanism of sulfonucleotide reduction, that formation of the S-sulfocysteine intermediate, and its subsequent reduction, are discrete chemical steps of the overall reaction. Two key states remaining to be characterized structurally are the PAPS reductase S-sulfocysteine intermediate in complex with Trx prior to cleavage and the S-sulfated peptide with the product, PAP, bound in the active site. This structure motivates experiments that target the observed peptide and protein recognition surfaces as sites for inhibition of sulfonucleotide reductase activity.
Supplementary Material
Acknowledgments
We thank Profs. Janet Smith, Rowena Matthews, and Carolyn Bertozzi for many helpful discussions. We thank the generous assistance of the staff at the Stanford Synchrotron Radiation Laboratory (SSRL). SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Footnotes
This work was supported by National Institutes of Health Grants GM-48870 and AI-51622. K.S.C. was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation (DRG-1783-03) and by funding from the University of Michigan Life Sciences Institute.
The structure has been deposited in the Protein Data Bank as entry 2O8V.
Abbreviations: PAPS, 3′-phosphoadenosine-5′-phosphosulfate; Cys-Sγ-SO3−, S-sulfocysteine; APS, adensoine-5′-phosphosulfate; SR, sulfonucleotide reductase; Trx, thioredoxin; HPLC, high-performance liquid chromatography; TFA, trifluoroacetic acid; P-loop, phosphate-binding loop; Cys239-peptide, PAPS reductase residues 235–244; DTT, dithiothreitol; GSH, glutathione; apo-PAPS reductase, substrate-free PAPS reductase; E, enzyme.
SUPPORTING INFORMATION AVAILABLE
Relevant data for nonreducing SDS–PAGE analysis and the dimer interface (Figures S1–S3). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kredich NM. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Vol. 1. American Society for Microbiology; Washington, DC: 1996. [Google Scholar]
- 2.Schwenn JD. Photosynthetic Sulphate Reduction. Z Naturforsch. 1994;49c:531–539. [Google Scholar]
- 3.Carroll KS, Gao H, Chen H, Stout CD, Leary JA, Bertozzi CR. A conserved mechanism for sulfonucleotide reduction. PLoS Biol. 2005;3:e250. doi: 10.1371/journal.pbio.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopriva S, Buchert T, Fritz G, Suter M, Weber M, Benda R, Schaller J, Feller U, Schurmann P, Schunemann V, Trautwein AX, Kroneck PM, Brunold C. Plant adenosine 5′-phosphosulfate reductase is a novel iron-sulfur protein. J Biol Chem. 2001;276:42881–42886. doi: 10.1074/jbc.M107424200. [DOI] [PubMed] [Google Scholar]
- 5.Bick JA, Dennis JJ, Zylstra GJ, Nowack J, Leustek T. Identification of a new class of 5′-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J Bacteriol. 2000;182:135–142. doi: 10.1128/jb.182.1.135-142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berendt U, Haverkamp T, Prior A, Schwenn JD. Reaction mechanism of thioredoxin: 3′-Phospho-adenylylsulfate reductase investigated by site-directed mutagenesis. Eur J Biochem. 1995;233:347–356. doi: 10.1111/j.1432-1033.1995.347_1.x. [DOI] [PubMed] [Google Scholar]
- 7.Schwenn JD, Krone FA, Husmann K. Yeast PAPS reductase: Properties and requirements of the purified enzyme. Arch Microbiol. 1988;150:313–319. doi: 10.1007/BF00408300. [DOI] [PubMed] [Google Scholar]
- 8.Chartron J, Carroll KS, Shiau C, Gao H, Leary JA, Bertozzi CR, Stout CD. Substrate recognition, protein dynamics, and iron-sulfur cluster in Pseudomonas aeruginosa adenosine 5′-phosphosulfate reductase. J Mol Biol. 2006;364:152–169. doi: 10.1016/j.jmb.2006.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage H, Montoya G, Svensson C, Schwenn JD, Sinning I. Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: A new family of adenine nucleotide α hydrolases. Structure. 1997;5:895–906. doi: 10.1016/s0969-2126(97)00244-x. [DOI] [PubMed] [Google Scholar]
- 10.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. The growing burden of tuberculosis: Global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 11.Moore M, Onorato IM, McCray E, Castro KG. Trends in drug-resistant tuberculosis in the United States, 1993–1996. J Am Med Assoc. 1997;278:833–837. [PubMed] [Google Scholar]
- 12.Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Post-Martens K, Denkin S. New drug candidates and therapeutic targets for tuberculosis therapy. Drug Discovery Today. 2006;11:21–27. doi: 10.1016/S1359-6446(05)03626-3. [DOI] [PubMed] [Google Scholar]
- 14.Williams SJ, Senaratne RH, Mougous JD, Riley LW, Bertozzi CR. 5′-Adenosinephosphosulfate lies at a metabolic branch point in mycobacteria. J Biol Chem. 2002;277:32606–32615. doi: 10.1074/jbc.M204613200. [DOI] [PubMed] [Google Scholar]
- 15.Newton GL, Fahey RC. Mycothiol biochemistry. Arch Microbiol. 2002;178:388–394. doi: 10.1007/s00203-002-0469-4. [DOI] [PubMed] [Google Scholar]
- 16.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senaratne RH, De Silva AD, Williams SJ, Mougous JD, Reader JR, Zhang T, Chan S, Sidders B, Lee DH, Chan J, Bertozzi CR, Riley LW. 5′-Adenosine-phosphosulphate reductase (CysH) protects Mycobacterium tuberculosis against free radicals during chronic infection phase in mice. Mol Microbiol. 2006;59:1744–1753. doi: 10.1111/j.1365-2958.2006.05075.x. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez Porque P, Baldesten A, Reichard P. The involvement of the thioredoxin system in the reduction of methionine sulfoxide and sulfate. J Biol Chem. 1970;245:2371–2374. [PubMed] [Google Scholar]
- 19.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 20.Martin JL. Thioredoxin: A fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 21.Gao H, Leary J, Carroll KS, Bertozzi CR, Chen H. Noncovalent complexes of APS reductase from M. tuberculosis: Delineating a mechanistic model using ESI-FTICR MS. J Am Soc Mass Spectrom. 2007;18:167–178. doi: 10.1016/j.jasms.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber M, Suter M, Brunold C, Kopriva S. Sulfate assimilation in higher plants: Characterization of a stable intermediate in the adenosine 5′-phosphosulfate reductase reaction. Eur J Biochem. 2000;267:3647–3653. doi: 10.1046/j.1432-1327.2000.01394.x. [DOI] [PubMed] [Google Scholar]
- 23.Orr MD, Vitols E. Thioredoxin from Lactobacillus leichmannii and its role as hydrogen donor for ribonucleoside triphosphate reductase. Biochem Biophys Res Commun. 1966;25:109–115. doi: 10.1016/0006-291x(66)90647-4. [DOI] [PubMed] [Google Scholar]
- 24.Boschi-Muller S, Azza S, Sanglier-Cianferani S, Talfournier F, Van Dorsselear A, Branlant G. A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J Biol Chem. 2000;275:35908–35913. doi: 10.1074/jbc.M006137200. [DOI] [PubMed] [Google Scholar]
- 25.Maeda K, Hagglund P, Finnie C, Svensson B, Henriksen A. Structural basis for target protein recognition by the protein disulfide reductase thioredoxin. Structure. 2006;14:1701–1710. doi: 10.1016/j.str.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. W. H. Freeman and Co; New York: 1999. [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project No4 . The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. 2004;D60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 31.McRee DE. XtalView/Xfit: A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 34.Jeng MF, Reymond MT, Tennant LL, Holmgren A, Dyson HJ. NMR characterization of a single-cysteine mutant of Escherichia coli thioredoxin and a covalent thioredoxin-peptide complex. Eur J Biochem. 1998;257:299–308. doi: 10.1046/j.1432-1327.1998.2570299.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim SK, Rahman A, Mason JT, Hirasawa M, Conover RC, Johnson MK, Miginiac-Maslow M, Keryer E, Knaff DB, Leustek T. The interaction of 5′-adenylylsulfate reductase from Pseudomonas aeruginosa with its substrates. Biochim Biophys Acta. 2005;1710:103–112. doi: 10.1016/j.bbabio.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Fetrow JS. Omega loops: Nonregular secondary structures significant in protein function and stability. FASEB J. 1995;9:708–717. [PubMed] [Google Scholar]
- 37.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 38.Lillig CH, Prior A, Schwenn JD, Aslund F, Ritz D, Vlamis-Gardikas A, Holmgren A. New thioredoxins and glutaredoxins as electron donors of 3′-phosphoadenylylsulfate reductase. J Biol Chem. 1999;274:7695–7698. doi: 10.1074/jbc.274.12.7695. [DOI] [PubMed] [Google Scholar]
- 39.Nordstrand K, Åslund F, Holmgren A, Otting G, Berndt KD. NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: Implications for the enzymatic mechanism. J Mol Biol. 1999;286:541–552. doi: 10.1006/jmbi.1998.2444. [DOI] [PubMed] [Google Scholar]
- 40.Berardi MJ, Bushweller JH. Binding specificity and mechanistic insight into glutaredoxin-catalyzed protein disulfide reduction. J Mol Biol. 1999;292:151–161. doi: 10.1006/jmbi.1999.3067. [DOI] [PubMed] [Google Scholar]
- 41.Bushweller JH, Billeter M, Holmgren A, Wuthrich K. The nuclear magnetic resonance solution structure of the mixed disulfide between Escherichia coli glutaredoxin(C14S) and glutathione. J Mol Biol. 1994;235:1585–1597. doi: 10.1006/jmbi.1994.1108. [DOI] [PubMed] [Google Scholar]
- 42.Fontecave M, Choudens SO, Py B, Barras F. Mechanisms of iron-sulfur cluster assembly: The SUF machinery. J Biol Inorg Chem. 2005;10:713–721. doi: 10.1007/s00775-005-0025-1. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 44.Zheng L, White RH, Cash VL, Dean DR. Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- 45.Ollagnier-de-Choudens S, Lascoux D, Loiseau L, Barras F, Forest E, Fontecave M. Mechanistic studies of the SufS-SufE cysteine desulfurase: Evidence for sulfur transfer from SufS to SufE. FEBS Lett. 2003;555:263–267. doi: 10.1016/s0014-5793(03)01244-4. [DOI] [PubMed] [Google Scholar]
- 46.Tirupati B, Vey JL, Drennan CL, Bollinger JM., Jr Kinetic and structural characterization of Slr0077/SufS, the essential cysteine desulfurase from Synechocystis sp. PCC 6803. Biochemistry. 2004;43:12210–12219. doi: 10.1021/bi0491447. [DOI] [PubMed] [Google Scholar]
- 47.Lennon BW, Williams CH, Jr, Ludwig ML. Twists in catalysis: Alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000;289:1190–1194. doi: 10.1126/science.289.5482.1190. [DOI] [PubMed] [Google Scholar]
- 48.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 22 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.