SUMMARY
The spindle assembly checkpoint (SAC) delays anaphase onset until kinetochores accomplish bioriented microtubule attachments [1]. While several centromeric and kinetochore kinases, including Aurora B, regulate kinetochore-microtubule attachment and/or SAC activation [2–4], the molecular mechanism that translates bioriented attachment into SAC silencing remains unclear [5]. Employing a method to rapidly induce exact gene replacement in budding yeast [6], we show here that the binding of protein phosphatase 1 (PP1/Glc7) to the evolutionarily conserved RVSF motif of the kinetochore protein Spc105 (KNL1/Blinkin/CASC5) is essential for viability by silencing the SAC, while it plays an auxiliary nonessential role for physical chromosome segregation. Although Aurora B may inhibit this binding, persistent PP1-Spc105 interaction does not affect chromosome segregation and is insufficient to silence the SAC in the absence of microtubules, indicating that dynamic regulation of this interaction is dispensable. However, the amount of PP1 targeted to kinetochores must be finely tuned, since recruitment of either none or one extra copy of PP1 to Spc105 is detrimental, illustrating the vital impact of targeting an exiguous fraction of PP1 to the kinetochore. We propose that the PP1-Spc105 interaction enables local regulation of dynamic phosphorylation and dephosphorylation at the kinetochore to couple microtubule attachment and SAC silencing.
Results and Discussion
PP1 (Glc7 in budding yeast) is the essential phosphatase that counteracts Aurora B (Ipl1) [7–9] and silences the SAC [10, 11]. The kinetochore protein KNL1 (Spc105) recruits PP1 through a conserved RVSF motif (Figure S1A), and abrogation of this interaction compromises kinetochore microtubule stability in HeLa cells [12, 13]. It was also suggested that Aurora B weakens this interaction at unattached kinetochores by phosphorylating KNL1 [12]. However, the critical function of this interaction and the physiological significance of its regulation remain speculative. We therefore sought to examine the Spc105-Glc7 interaction using the genetic tools of Saccharomyces cerevisiae.
A PP1 binding mutant of spc105 is lethal
We generated two mutants of the Spc105 RVSF motif, a PP1-binding mutant, RASA, and a potential phosphorylation site mutant, RVAF (see Figures S2A and B), by employing a method to rapidly introduce a site-specific mutation on the genome without selection (HO-induced Gene Replacement, or HGR) [6] (Figure 1A). At the promoter region of SPC105, we first inserted a cassette (spc105-NT), containing a partial gene encoding the N-terminal region of Spc105 with desired mutations, followed by an HO endonuclease cut site (HOcs). Cleavage at the HOcs by induction of GAL-HO stimulates homologous recombination between the truncated spc105 and the full length SPC105. This results in essentially all cells in the culture undergoing recombination to produce either wild-type or mutant spc105, depending on the site of crossover.
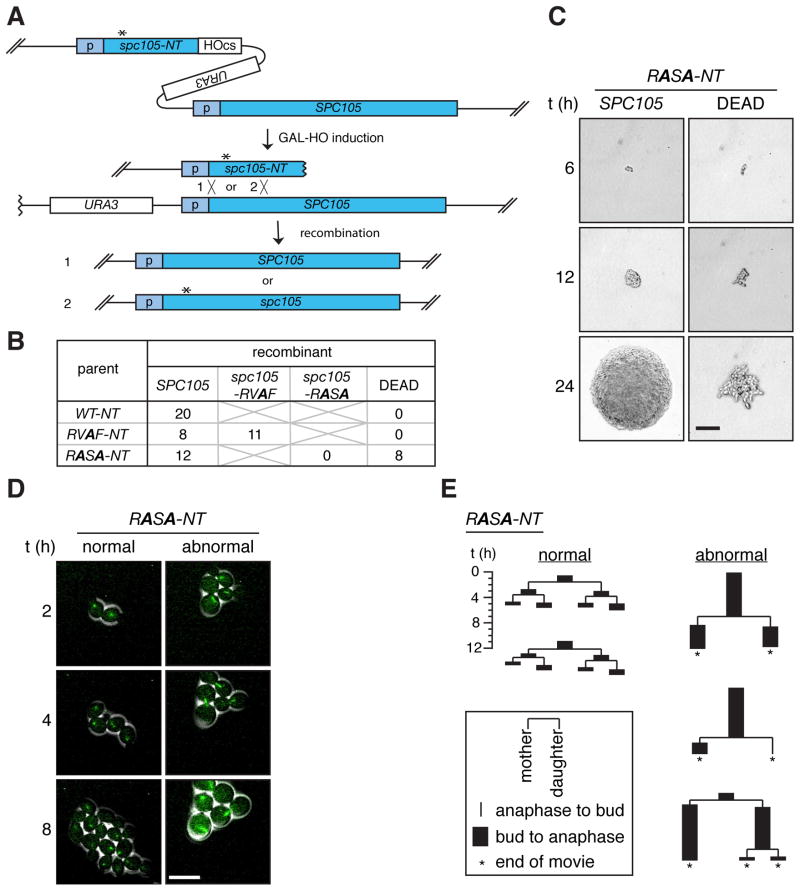
Figure 1. A PP1 binding mutant, spc105-RASA, is lethal with mitotic arrest.
(A) Schematic of the HGR method. Asterisk indicates the desired mutation. Homologous recombination after HO-induced DNA breaks generates full-length wild-type SPC105 (1) or mutant spc105 (2) (B) Single cell colony assay of cells harboring the WT-NT, RVAF-NT, and RASA-NT cassettes. Six hours after GAL-HO induction, single cells were isolated, allowed to grow into isogenic colonies, and genotyped. Number of colonies with the indicated genotypes or those that failed to form macroscopic colonies (DEAD) is shown. (C) Representative colonies of the two classes of recombinants resulting from the RASA-NT cassette were imaged at the times indicated after single cell isolation. The colony on the left harbors wild-type SPC105 as confirmed by genotyping analysis. Scale bar, 50 μm. (D) Time-lapse microscopy of GFP-Tub1 (green) was performed on RASA-NT cells beginning 6 hours after GAL-HO induction. Scale bar, 10 μm. (E) Pedigree analysis of recombinants generated from the cells harboring the RASA-NT cassette during live cell imaging (Movies S2, S3). Each lineage starts from a single unbudded cell and the duration of budding to anaphase (black rectangle) and anaphase to budding (line) were measured for three generations or until the end of the movie (asterisks). At each division, fates of the mother cell and the daughter cell are shown on the left and right, respectively. Representative lineages showing normal cell divisions (left 2 examples) and abnormal cell divisions (right 3 examples) are shown. See also Movies S1–3 and Figure S1.
Six hours after recombination induction, individual cells were isolated to monitor colony formation. Hereafter, this assay will be referred to as a single cell colony assay. From the wild-type- and RVAF-inducing parents (WT-NT and RVAF-NT, respectively), all of the single cells uniformly formed colonies. Genotype analysis revealed that the wild-type SPC105 recombinant was generated in all 20 tested cells from WT-NT cells, whereas both wild-type and spc105-RVAF recombinants were isolated at comparable frequency from RVAF-NT cells (Figure 1B, S1B). The recovered spc105-RVAF mutant did not show any proliferation defect (Figure S1C).
From the RASA-inducing parents (RASA-NT), all of the cells that produced normal colonies were wild-type SPC105 (Figures 1B and S1B), but a similar number of cells failed to make macroscopic colonies, arresting with elongated cells after several cell divisions (Figure 1C). Although we could not genotype these microcolonies, we confirmed that mutant spc105-RASA genes were generated in bulk culture directly after recombination (Figure S1D). Thus, we infer that this lethal phenotype is due to the spc105-RASA mutation.
The HGR method allowed us to monitor cell cycle progression of recombinant cells right after introduction of the point mutation by imaging microtubules using Tub1-GFP. Recombinants generated from cells containing the wild-type control (data not shown) or the spc105-RVAF cassette (Figure S1E and Movie S1) all divided normally. From the spc105-RASA cassette, about half of the cells behaved similarly to wild-type (“normal”), while the rest showed long periods of delay (> 6 h) with large buds and a short spindle (“abnormal”) (Figures 1D, E and Movies S2, S3).
Lethality of spc105-RASA mutant can be rescued by an ipl1 mutation or by elimination of the SAC
We assume that the spc105-RASA phenotype is caused by impaired local dephosphorylation at kinetochores due to a failure to recruit Glc7. We therefore tested whether dampening the antagonizing kinase Ipl1 might rescue the lethality of spc105-RASA. Indeed, in the ipl1-1 background, viable spc105-RASA cells were recovered at the semipermissive temperature for ip1-1 (30°C), but these spc105-RASA ipl1-1 cells did not grow at the permissive temperature (23°C) (Figure 2A). Conversely, the spc105-RASA mutation partially rescued the temperature sensitivity of ipl1-1 at 37°C. These results support the idea that the function of the Spc105 RVSF motif is to counteract Ipl1 by recruiting Glc7 to the kinetochore.
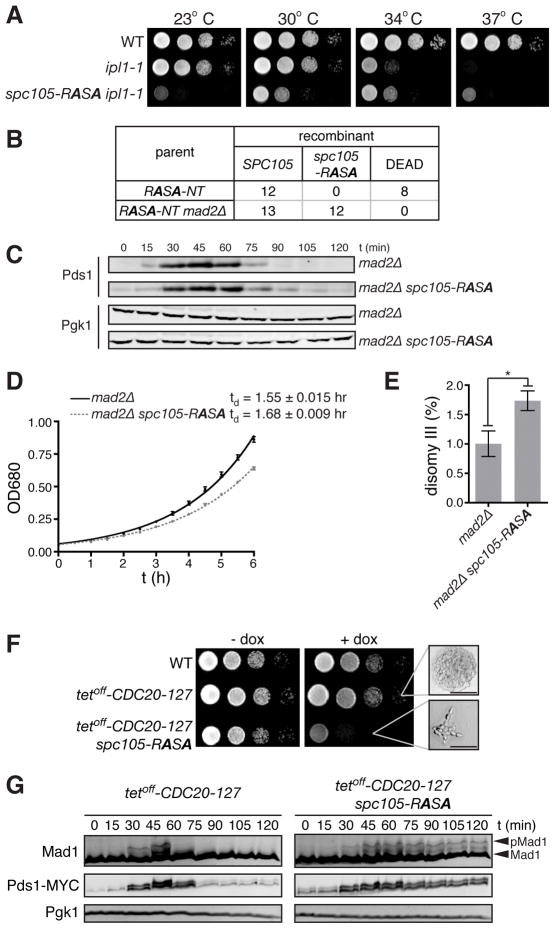
Figure 2. spc105-RASA lethality is rescued by dampened Ipl1 activity or impaired SAC.
(A) Ten-fold serial dilutions of WT, ipl1-1, and ipl1-1 spc105-RASA were plated on YEPD at 23, 30, 34, and 37°C. (B) Number of colonies with indicated genotypes or those that failed to form macroscopic colonies (DEAD) derived from single cells isolated after HGR on the strain harboring the RASA-NT cassettes in the background of wild-type or mad2Δ. (C) G1 synchronized mad2Δ and mad2Δ spc105-RASA cells were released, and Pds1 and Pgk1 (loading control) levels were monitored by Western blot. (D) Growth curve of mad2Δ and mad2Δ spc105-RASA at 30°C in YEPD medium. Average ± SEM of the doubling time of three independent experiments are also shown. (E) Disomy III formation in mad2Δ and mad2Δ spc105-RASA cells containing a chromosome III marked with a leu2 locus disrupted by URA3. The mean frequency ± SEM of disomy formation (assessed by generation Leu+, Ura+ colonies) from ten independent cultures are shown. Asterisk, p = 0.0149. (F) Ten-fold serial dilutions of WT, tetoff-CDC20-127, and tetoff-CDC20-127 spc105-RASA were plated on YEPD with or without 5 μg/ml doxycycline at 30°C. High magnification images of microcolonies are also shown. Scale bar, 50 μm. (G) G1 synchronized tetoff-CDC20-127 and tetoff-CDC20-127 spc105-RASA cells were released in the presence of doxycycline. Pds1-18MYC, Mad1, and Pgk1 (loading control) levels were analyzed by Western blots at the indicated timepoints after release. See also Figure S2.
It has been suggested that PP1/Glc7 stabilizes kinetochore-microtubule attachment [12, 14] and silences the SAC [10, 11]. We therefore asked whether the lethality of spc105-RASA is due to unstable kinetochore-microtubule attachment or to the persistence of SAC activity. If the lethality of spc105-RASA is caused by the kinetochore-microtubule attachment defect, deletion of MAD2, which is essential for the SAC but not for mitotic chromosome segregation [15], should not restore viability to spc105-RASA cells. On the other hand, if spc105-RASA lethality is caused by a failure to silence the SAC, the spc105-RASA mutant might become viable by deleting MAD2. The latter hypothesis was borne out: viable spc105-RASA cells were recovered in a mad2Δ background (Figure 2B). This is a stark contrast to many kinetochore mutants, which are synthetic lethal with mad2Δ [16–20].
Although cell cycle progression of mad2Δ spc105-RASA is similar to that of mad2Δ (Figures 2C and data not shown), mad2Δ spc105-RASA showed a slightly reduced growth rate (Figure 2D). An increase in ploidy (IPL) assay [21] revealed a 1.7-fold increase in disomy III in mad2Δ spc105-RASA compared to mad2Δ cells (Figure 2E), indicating that the spc105-RASA mutation causes a minor, non-lethal effect on physical chromosome segregation.
To confirm that the cell cycle delay of spc105-RASA is due to persistent SAC activity, we constructed an spc105-RASA strain in which the SAC can be conditionally inactivated by expressing a dominant mutant of CDC20 under a tetracycline-repressible promoter (tetoff-CDC20-127) [22]. With expression of CDC20-127, which drives anaphase progression even when the upstream SAC pathway is activated, spc105-RASA was viable, but when the SAC was restored by repressing CDC20-127 with doxycycline, spc105-RASA cells died with the same morphological phenotype observed with the single spc105-RASA mutation (Figure 2F). To analyze the cell cycle state of these cells, G1 synchronized cells were released in the presence of doxycycline. In SPC105 tetoff-CDC20-127 cells, Mad1 phosphorylation (a marker for SAC activation [23]) was transiently induced followed by the decrease of Pds1 levels. In contrast, in spc105-RASA tetoff-CDC20-127 cells, both Mad1 phosphorylation and Pds1 levels increased and then remained high, indicative of sustained SAC signaling.
Altogether, these data suggest that lethality of spc105-RASA is primarily caused by persistent activation of the SAC. Without SAC execution, these cells can still support chromosome segregation, which indicates proper establishment of bipolar attachments. As the SAC is always activated prior to bipolar attachments [24, 25], it appears that this signal cannot be silenced in spc105-RASA cells even when bioriented attachments are accomplished.
Phosphorylation at the RVSF sequence in Spc105 is dispensable for mitosis
The N-terminal region of vertebrate KNL1 harbors three conserved Aurora B phosphorylation sites (Figure S1A). In human [12, 26] and Xenopus (Figures S2A-D and data not shown), phosphorylation of these residues by Aurora B weakens the PP1-KNL1 interaction, potentially providing a feedback mechanism to control this interaction in response to microtubule attachment status [12]. To assess the functional significance of this regulation, we further examined the isolated spc105-RVAF strain (Figures 1B and S1C), in which the sole conserved Aurora B/Ipl1 site in Spc105 was mutated. Without RVSF phosphorylation, Glc7 may be constitutively recruited to Spc105 and prematurely silence the SAC. Contrary to this prediction, however, unlike the SAC-deficient mad2Δ, the spc105-RVAF mutation did not affect sensitivity to benomyl (Figure S2E), and the majority of spc105-RVAF cells arrested with large buds in the presence of nocodazole (Figure S2F). We also examined the effect of spc105-RVAF on the SAC when it is induced by lack of tension at kinetochores. In the cohesin mutant scc1-73, tension cannot be established due to a defect in sister chromatid cohesion at the restrictive temperature and thus the SAC is activated [27]. Anaphase delay in scc1-73, marked by delayed Pds1 degradation, was rescued by mad2Δ but not by the spc105-RVAF mutation (Figure S2G), indicating that lack of RVSF phosphorylation is insufficient to prematurely silence the SAC. Finally, spc105-RVAF cells showed no significant increase in disomy III formation (Figure S2H). Thus, we conclude that phosphorylation of this site is dispensable for mitosis.
Tethering Glc7 to Spc105-RASA rescues lethality but tethering Glc7 to wild-type Spc105 is lethal
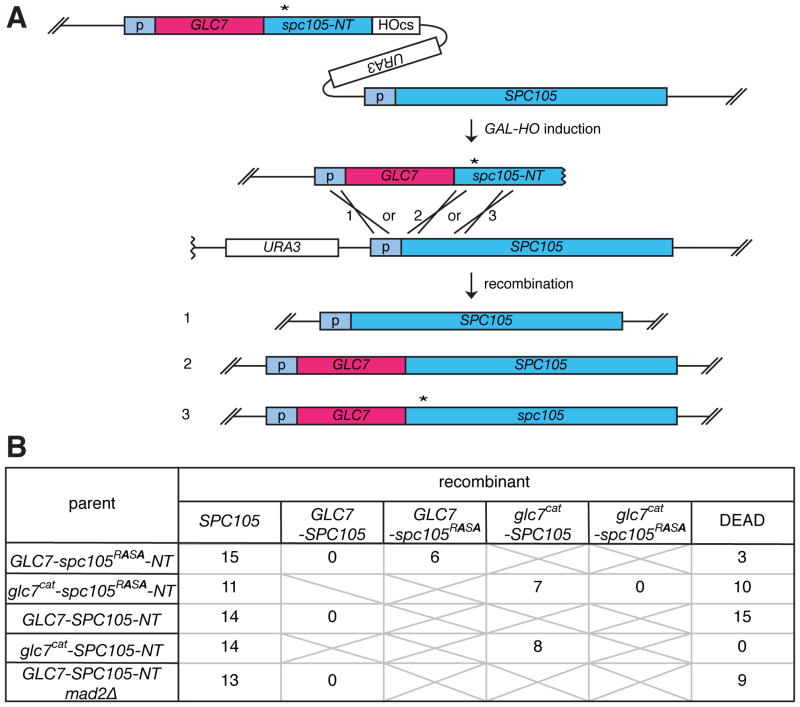
To determine whether the spc105-RASA phenotype is solely caused by failure to interact with Glc7 and whether there are any other mechanisms, such as other phosphorylation sites, which effect the dynamicity of this interaction, we sought to obtain a strain where Glc7 is linearly fused to the Spc7 N-terminus. Using HGR with GLC7 fused to the N-terminal truncation of spc105 containing the RASA mutation (GLC7-spc105RASA-NT), three types of recombinants were expected to be generated: (1) wild-type SPC105, (2) GLC7-SPC105, and (3) GLC7-spc105RASA (Figure 3A). As a control, a fusion of a catalytically dead D94A mutant of Glc7 (Glc7cat) [28] was also examined. The single cell colony assay demonstrated that recombinants encoding SPC105 and GLC7-spc105RASA, but not glc7cat -spc105RASA, can form normal colonies (Figure 3B). Sequence analysis confirmed that isolated GLC7-spc105RASA cells encode the expected fusion gene without any additional mutations. This result establishes that the sole essential role of the RVxF motif of Spc105 is to recruit catalytic activity of Glc7.
Figure 3. Generation of GLC7 fused to wild-type or mutant SPC105.
(A) Schematic of HGR, used to generate GLC7-spc105 fusion genes. (B) Number of colonies with the indicated genotypes or those that failed to form macroscopic colonies (DEAD) derived from single cells isolated after HGR on the strain harboring the indicated cassettes is shown.
Strikingly, we recovered no viable recombinants encoding GLC7-SPC105, even when the GLC7-SPC105-inducing cassette (GLC7-SPC105-NT) was used (Figure 3B). Since viable glc7cat–SPC105 cells were frequently recovered, the lethality of GLC7-SPC105 cells depends on the catalytic activity of GLC7, and thus it should not be caused by a structural problem of the Glc7-Spc105 fusion protein. Live microscopy analysis revealed that, in addition to normal growing cells, the GLC7-SPC105-inducing cassette generated cells that cause pleiotropic cell proliferation defects, many of which were associated with a cell cycle delay (> 5 h) with a short spindle (Figure S3 A, B and movies S4, S5). However, unlike spc105-RASA, the lethality of GLC7-SPC105 was not rescued by mad2Δ (Figure 3B), indicating that GLC7-SPC105 causes detrimental effects beyond SAC activation, perhaps by suppressing function of Ipl1 and/or other kinases that control kinetochore functions. The lethality of both spc105-RASA cells and GLC7-SPC105 indicates that the amount of Glc7 recruited at the kinetochore must be finely tuned. It was estimated that about 15,000 copies of Glc7 exists in a cell [29], while only 5 copies of Spc105 are recruited to each kinetochore [30]. Therefore, these results highlight the vital impact of targeting an exiguous fraction of Glc7 to the kinetochore.
Constitutive recruitment of Glc7 is insufficient to silence the SAC
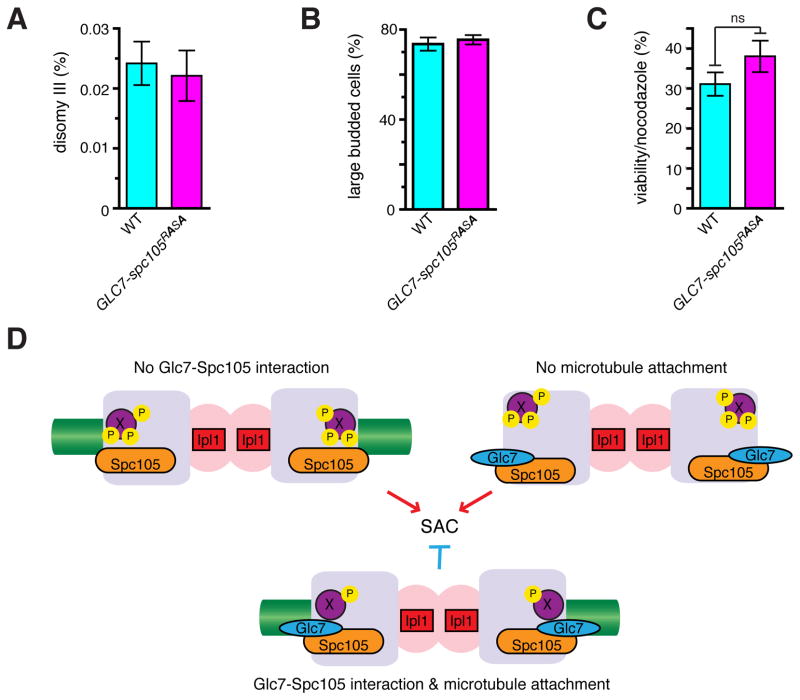
In contrast to spc105-RASA or GLC7-SPC105 cells, GLC7-spc105RASA cells showed no proliferation defect (Figure S4A) and no increase in chromosome missegregation (Figure 4A). In addition, GLC7-spc105RASA cells were normally arrested at M phase with large buds in response to nocodazole (Figure 4B), did not display increased lethality after nocodazole treatment (Figure 4C), and did not differ from wild type cells in sensitivity to benomyl (figure S4B). These results demonstrate that dynamic regulation of Glc7-Spc105 interaction is not critical for mitosis, and that the Glc7-Spc105 interaction is insufficient to trigger anaphase in the absence of microtubules. Microtubule attachment alone, however, is also insufficient for SAC silencing in the spc105-RASA mutant. In vertebrate cells, Aurora B-dependent phosphorylation at the kinetochore is high on unattached kinetochores, but decreases upon bipolar attachment in a manner dependent on the PP1-KNL1 interaction [12, 26]. We therefore propose that the Glc7-Spc105 interaction effectively couples the microtubule-kinetochore attachment to dephosphorylation of critical substrates, such as Ndc80 [31], for SAC silencing (Figure 4D). Our study also illustrates that the presence of the SAC, a nonessential surveillance mechanism, makes its silencing mechanism an essential process and necessitates the acquisition of the PP1-targetting module to a kinetochore protein.
Figure 4. Constitutive recruitment of Glc7 is insufficient to silence the SAC.
(A) Disomy III formation in WT and GLC7-spc105RASA cells containing a chromosome III marked with a leu2 locus disrupted by URA3. The mean frequency ± SEM of disomy formation (assessed by generation Leu+, Ura+ colonies) from 15 independent cultures are shown. (B, C) WT and GLC7-spc105RASA strains were treated with nocodazole and benomyl for 3 hours, cell morphology was counted (B), and cells were washed and plated on YEPD to count colony formation (C). Average and SEM for 3 separate experiments are shown; n > 400 cells each (B); ns: not significant, p = 0.23 (C). (D) Model. In the absence of Glc7-Spc105 interaction (top left), or without microtubule attachment (top right), putative kinetochore proteins (X) are efficiently phosphorylated in an Ipl1-dependent manner and the SAC is turned on. Only when Glc7 is recruited to Spc105 and microtubules are attached to the kinetochore, X is dephosphorylated and the SAC becomes silenced. See also Figure S4.
Supplementary Material
Acknowledgments
We thank D. Botstein, C. Chan, K. Hardwick, A. Kelly, A. Murray and A. Rudner for reagents, and members of Funabiki lab for critical reading of the manuscript. This study was supported by grants from NIH to F. R. C. (GM47238) and H.F. (GM075249-05S1).
Footnotes
Supplemental Information includes four figures, two tables, five movies, experimental procedures, and references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Kelly AE, Funabiki H. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol. 2009;21:51–58. doi: 10.1016/j.ceb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang J, Yu H. Kinase signaling in the spindle checkpoint. J Biol Chem. 2009;284:15359–15363. doi: 10.1074/jbc.R900005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanoosthuyse V, Hardwick KG. Overcoming inhibition in the spindle checkpoint. Genes Dev. 2009;23:2799–2805. doi: 10.1101/gad.1882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross FR, Pecani K. Efficient and rapid exact gene replacement without selection. Yeast. 2011;28:167–179. doi: 10.1002/yea.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francisco L, Wang W, Chan CS. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 9.Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol. 2009;19:1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Sassoon I, Severin FF, Andrews PD, Taba MR, Kaplan KB, Ashford AJ, Stark MJ, Sorger PK, Hyman AA. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick KG, Li R, Mistrot C, Chen RH, Dann P, Rudner A, Murray AW. Lesions in many different spindle components activate the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics. 1999;152:509–518. doi: 10.1093/genetics/152.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 19.Montpetit B, Thorne K, Barrett I, Andrews K, Jadusingh R, Hieter P, Measday V. Genome-wide synthetic lethal screens identify an interaction between the nuclear envelope protein, Apq12p, and the kinetochore in Saccharomyces cerevisiae. Genetics. 2005;171:489–501. doi: 10.1534/genetics.105.045799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniel JA, Keyes BE, Ng YP, Freeman CO, Burke DJ. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 2006;172:53–65. doi: 10.1534/genetics.105.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- 23.Hardwick K, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khodjakov A, Rieder CL. The nature of cell-cycle checkpoints: facts and fallacies. J Biol. 2009;8:88. doi: 10.1186/jbiol195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welburn JPI, Vleugel M, Liu D, Iii JRY, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B Phosphorylates Spatially Distinct Targets to Differentially Regulate the Kinetochore-Microtubule Interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang HB, Horiuchi A, Goldberg J, Greengard P, Nairn AC. Site-directed mutagenesis of amino acid residues of protein phosphatase 1 involved in catalysis and inhibitor binding. Proc Natl Acad Sci USA. 1997;94:3530–3535. doi: 10.1073/pnas.94.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 30.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.