Abstract
Chronic infection with the hepatitis B virus (HBV) occurs in approximately 6% of the world's population and carriers of the virus are at risk for complicating hepatocellular carcinoma. Current treatment options have limited efficacy and chronic HBV infection is likely to remain a significant global medical problem for many years to come. Silencing HBV gene expression by harnessing RNA interference (RNAi) presents an attractive option for development of novel and effective anti HBV agents. However, despite significant and rapid progress, further refinement of existing technologies is necessary before clinical application of RNAi-based HBV therapies is realized. Limiting off target effects, improvement of delivery efficiency, dose regulation and preventing reactivation of viral replication are some of the hurdles that need to be overcome. To address this, we assessed the usefulness of the recently described class of altritol-containing synthetic siRNAs (ANA siRNAs), which were administered as lipoplexes and tested in vivo in a stringent HBV transgenic mouse model. Our observations show that ANA siRNAs are capable of silencing of HBV replication in vivo. Importantly, non specific immunostimulation was observed with unmodified siRNAs and this undesirable effect was significantly attenuated by ANA modification. Inhibition of HBV replication of approximately 50% was achieved without evidence for induction of toxicity. These results augur well for future application of ANA siRNA therapeutic lipoplexes.
Key words: altritol, siRNA, RNAi, HBV, lipoplex, non viral vector
Introduction
Globally it is estimated that there are 387 million carriers of hepatitis B virus (HBV) and persistent infection with the virus places individuals at very high risk for the life-threatening complications of cirrhosis and hepatocellular carcinoma (HCC).1,2 Licensed anti HBV agents, which include interferon-alpha (IFNα), nucleoside (lamivudine) and nucleotide (adefovir) analogues, are only partially effective.3 Serious complications of infection with HBV are thus likely to remain significant global medical problems for several years and development of new effective therapy continues to be an important objective. Recent demonstration that HBV replication is susceptible to RNA interference-(RNAi-)mediated silencing4–9 has prompted investigations aimed at harnessing this pathway for therapeutic application. Exogenous activators of RNAi are typically expressed pri/pre microRNA (miR) mimics or synthetic small interfering RNAs (siRNAs).10 Both classes have been used to silence HBV replication and each has distinct advantages. Importantly, it is easier to achieve correct dosing and delivery of synthetic siRNAs, but undesirable immunostimulatory properties of siRNAs represent a major obstacle to achieving safe therapy.
A variety of chemical modifications of siRNAs has been used to improve stability, efficacy and attenuate the immunostimulation by siRNAs (reviewed in ref. 11). Included amongst these are 2′-OMe, 2′-F and 2′-H changes to the 2′ ribose carbon of the nucleotides within the siRNAs. Favorable effects on specificity, stability and diminished immunostimulation have been described. Recently modification of siRNAs by incorporation of altritol carbohydrate moieties has been reported.12,13 These altritol nucleic acids (ANAs) have a six membered sugar ring and have been derived from hexitol nucleic acids. The additional hydroxyl group at the 3′ position, which is directed to the minor groove of the siRNA duplex, contributes to helix stability. A-form structures of ANAs are favored and this is thought to improve silencing by siRNAs in cell culture. Compared to natural siRNAs, efficacy was improved when altritol modifications were incorporated, and their positioning at the 3′ end of the siRNA strands is important for optimal silencing. These encouraging observations suggest that ANA siRNAs are potentially useful for therapeutic application. To investigate pharmacological effectiveness of ANA siRNAs in a murine model of HBV infection, we have assessed antiviral efficacy of siRNA lipoplex formulations that target the conserved HBx open reading frame (ORF). The data indicated that systemically administered ANA siRNA lipoplexes are capable of inhibiting HBV replication in a stringent HBV transgenic mouse model of the human condition. Importantly, compared to unmodified siRNAs, the immunostimulatory properties of ANA siRNAs are attenuated. These ANA modifications therefore confer an improved safety profile on potentially therapeutic siRNAs that require systemic administration.
Results
Optimising selection of siRNAs based on inhibition of HBV surface antigen secretion from transiently transfected liver-derived cells.
The HBx ORF of HBV has been shown to be a good target for RNAi-mediated silencing of HBV replication.4,14–17 This sequence is common to all HBV transcripts and is conserved within the compact viral genome. To select optimal HBV targets for ANA siRNAs, the most highly conserved regions within the HBx ORF were identified. Thereafter, four specific sequences that were predicted to be favorable siRNA targets were chosen for the synthesis of siRNA 1, siRNA 2, siRNA 3 and siRNA 4. Initially, to assess inhibitory efficacy of the four siRNAs against HBV in vitro, Huh7 cells were cotransfected with pCH-9/3091 HBV replication competent target plasmid18 (Fig. 1a) an eGFP reporter plasmid (pCi-GFP) and one of each antiviral siRNA or a mismatched siRNA control (siRNAmm). An additional control included a U6 shRNA-encoding plasmid (U6 shRNA 5), which we have previously shown to be effective against HBV.4 Consistently, approximately 60% of transfected cells were also positive for eGFP, which confirmed good transfection efficiency that was equivalent for all wells. Compared to mock treated cells, knockdown of up to 80% of viral antigen secretion was achieved by siRNA 3 when measured 48 hours after transfection (Fig. 1b). This knockdown was achieved using a siRNA concentration of 10 nM. The vector encoding U6 shRNA 5 caused most efficient silencing of greater than 90%. The HBsAg secretion data were corroborated using a reporter gene plasmid (pCH-FLuc4) to measure knockdown efficiency in vitro (Fig. 1c). Based on the observation that siRNA 3 was the most effective, this sequence was selected for further analysis and efficacy testing in vivo.
Figure 1.
Selection of anti HBV siRNA. (A) Organization of the hepatitis B virus genome in the linear HBV-encoding component of the pCH-9/3091 plasmid or the pCH-FLuc reporter vector. Sites targeted by siRNA sequences are shown. (B) Relative HBsAg concentrations in cell culture supernatants following transfection with pCH-9/3091 HBV replication competent plasmid followed by siRNA transfection. Assay of HBsAg was carried out 48 hours after siRNA transfection. (C) Luciferase reporter gene-based assay of siRNA-mediated knockdown efficacy in situ. pCH Firefly Luc was cotransfected with indicated siRNAs together with a plasmid constitutively expressing Renilla luciferase. Results are given as ratios of Firefly to Renilla luciferase activity. Statistical significance was determined by comparing HBsAg or Firefly measurements to those obtained after transfection of the mismatched control siRNA (*p < 0.05, **p < 0.01 and ***p < 0.0001).
Lipoplex siRNA formulations.
Lipoplex non viral vectors were used to deliver the ANA siRNAs to the liver in vivo. Three lipid components of the vectors were combined to formulate the siRNA-lipoplexes. These were a galactose derivative of cholesterol that was generated using ‘click’ chemistry (Fig. 2A) (manuscript describing synthesis is in preparation), the fusogenic helper lipid DOPE (Fig. 2B) and a nucleic acid-binding cationic lipid component contains two cholesteryl moieties covalently linked via an aza-macrocycle (Fig. 2C). Formulations were prepared using standard procedures of resuspending dried lipids with sonication followed by combination with siRNAs. A mass ratio of 6:4 of cholesterol-containing lipid to helper lipid was used. Since the cationic component included two cholesterol moieties, the ratio of cationic lipid:galactose cholesterol:DOPE was 11:8:20. These formulations generated particle sizes that ranged from 100–200 nm in diameter, and the siRNA components were protected from nuclease degradation.19 Assessment of delivery efficiency in vivo, showed that approximately 50% of hepatocytes were transfected when assessed 2 hours after low pressure intravenous injection of formulations containing Cy3-labelled siRNAs (Fig. 2D). These promising results prompted further investigation aimed at assessing knockdown of viral replication in HBV transgenic mice20 using ANA-modified siRNAs.
Figure 2.
Lipid components of lipoplex formulations used to deliver anti HBV siRNAs in vivo. (A) Acetylated galactose-conjugated cholesterol, generated using ‘click’ chemistry, was incorporated to confer hepatotropism on vectors. (B) DOPE helper fusion lipid. (C) Cationic nucleic acid binding cholesterol derivative. (D) Fluorescence microscopy visualisation (frozen section) of hepatic Cy3-labelled siRNAs detected in frozen sections taken from livers of mice 2 hours following injection of formulations.
ANA-modified anti HBV siRNAs.
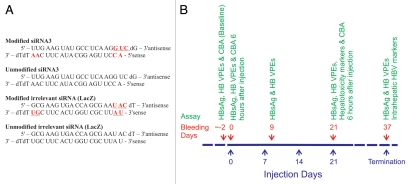
Use of ANA siRNAs to inhibit gene expression in cultured cells has been described.12 These RNAi activators are efficient and more stable than their native counterparts. Importantly, the altritol component facilitates formation of A-form dsRNA and the modified strand hybridizes strongly to its complement.21 Modifications of the siRNA that targeted HBx were incorporated at the 3′ ends of the sense and antisense strands and at the 5′ end of the sense strand (Fig. 3A), and this was according to design rules that were originally described for this class of siRNAs.12 In addition to ANA siRNAs, native siRNAs that did not include modifications as well as a control siRNA targeted to a reporter gene (anti LacZ) were included in the panel of tested synthetic RNAi effecters.
Figure 3.
Control and anti HBV siRNAs with protocol for their administration. (A) Sequences of anti HBV siRNAs together with control siRNAs. ANA modifications are indicated in red font, and are underlined in the antisense strand. (B) Schematic illustration of experimental program carried out on HBV transgenic mice. Twelve groups of eight mice were included in the investigation, and groups received a maximum of four injections at weekly intervals at the indicated time points. Some groups received a single injection of siRNA formulations at commencement of the investigation. Blood samples were collected at day 0, 2, 9, 23 and 37 days for the measurement of HBV replication markers. Assessment of release of proinflammatory cytokines was carried out after the first and fourth injections, and measurement of serum transaminases was determined two days after receiving the fourth and final injection.
Assessment of efficacy in vivo of anti HBV ANA siRNA lipoplex formulations.
HBV transgenic mice were injected with various formulations according to the scheme described in Figure 3B. Blood samples were collected from the mice using retro-orbital puncture and assays for measuring HBV replication, immunostimulation and toxicity were also performed (Fig. 3B). There were a total of ten groups of eight mice each, that each received intravenous administration of different formulations, naked siRNA or saline. The dose of siRNA administered to each animal was 1 mg/kg body weight.
A quantitative real time PCR (qPCR) assay was used to measure circulating viral particle equivalents (VPEs).4 Results showed that viral replication in animals receiving the ANA and native siRNA formulations, which were administered as a weekly injection, were significantly diminished (Student's unpaired t-test, p < 0.05) in both of these groups of mice. Moreover, the effect was sustained over the 3-week period of testing (Fig. 4). A comprehensive analysis of the efficacy of the full panel of naked and formulated siRNAs however revealed that no inhibitory effects of the siRNA targeted to LacZ. Observations based on assay of circulating VPEs were corroborated by measurement of serum HBsAg and detection of intrahepatic HBcAg (Fig. 5). However, measurement of the viral antigen suggested a less marked silencing effect of the siRNAs on this marker of viral replication. Importantly, HBsAg is expressed at a high level in HBV transgenic mice and may be less sensitive than circulating VPEs as an indicator of siRNA-mediated silencing. Since immunostimulation is potentially toxic and is also a confounding variable for interpretation of silencing efficacy, we assessed effects of lipoplex formulations on release of proinflammatory cytokines, markers of hepatotoxicity and induction of the interferon response.
Figure 4.
Measurement of circulating VPEs in mice receiving indicated antiviral siRNA, which were formulated in lipoplexes (L) or administered as naked nucleic acids (N). (A) Graphical representation of inhibition of circulating VPEs in mice receiving weekly administration of saline (control) or formulations that included ANA-modified or native siRNAs. (B) Bar graph representation of circulating VPEs for all of the ten groups of mice. Each bar from left to right represents a specific time point of 0, 2, 9, 23 and 37 days after injection. Bars represent mean values for the concentrations from eight animals in each group and error bars show the SEM. Statistical significance was assessed by comparing values obtained at indicated time points to the baseline values at time 0 (*p < 0.05).
Figure 5.
Serum HBsAg concentrations in mice receiving control and antiviral siRNA, which were with formulated in lipoplexes (L) or administered as naked nucleic acids (N). (A) Graphical representation of effects on circulating HBsAg (relative concentrations ELISA OD readings) in mice receiving weekly administration of saline (control) or formulations that included ANA-modified or native siRNAs. (B) Bar graph representation of HBsAg concentrations for all of the ten groups of mice. Representation of data is as described in Figure 4. (C) Representative low power fields after carrying out immunohistochemistry to detect the HBcAg. Livers were harvested from animals at the termination of the experiments at day 31 after weekly intravenous injection with saline or the ANA lipoplex formulations. Scale bar indicates 200 mm.
Measurement of siRNA-induced hepatoxicity and immunostimulation.
Hepatotoxic effects of the ANA siRNA lipoplexes were determined by measurement of aspartate aminotransferase (AST), alanine aminotransferase (ALT) and lactate dehydrogenase (LDH) activities in the serum of mice at day 23 after receiving single or repeated administrations of the formulated or naked siRNAs (Fig. 6). These enzyme markers are released following hepatocyte injury and are useful and widely used indicators of hepatotoxicity. Comparison of enzyme activities detected in the serum samples of each of the ten groups of mice revealed that there was minor elevation in ALT, AST and LDH in animals that had received complexed and naked ANA siRNAs. As the serum activities for AST and ALT were below 100 IU/ml and LDH levels were less than 1,000 IU/L, the inflammation is likely to be minimal.
Figure 6.

Serum activities of (A) AST, (B) ALT and (C) LDH in mice receiving each of the siRNA lipoplex formulations. Blood samples were collected at day 23 of the experiment, which was two days after animals received the fourth and final siRNA injections. Bars represent mean values for the concentrations from eight animals in each group and error bars show the SEM. No statistical difference was detected when comparing values from experimental animals to those of controls.
To analyze release of proinflammatory cytokines a cytometric bead array (CBA) assay was carried out. A panel of proinflammatory cytokines was measured at 6 hours and 48 hours after intravenous administration of the first and fourth dose of the siRNAs to mice. Six hours after receiving the first dose of the siRNA formulations, there was marked elevation of IFNγ, interleukin-6 (IL-6), monocyte chemoattractant protein-1 (MCP-1) and tumor necrosis factor-α (TNFα) in mice that received the non viral vectors containing the unmodified siRNAs (Fig. 7A). However, serum concentrations of these cytokines were significantly lower and not different to the background concentrations in the mice that received ANA siRNA lipoplex formulations. Representative flow cytometry histograms for two individual animals from each of the groups are shown in Figure 7B. These results indicate that injection of formulated unmodified siRNAs cause more significant innate immune response induction than is evident after administration of ANA siRNA lipoplexes. When tested at 6 hours after the fourth injection, concentrations of IFNγ, IL-6, MCP-1 and TNFα were again more increased in the mice that had received the unmodified siRNAs (Fig. 7C). Compared to serum proinflammatory cytokine concentrations measured after the first injection of the ANA siRNA lipoplexes, there was an increase in circulating IFNγ, IL-6 and TNFα after the fourth injection of these complexes. This suggests that ANA siRNA formulations induce an immune response after repeated administration. Although attenuated, the significance of this immunostimulation, as a factor that may compromise efficacy of long term administration of ANA siRNAs, will require further analysis.
Figure 7.
Use of CBA to measure proinflammatory cytokine released from mice receiving lipoplexed (L) unmodified and ANA-modified siRNAs. (A) Measurements of serum concentrations of IFNγ, IL-6, MCP-1 and TNFα were determined six hours after the first injection of the formulations. Values are given as the mean ± SEM. (B) Representative flow cytometry data from CBA assay of IL-6, IL-10, MCP-1, IFNγ, TNFα and IL12 concentrations at the time point of six hours after the first injection with lipoplexed unmodified and ANA-modified siRNAs. (C) Measurements of serum concentrations of IFN., IL-6, MCP-1 and TNFα were determined six hours and 48 hours after the fourth injection of the formulations. Bars represent mean values for the concentrations from eight animals in each group and error bars show the SEM. Statistical significance was assessed by comparing values obtained after injection with lipoplexes containing the ANA modified siRNAs with those formulated with unmodified RNA duplexes (*p < 0.05, **p < 0.01 and ns, not significant).
Intrahepatic induction of IFN response genes was also measured as a sensitive indicator of immunostimulation. A separate group of BALBc mice was treated with poly (I:C), which is a synthetic analogue of double-stranded RNA (positive control administered using hydrodynamic injection), as well as a the lipoplexes containing ANA or unmodified siRNAs (Fig. 8). Concentrations of RNA from IFNβ, oligoadenylate synthetase-1 (OAS-1) and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) genes, measured using a real time qPCR method and calculated as a ratio to the GAPDH housekeeping gene mRNA concentration,17 were used as indicators of induction of the IFN response. As expected, administration of poly (I:C) resulted in marked elevation of RNA from all three genes. None of the formulations resulted in the increased expression of IFNβ, but lipoplexes containing unmodified and ANA siRNAs both caused increases in concentrations of OAS-1 and IFIT1 mRNA. However, compared to RNA extracts from livers of mice that received formulations containing unmodified siRNAs, concentrations of OAS-1 and IFIT1 transcripts were significantly diminished in animals that received ANA siRNA lipoplexes. Collectively, these data support the interpretation that ANA modification attenuates immunostimulatory properties of siRNAs and is likely to result in an improved safety profile of this new class of RNAi effecters.
Figure 8.

Interferon response following injection of BALBc mice with lipoplex siRNA formulations. Mice were injected with poly (I:C) using the hydrodynamic procedure or with liposomal formulations (L), naked siRNA (N) using low pressure injection. Six hours later, RNA was extracted from the livers and then subjected to quantitative real time PCR to determine intrahepatic concentrations of IFNβ (A), IFIT1 (B) and OAS-1 (C). Values were calculated relative to the concentration of GAPDH mRNA. Bars represent mean values for the concentrations from 6 animals in each group and error bars show the SEM. Statistical significance was assessed by comparing values to the poly (I:C) positive control (*p < 0.05, **p < 0.01 and ns, not significant).
Discussion
The progress that has been made in advancing RNAi-based therapeutics for the management of chronic HBV infection has been rapid and impressive (reviewed in ref. 22). Both expressed and synthetic RNAi activators have been used and each class of silencing sequence has advantages and disadvantages. Although expressed RNAi activators potentially achieve a more sustained antiviral effect, their use is complicated by difficulties with delivery and regulation of dose. Particularly important to advancing use of expressed anti HBV sequences has been the demonstration that over production of anti HBV shRNAs has serious side effects in vivo that may result in fatality.23 This has prompted development of chemically modified synthetic siRNAs with better stability and safety profiles.24 ANA-modified siRNAs targeting the MDR1 gene, which codes for the P-glycoprotein transmembrane ATPase efflux pump, have been evaluated. ANA modifications in both sense and antisense strands of siRNA, resulted in greater knockdown as well as more sustained efficacy when compared to unmodified controls.12 However, these data were generated after analysis of cells that were transfected in culture, and no evidence for the efficacy of ANA-modified siRNAs in vivo has been available to date. In this study we have shown that lipoplexes containing ANA-modified siRNAs, which are a new class of silencing sequence, are capable of inhibiting viral replication in vivo in a murine transgenic model of HBV infection. A decrease of circulating VPEs of approximately 80% was achieved with ANA siRNAs, however the inhibitory effect on serum HBsAg concentration was less marked, although staining for HBcAg in situ suggested effective silencing of intrahepatic HBV replication. These effects were observed without causing unwanted harmful immunostimulation that characterized use of the unmodified siRNAs and silencing by ANA siRNAs compares favorably to that reported in previous studies. The only other study using lipoplexes to deliver unmodified anti HBV siRNAs sequences to transgenic mice reported inhibition of markers of viral replication of approximately 50%.25 In another study, delivery of modified anti HBV siRNAs using small nucleic acid lipid particles (SNALPs) was reported to cause a 10-fold decrease in circulating VPEs. Although this effect is impressive, the effect was observed in mice that were subjected to hydrodynamic injection. Importantly this model only causes transient replication of HBV, unlike the stringent HBV transgenic mouse model, which simulates the human condition of persistent viral infection more accurately.
The highly charged nature of nucleic acids, their high molecular weight and hydrophilicity makes use of vectors essential for their cellular delivery. In addition, an important consideration is the avoidance of induction of immunostimulatory effects and minimising toxicity of potentially therapeutic siRNAs. Formulating siRNAs in lipid nanoparticles has been widely used and several different methods have been utilized to deliver siRNAs in vivo with varying success. Approaches have included use of SNALPs24 and modular nanoparticles that can be assembled from a variety of lipid components.25 Recently, rational design of cationic lipid components of SNALPs has been employed to improve efficiency of these vectors.26 siRNA doses as low as 0.01 mg/kg body weight were capable of silencing factor VII expression in rodent liver. Despite using a dose of siRNAs that was 100-fold higher, the lipoplexes used in this study do not show evidence of causing unwanted effects. Importantly, a liver targeting cholesterol galactoside was incorporated to improve efficiency of hepatotropism. This novel lipid derivative was synthesized using click chemistry and is amenable to convenient large scale preparation, which would be important for clinical application. Compatibility of this cholesterol galactoside moiety with other cationic siRNA-binding lipids is currently being assessed with a view to further improvement of therapeutic index, safety profile, potency and pharmacokinetics of ANA siRNA lipoplexes.
Unintended knockdown of cellular genes that results from seed region interaction is another concern of utilising RNAi for therapeutic application. Recent demonstration that chemical modification of synthetic siRNAs improves guide strand selectivity and unintended translational suppression is an important development.27 ANA modifications may have similar properties and current investigations aim to optimize specificity by introducing seed sequence modifications to diminish inadvertent inhibition of cellular gene targets (Bramsen J et al., in preparation).
A recent study demonstrating that RNAi-mediated suppression of HBV targets did not alter established cccDNA adds another difficulty to advancing RNAi therapy for chronic HBV infection.28 The persistence of HBV cccDNA in infected hepatoytes, which exists as a stable minichromosome in infected hepatocytes, has been a major obstacle to eliminating this virus infection. Treatment regimens that use RNAi effecters in combination with established licensed drugs will be interesting to assess and augmentation of RNAi-based anti HBV efficacy when used together with licensed treatments will be a focus of future research.
Materials and Methods
Synthesis of unmodified and ANA-modified siRNAs.
Unmodified siRNAs were synthesized using standard procedures (Dharmacon, CO, USA). Four specific sequences that were predicted to be favorable siRNA targets (www1.qiagen.com/Products/RNAi/) were chosen for the generation of siRNA 1, siRNA 2, siRNA 3 and siRNA 4. Sequences of the complementary oligonucleotides that were used to constitute the siRNAs were the following: siRNA1: 5′ UAU UUG GUG GGC GUU CAC GGT 3′ (antisense) and 5′ CGU GAA CGC CCA CCA AAU ATT 3′ (sense); siRNA2: 5′ UAA ACA AAG GAC GUC CCG CGC 3′ (antisense) and 5′ GCG GGA CGU CCU UUG UUU ATT 3′ (sense); siRNA3: 5′ UUG AAG UAU GCC UCA AGG UCG 3′ (antisense) and 5′ ACC UUG AGG CAU ACU UCA ATT 3′ (sense); siRNA4: 5′ UAG UAC AAA GAC CUU UAA CCT 3′ (antisense) and 5′ GUU AAA GGU CUU UGU ACU ATT 3′ (sense); siRNA mismatch: 5′ UAU UGG GUG UGC GGU CAC GGT 3′ (antisense) and 5′ CGU GAC CGC ACA CCC AAU ATT 3′ (sense). The synthesis and purification of the ANA amidites29 and of ANA oligonucleotides has been described previously.12,21
Transfection of Huh7 cells in culture.
Huh7 cells were maintained in RPMI medium supplemented with 2.5% fetal calf serum (FCS), penicillin (50 IU/ml) and streptomycin (50 µg/ml) (Gibco BRL, UK). Fifty thousand Huh7 liver-derived cells were seeded in each well of a 24 well plate (approx 1 cm diameter) on the day prior to siRNA and target DNA administration. HBV target DNA plasmids (pCH9/3091,18 or pCH-FLuc16) were transfected using lipofectamine 2000 at a 1:1 ratio according to the manufacturer's instructions. A plasmid vector that constitutively produces enhanced green fluorescent protein (eGFP)30 was also included in each cotransfection to verify equivalent transfection efficiencies. Five hours thereafter, cells were transfected with siRNAs (10 nM) and the ratio of siRNA to lipofectamine was 3:1. Cells were then incubated overnight and the medium changed on the following day. The culture supernatant was harvested for HBsAg determination after 48 hours. Concentrations of HBsAg secreted into the culture supernatants were measured using the Monolisa (ELISA) immunoassay kit (BioRad, CA, USA). Equivalent transfection efficiency was confirmed using eGFP fluorescence microscopy.
Formulation of siRNA-containing lipoplexes.
The bis-cholesterol-substituted cyclen derivative was synthesized by reacting cholesteryl chloroformate with cyclen using a convenient acylation procedure that has been described.19 In addition, the galactose-cholesterol conjugate was synthesized by a copper-mediated ‘click’ reaction between the 2-propynylcarbamate derivative of cholesterol and O-tetraacetate galactose azide (manuscript describing synthesis is in preparation). All compounds were characterized by NMR, IR and mass spectroscopy and gave acceptable data. Three lipid components of the vectors were then combined to make the formulations. These were a galactose derivative of cholesterol that was generated using ‘click’ chemistry (Fig. 2a), DOPE (Fig. 2b) and a nucleic acid-binding cationic lipid 19 (Fig. 2c), which were combined in a molar ratio of 8:20:11.. Formulation of complexes with siRNAs was carried out using standard methodology and siRNA to liposome mass ratio was 1:17.31 To assess hepatotropic delivery of labelled siRNAs in vivo, unlabelled or Cy3-labelled duplex RNA was incorporated into lipoplexes before tail vein injection of HBV transgenic mice (see below).20 Two hours thereafter, mice were killed, livers removed and subjected to frozen section and fluorescence microscopy according to standard methods.
Assessment of anti HBV efficacy of ANA siRNA lipoplexes.
HBV transgenic mice20 were used to determine the effects of ANA siRNA lipoplexes on markers of HBV replication in vivo. All animal experimentation protocols were approved by the University of the Witwatersrand Animal Ethics Screening Committee. There were a total of ten groups of eight mice each, that each received intravenous administration of different formulations, naked siRNAs or saline. A constant dose of 1 mg/kg body weight of siRNA was used throughout and the volume of injectate ranged from 0.1–0.2 ml. These groups were the following:
Altritol modified anti-HBV, single dose (1 mg/kg) as lipoplex,
Altritol modified anti-HBV, weekly dose ×4 (1 mg/kg) as lipoplex,
Altritol modified anti-HBV, single dose (1 mg/kg) naked,
Unmodified anti-HBV, single dose (1 mg/kg) as lipoplex,
Unmodified anti-HBV weekly dose ×4 (1 mg/kg) as lipoplex,
Unmodified anti-HBV single dose (1 mg/kg) naked,
Altritol modified anti LacZ siRNA weekly dose ×4 (1 mg/kg) as lipoplex,
Unmodified anti LacZ siRNA weekly dose ×4 (1 mg/kg) as lipoplex,
Liposome only injection weekly ×4 and
Saline injection weekly ×4.
Blood samples were collected from the mice using retro-orbital puncture. Circulating viral particle equivalents (VPEs) and HBsAg were determined using qPCR and ELISA according to previously described methods.4 A rabbit polyclonal antibody against HBcAg (Signet Laboratories Inc., MA, USA) and horseradish peroxidase-conjugated secondary antibody (Dako, Denmark) were used to detect the viral antigen in paraffin embedded sections according to standard procedures.
Assessment of siRNA-induced hepatoxicity and immunostimulation.
Hepatotoxic effects of lipoplexes were determined by measurement of AST, ALT and LDH activities in the serum of mice after receiving single or repeated administrations of formulated or naked siRNAs. Assays were carried out using accredited diagnostic procedures of the South African National Health Laboratory Service (NHLS).
To analyze induction of release of proinflammatory cytokines after the first and fourth intravenous injections of the siRNA formulations, a CBA Mouse Inflammation Kit (BD Biosciences San Jose, CA, USA) was utilized. Measurements were made of IL-6, IL-10, MCP-1, IFNγ, TNFα and IL-12p70. Recombinant standards were also included to calibrate the assay. Assays were performed on blood collected immediately before injection, six hours after and then 48 hours after injection of the siRNA formulations.
Effect of ANA siRNA lipoplex formulations on intrahepatic induction of the IFN response was measured using a sensitive real time qPCR assay that has been described previously.17 Briefly separate groups of non transgenic BALBc mice were treated with poly (I:C) (positive control) as well as lipoplexes containing ANA or unmodified siRNAs. Poly (I:C) was administered using hydrodynamic injection and the lipoplexes were given intravenously under low pressure. RNA expressed from IFNβ, OAS-1 and IFIT1 genes, was measured as a ratio to the GAPDH mRNA.
Statistical analysis.
Data are expressed as the mean ± standard error of the mean. Statistical difference was considered significant when p < 0.05 and was determined according to the Student's two tailed unpaired t-test using the GraphPad Prism software package (GraphPad Software Inc., CA, USA).
Acknowledgements
This work was supported by funding under the Sixth Research Framework Program of the European Union, Project RIGHT (LSHB-CT-2004-005276), from CANSA, the South African National Research Foundation (NRF GUNs 68339, 2053652, 65495, Institutional Research Development Program and Research Niche Areas Program), ESASTAP and the South African Poliomyelitis Research Foundation. The technical support of Gladys Gagliardi and Margaret Badenhorst is gratefully acknowledged.
Abbreviations
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- siRNA
short interfering RNA
- RNAi
RNA interference
- ANA
altritol nucleic acid
- ORF
open reading frame
- shRNA
short hairpin RNA
- HBx
HBV X gene
- HBsAg
HBV surface antigen
- HBcAg
HBV core antigen
- PCR
polymerase chain reaction
- qPCR
quantitative polymerase chain reaction
- RT
reverse transcription
- VPE
viral particle equivalent
- ALT
alanine transaminase
- AST
aspartate transaminase
- LDH
lactate dehydrogenase
- IFNα
interferon-alpha
- IFNβ
interferon-beta
- IFNγ
interferon-gamma
- MCP-1
monocyte chemoattractant protein-1
- IL-6
interleukin-6
- IL-10
interleukin-10
- IL-12
interleukin-12
- DOPE
dioleylphosphatidyl ethanolamine
- OAS-1
oligoadenylate synthetase-1
- IFIT1
interferon-induced protein with tetratricopeptide repeats 1
- CBA
cytometric bead array
- GAPDH
glyceraldehyde phosphate dehydrogenase
- MDR1
multidrug resistance1
- SNALPs
small nucleic acid lipid particles
- OD
optical density
Footnotes
Previously published online: http://www.landesbioscience.com/journals/artificialdna/article/11981
References
- 1.Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77–100. doi: 10.1111/j.1365-2613.2001.iep0082-0077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Hanazaki K. Antiviral therapy for chronic hepatitis B: a review. Curr Drug Targets Inflamm Allergy. 2004;3:63–70. doi: 10.2174/1568010043483908. [DOI] [PubMed] [Google Scholar]
- 4.Carmona S, Ely A, Crowther C, Moolla N, Salazar FH, Marion PL, et al. Effective inhibition of HBV replication in vivo by anti-HBx short hairpin RNAs. Mol Ther. 2006;13:411–421. doi: 10.1016/j.ymthe.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 6.Klein C, Bock CT, Wedemeyer H, Wustefeld T, Locarnini S, Dienes HP, et al. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- 7.McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- 8.Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- 9.Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykxhoorn DM, Palliser D, Lieberman J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 11.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 12.Fisher M, Abramov M, Van Aerschot A, Xu D, Juliano RL, Herdewijn P. Inhibition of MDR1 expression with altritol-modified siRNAs. Nucleic Acids Res. 2007;35:1064–1074. doi: 10.1093/nar/gkl1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowther C, Ely A, Hornby J, Mufamadi S, Salazar F, Marion P, et al. Efficient inhibition of hepatitis B virus replication in vivo, using polyethylene glycol-modified adenovirus vectors. Hum Gene Ther. 2008;19:1325–1331. doi: 10.1089/hum.2008.066. [DOI] [PubMed] [Google Scholar]
- 15.Ely A, Naidoo T, Arbuthnot P. Efficient silencing of gene expression with modular trimeric Pol II expression cassettes comprising microRNA shuttles. Nucleic Acids Res. 2009;37:91. doi: 10.1093/nar/gkp446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ely A, Naidoo T, Mufamadi S, Crowther C, Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol Ther. 2008;16:1105–1112. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- 17.Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, et al. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- 18.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islam RU, Hean J, van Otterlo WAL, de Koning CB, Arbuthnot P. Efficient nucleic acid transduction with lipoplexes containing novel piperazine- and polyamine-conjugated cholesterol derivatives. Bioorg Med Chem Lett. 2009;19:100–103. doi: 10.1016/j.bmcl.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Marion PL, Salazar FH, Liittschwager K, Bordier BB, Seegers C, Winters MA, et al. A transgenic mouse lineage useful for testing antivirals targeting hepatitis B virus. Frontiers in Viral Hepatitis: Elsevier Science Amsterdam. 2003:197–202. [Google Scholar]
- 21.Allart B, Khan K, Rosemeyer H, Schepers G, Hendrix C, Rothenbacher K, et al. D-Altritol Nucleic Acids (ANA): Hybridisation properties, stability and initial Structural Analysis. Chem Eur J. 1999;5:2424–2431. [Google Scholar]
- 22.Arbuthnot P, Ely A. Advances in the Use of RNAi to Treat Chronic Hepatitis B Virus Infection. In: Martinez MA, editor. RNA Interference and Viruses. Norfolk, UK: Caister Academic Press; 2010. pp. 143–160. [Google Scholar]
- 23.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 24.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 25.Carmona S, Jorgensen MR, Kolli S, Crowther C, Salazar FH, Marion PL, et al. Controlling HBV replication in vivo by intravenous administration of triggered PEGylated siRNA-nanoparticles. Mol Pharm. 2009;6:706–717. doi: 10.1021/mp800157x. [DOI] [PubMed] [Google Scholar]
- 26.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 27.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starkey JL, Chiari EF, Isom HC. Hepatitis B virus (HBV)-specific short hairpin RNA is capable of reducing the formation of HBV covalently closed circular (CCC) DNA but has no effect on established CCC DNA in vitro. J Gen Virol. 2009;90:115–126. doi: 10.1099/vir.0.004408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abramov M, Herdewijn P. Synthesis of altritol nucleoside phosphoramidites for oligonucleotide synthesis. Curr Protoc Nucleic Acid Chem. 2007;1:1–18. doi: 10.1002/0471142700.nc0118s30. [DOI] [PubMed] [Google Scholar]
- 30.Passman M, Weinberg M, Kew M, Arbuthnot P. In situ demonstration of inhibitory effects of hammerhead ribozymes that are targeted to the hepatitis Bx sequence in cultured cells. Biochem Biophys Res Commun. 2000;268:728–733. doi: 10.1006/bbrc.2000.2209. [DOI] [PubMed] [Google Scholar]
- 31.Singh M, Kisoon N, Ariatti M. Receptor-mediated gene delivery to HepG2 cells by ternary assemblies containing cationic liposomes and cationized asia-loorosomucoid. Drug Deliv. 2001;8:29–34. doi: 10.1080/107175401300002739. [DOI] [PubMed] [Google Scholar]