Abstract
Cyclohexene Nucleic Acids (CeNA), in which the 2′-deoxyribofuranose ring of the DNA building blocks is substituted by a cyclohexenyl ring, were designed as potential mimics of natural nucleic acids for antisense and, later, for siRNA applications. CeNA units, in contrast to HNA (hexitol nucleic acid) building blocks, show more flexibility at the level of the C2′–C3′ bond due to the possibility of the cyclohexenyl moiety to adopt different conformations. In order to analyze the influence of CeNA residues onto the helix conformation and hydration of natural nucleic acid structures and to verify the cyclohexenyl ring conformation, a cyclohexenyl-thymine building block was incorporated into the non-self-complementary sequence d(GCG(xT)GCG)/d(CGCACGC) with (xT) a cyclohexene residue. The crystal structure of this sequence has been determined to a resolution of 1.17 Å and contains two duplexes in the asymmetric unit. The global helices belong to the B-type family and the conformations of the cyclohexenyl rings in both duplexes are different. The cyclohexene ring adopts as well the 2H3-conformation (similar to C2′-endo) as the 3H2-conformation (similar to C3′-endo). The crystal packing is stabilized by cobalt hexamine residues and triplet formation.
Key words: cyclohexene nucleic acids, CeNA, cobalt hexamine, triplet formation, ring conformation
Introduction
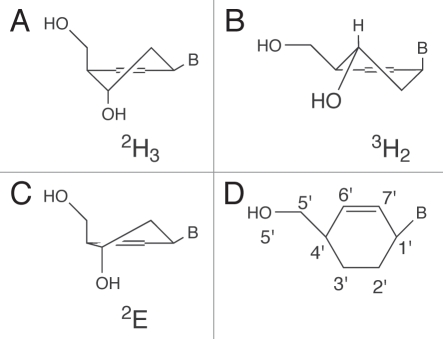
The 2′-deoxyribofuranose ring of the nucleotides, as occurring in DNA, can adopt two extreme conformations (C3′-endo and C2′-endo). Previously, we have synthesized cyclohexenyl nucleic acids (CeNA) in which the 2′-deoxyribofuranose ring of the DNA building blocks has been substituted with a cyclohexenyl ring. It was hypothesized that the cyclohexenyl nucleotides might adopt different conformations when incorporated in different DNA or RNA sequences, due to the flexibility of the cyclohexenyl moiety. The existence of different conformers of cyclohexenyl nucleotides when incorporated in DNA has been indirectly observed using NMR spectroscopy (based on NOE effects)1 but has not been confirmed by high-resolution X-ray studies. We have now demonstrated that cyclohexenyl nucleotides can occur in the 3H2− and 2H3-half chair conformation when incorporated in a non-self-complementary DNA duplex (of the same sequence) (Fig. 1).
Figure 1.
CeNA in its two stable conformations: (A) the 2H3 half-chair conformation which mimics the C2′-endo conformation of the natural ribose sugar and (B) the 3H2 half-chair conformation mimicking the C3′-endo conformation. (C) The CeNA in the 2E-envelope conformation. (D) Numbering scheme used for the CeNA cyclohexene ring.
This structural investigation, where one CeNA residue is incorporated into a double helix consisting of two non-self-complementary DNA stands, is to be compared with a previous study2 where two CeNA residues were present in opposite self-complementary strands in the double helix. Any possible interaction between two CeNA residues is thus ruled out, as well as any cooperative action to stabilize or destabilize the global helical structure. This one-residue incorporation allows therefore the exploration of the global and local influence of the cyclohexene moiety, as well as the conformational behavior of the CeNA sugar ring itself.
The B-type helices are stabilized by interactions with cobalt hexamine residues, which is quite a rare occurrence in non Z-type double helices (to date only 23 double helices of the A- or B-type), but which was also present in the previously reported structure containing CeNA.2
Furthermore this structural investigation can be placed in perspective of the synthetic and enzymatic production of xenonucleic acids (XNA), whose chemical backbone motive differs from deoxyribose and ribose, as an (non-interfering) alternative to the DNA and RNA biosynthesis.3
Results
The oligonucleotide sequence d(CGCACGC) forms a right-handed, anti-parallel double helix with its base pair complement d[GCG(xT)GCG], where the natural occurring thymine nucleotide is replaced by its cyclohexene analogue. Two helices are present in the asymmetric unit. Only six out of seven bases are engaged in standard Watson-Crick base pairs, with hydrogen bonding distances close to the standard values (Fig. 2), the remaining bases are flipped away and do not participate in base pair hydrogen bond interactions with the complementary base.
Figure 2.
Stick representation showing the two duplexes present in the crystal structure, duplex 1 on the left, duplex 2 on the right.
The average C1′—C1′ distance for the six base pairs is in both duplexes 10.7 Å and the average λ angles (angle between the vectors C1′—C1′ and C1′—N1/N9) are very comparable, 52.9° and 52.7°.
In the further discussion the duplex formed by chain A and chain B will be named duplex 1, the duplex formed by chains C and D, duplex 2 (chain IDs referring to NDB code BD0108, within each chain the residues are numbered 1 to 7); chains A and C contain the CeNA modification (xT). The numbering scheme for the CeNA sugar ring is given in Figure 1D.
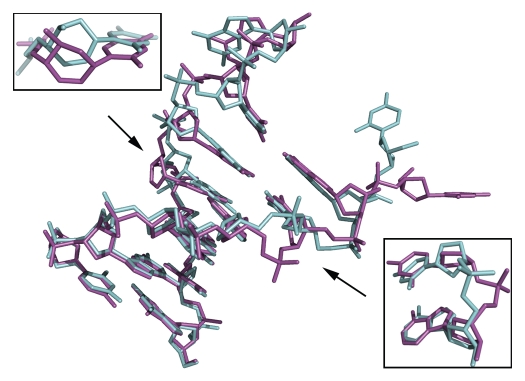
Both helices in the asymmetric unit show similar characteristics: the two nucleotides of the terminal G•C base pair are flipped away and in both duplexes the flexibility of the CeNA residue is visible in the sugar-phosphate backbone. When superposing the six standard base pairs present in both helices (ignoring the flipped out bases) the RMS deviation is 1.01 Å. The largest overall deviation is observed for the CeNA residues (Fig. 3) but the phosphate group of the adenine residue (base pair complement of the CeNA residue) shows the maximum positional deviation (4.77 Å for the O1P atom). As can be seen in Figure 3 the CeNA residue of duplex 2 is shifted towards the minor groove with respect to the CeNA residue in duplex 1. This can mainly be attributed to a larger shift for the CeNA residue (not for the complete base pair), 2.03 Å compared to 0.38 Å accompanied by an increased tilt for the CeNA residue in duplex 2 (8.6° opposed to −0.4°). The phosphate groups of the adenine residues (base pair complement) are found on opposite sides of the sugar-phosphate backbone (Fig. 3). In duplex 1 the phosphate torsion angles (α/ζ) are in the −gauche/+ac conformation, in duplex 2 the torsion angle configuration is practically reversed and is observed to be trans/−gauche.
Figure 3.
Stick representation of the two CeNA substituted DNA duplexes, duplex 1 (green) and 2 (magenta), superposed onto each other. Both duplexes closely resemble each other apart from the flipped out cytosine residue. Upper left, enlargement showing the shift of the CeNA residue of duplex 2 towards the minor groove with respect to the CeNA residue in duplex 1. Lower right, enlargement showing the phosphate groups of the adenine residues, which are found at opposite sides of the sugar-phosphate backbone.
Also for the flipped out bases both similarities and large differences are present. While the flipped out guanine residues are within the same region, the cytosine residues on the other hand are rotated almost 180° with respect to each other when superposing duplex 1 onto duplex 2.
Influence of the incorporated CeNA residue.
For the 2′-deoxyribose sugars of the natural nucleotides the ring puckering ranges from C4′-endo over C2′-endo to O4′-endo for duplex 1, and from C3′-exo over C2′-endo to O4′-endo for duplex 2. The adenine residue in duplex 2 lies outside this range and is in the C3′-endo conformation to accommodate the displaced phosphate group (Fig. 3). In both duplexes the five-membered sugar rings of the flipped out guanine residues are in the C2′-exo conformation. The ring puckering of the CeNA residues can be classified as a 3H2-halfchair conformation for the CeNA residue in duplex 2, and a 2H3-halfchair conformation in duplex 1, slightly distorted towards the 2E conformation. The latter behavior, where the C3′ atom is pushed into the plane of the double bond, has already been observed in a previous crystallographic study on a Dickerson dodecamer with an incorporated CeNA residue,2 where the stretching of the inter-phosphate distances was not observed and the global B-type helical conformation was maintained. In the CeNA substituted heptamer duplex the stretching effect is less pronounced, indicating a possible cumulative effect in the modified Dickerson sequence where two CeNA residues are located in close proximity to each other, while in opposite strands. Also the sugar-phosphate backbone displays alternative conformations around the CeNA residues in both duplex 1 and duplex 2, which will help to relax the internal strains that could result in an overstretching of the inter-phosphate distance. The 3H2-halfchair conformation was noted before; however in a octamer sequence built entirely from (left-handed) CeNA building blocks and forming a (left-handed) A-type double helix.4
From NMR studies as well as the previous crystallographic work1,2,4 it was already evident that the CeNA sugar-ring is flexible and tolerated in both A- and B-type helices. Here this flexibility is confirmed by the presence of both the 3H2-halfchair and the 2H3-halfchair (2E) conformation (Fig. 1) in two distinct helices in the same asymmetric unit. This flexibility is however not confined to the six-membered CeNA sugar, but spreads to the sugar-phosphate backbone in its proximity, which is visible as alternative conformations in the sugar-phosphate backbone.
The helical parameters for the helical region of both duplexes (Table 3) were calculated with the 3DNA package5 and compared with values of canonical A- and B-form duplexes. The average values for both duplex 1 and 2 are fairly comparable and correspond relatively well with the values found in a classical B-type helix. There are however large fluctuations for the individual base pair and base-pair step parameters. Especially the inclination and propeller twist in duplex 2 and the buckle in both helices show high variation. The helical rise (3.2–3.3 Å) and helical twist (33.3°, duplex 1; 31.5°, duplex 2) are similar in both duplexes and the latter corresponds to about 11.1 base pairs in a full turn. The minor groove width is calculated as the closest separation between the O4′ atoms (or the C7′ atoms for the CeNA residues) and was corrected for the van der Waals radii of the corresponding atoms (oxygen atoms: 1.52 Å, carbon atoms: 1.70 Å). For duplex 1 the corrected average minor groove width is 5.48 Å, which is smaller than the recorded value for duplex 2 (5.92 Å). The larger value for duplex 2 is the result of an expansion of the minor groove, as a result of the flipped out bases. Also the major groove width for duplex 2 is larger towards the flipped out bases because of the same effect, the major groove width for duplex 2 (9.77 Å) is on average 2 Å broader than in duplex 1 (7.77 Å). For the calculation of the major groove widths, calculated as the distance between phosphate atom of residue P(n) and the phosphate atom P(n + 2) on the opposite strands, the distances were corrected for the sum of the van der Waals radii of the two phosphate groups (5.80 Å).
Table 3.
Helical parameters for the modified heptamer sequence d(GCG(xT)GCG)/d(CGCACGC) with (xT) a cyclohexene residue and comparison with classical B-type DNA
| Heptamer sequence | ||||
| A-DNAa | B-DNAa | Duplex 1 | Duplex 2 | |
| x-displacement (Å) | −4.17 | 0.05 | 0.08 (0.88) | −0.54 (2.40) |
| Inclination (°) | 14.7 | 2.1 | 3.8 (9.5) | 4.6 (18.2) |
| Rise (Å) | 3.32 | 3.32 | 3.23 (0.13) | 3.32 (0.40) |
| Twist (°) | 31.1 | 36.0 | 33.3 (6.0) | 31.5 (8.9) |
| Slide (Å) | −1.53 | 0.23 | 0.28 (0.47) | 0.08 (0.72) |
| Roll (°) | 8.0 | 0.6 | 2.0 (5.0) | 0.5 (8.7) |
| Propeller twist (°) | −11.8 | −11.4 | −5.2 (5.4) | −3.6 (12.4) |
| Buckle (°) | −0.1 | 0.5 | −4.9 (11.2) | −5.9 (12.4) |
Helical parameters for A- and B-type DNA are taken form (Olson et al. 2001). Values in parentheses represent the s.d.
The hydration patterns in the minor groove observed in both duplex 1 and duplex 2 are characteristic for a wide minor groove. Narrow minor grooves typically display a hydration spine, particularly in AATT regions,2,4 where water molecules bridge the opposite strands. Widening of the minor groove breaks up these bridging waters and the hydration patterns become less regular, with water molecules coordinating to the O4′ and N and O atoms of the bases. It was already shown that the CeNA residue disrupts hydrogen bonding patterns, especially because of the absence of the O4′ in the sugar ring.2 There are indeed no hydrogen bonding patterns in the minor groove of the CeNA residue, however in both duplexes part of the minor groove is occupied by the flipped out guanine residue of the other duplex. As a result there are additional hydrogen bonds between water molecules in the minor groove and the O3′ atom of these residues.
Most of the water molecules are located around the phosphate groups, more than 50% of all contacts (donor-acceptor distance between full occupancy atoms in the 2.5 Å–3.5 Å range) with water molecules involve the two free oxygen atoms on the phosphate atom and the O3′ and O5′ atoms. The major groove of duplex 1 displays twice as many water molecules compared to the major groove in duplex 2. Both minor grooves and the major groove of duplex 2 show about the same hydration level, despite the fact that there are more hydration sites in the major groove.6 In duplex 2 only one out of the two possible purine hydration sites in the major groove is populated or both are populated by a bidentate binding water molecule. Furthermore the major groove in duplex 2 is filled with Co(NH3)63+ residues and bases of neighboring helices. The sugar ring oxygen, O4′, is hydrated in half of the residues.
The temperature factors for both duplexes follow the classical trend where the B-values increase in the order bases < sugars < phosphates. The average B-values observed in duplex 1 are higher than those found in duplex 2 (bases 7.4/7.0 Å2, sugars 10.0/8.7 Å2, phosphates 12.9/10.8 Å2, for duplex 1/2 respectively). The B-values for the water molecules are on average 18.6 Å2 and for the Co(NH3)63+ residues 10.4 Å2. The degree of flexibility of the CeNA residue and the sugar-phosphate backbone in general is reflected in the temperature factors of the individual chains. Chain A and C which contain the CeNA residue show the highest B-values. The Co(NH3)63+ residues may have a stabilizing effect on the individual chains, which might explain why the average B-value for chain C is lower compared with chain A (chain A, 23 contacts, 10.7 Å2; chain B, 21 contacts, 8.5 Å2; chain C, 33 contacts, 9.1 Å2; chain D, 20 contacts, 7.8 Å2). A similar stabilizing effect of Co(NH3)63+ residues was found in Robeyns et al.2 where a cobalt hexamine residue stabilized a hydration spine.
No inter-strand overlap between the bases is observed, for the intra-strand overlap there is a great difference between the two duplexes for the base pair steps involving the CeNA modification. There is a difference in base overlap in step 3 (from G3•C5 to T4•A4), 3.13 Å2 compared to 4.00 Å2 for duplex 1 and 2 respectively and 0 Å2 compared to 1.81 Å2 in step 4 (from T4•A4 to G5•C3).
In both duplexes the CeNA phosphate group is observed in two conformations, one of which can be characterized as BI, the other as BII (duplex 1, ε-ζ difference: 75.7°; duplex 2, ε-ζ difference: 36.6°). The torsion angles around the CeNA phosphate (α and ζ) are in the −gauche/trans or −gauche/−ac conformation except for the BII conformation in duplex 2 where a +gauche/trans conformation is observed. In both duplex 1 and 2 the CeNA phosphate group is in close proximity of a Co(NH3)63+ residue, which helps to stabilize the energetically less favorable BII conformations.
Crystal packing.
The helical regions of duplex 1 and 2 stack alternating to form pseudo-continuous helices along the c-axis, with the flipped out guanine bases in duplex 1 and 2 trapped in the minor groove of the subsequent duplex within the pseudo-continuous helices. The guanine base makes a dihedral angle of 44° and 47° (duplex 1 and 2 respectively) with the G1 base of the neighboring duplex (Fig. 4). Both bases interact via two N2–N3 guanine-guanine interactions. This type of minor groove G*(G•C) triplet interactions is already reported in a decamer structure and occurs also in dodecamer structures of the type d(CGX8GC).7 In duplex 1 the O4′ of the guanosine residue interacts in a similar way with the guanine N2 (3.19 Å) and the cytosine O2 (3.13 Å) of the first base pair of the neighboring duplex as previously described.17 In duplex 2 however the O4′ interactions are replaced by a single interaction with the guanine N2 of the second base pair of the neighboring helix (Fig. 4). In addition the major groove side of the guanine bases interacts with a cobalt hexamine.
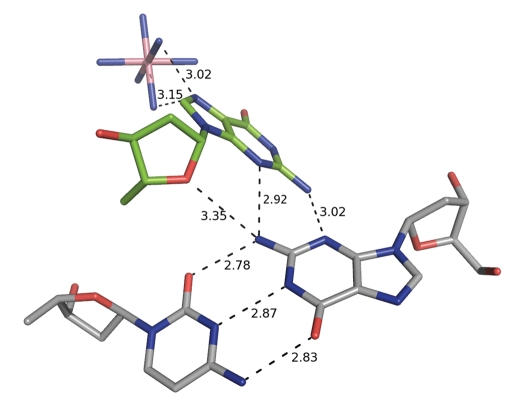
Figure 4.
The minor groove G*(G•C) triplet interactions found between the flipped out guanine base of duplex 2 (green) and the first G•C base pair of the neighboring duplex 1 (grey). The flipped out guanine is further stabilized by hydrogen bonding interactions with a Co(NH3)63+ residue. Hydrogen bonding interactions are indicated by dashed lines with distances given in Å.
The flipped out cytosine bases of the terminal base pair occupy the void space between the helical columns. The cytosine base in duplex 1 interacts with the major groove (interactions with the bases, phosphates and O4′ of the sugar) of neighboring helical columns. The corresponding cytosine residue in duplex 2, which displays a totally different orientation (Fig. 3), shows almost no interactions. This lack of interactions results in the ability to move; therefore the occupancy of the sugar-base entity has been refined to 73%. Furthermore both cytosine residues are also stabilized by interactions with cobalt hexamine residues.
Moving across the pseudo-continuous helix from duplex 1 to duplex 2, the inter-helical base pair step has a twist of −62.4° and a rise of 3.81 Å; when going from duplex 2 to duplex 1 there is a rise of 3.90 Å and the long axes of the base pairs are almost perpendicular to each other (twist −83.2°).
Cobalt hexamine interactions.
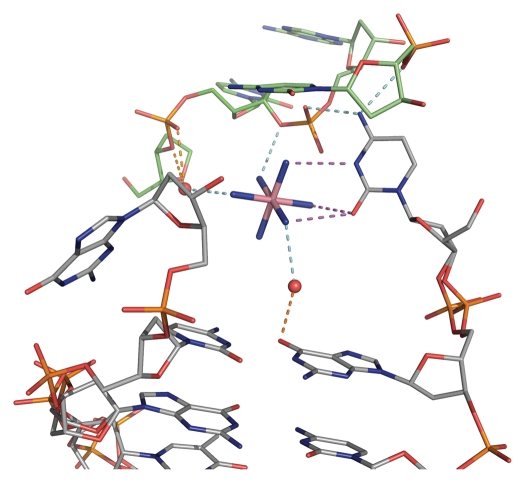
The cobalt hexamine residues played an important role in terms of solving the crystal structure, but are also inherent to the stability of the crystal structure. The cobalt hexamine complexes are found in the proximity of all four flipped out bases, reducing the flexibility of these bases and linking them to neighboring chains. The stabilization effect of the cobalt hexamine complex is comparable for both flipped out guanine bases but differs for the flipped out cytosine bases. In duplex 1 the cytosine base interacts through the O2 and N3 atoms with the cobalt hexamine complex and links the cytosine base through the cobalt hexamine complex with two neighboring guanine bases (Fig. 5). In duplex 2 only the cytosine O2 atom interacts with a cobalt hexamine complex. This is to the best of our knowledge the first observation of cobalt hexamine complexes interacting with the cytosine base.
Figure 5.
Hydrogen bonding interactions between the terminal flipped out cytosine base in duplex 1 with the cobalt hexamine residue. Three hydrogen bonds are present between the cobalt complex and the pyrimidine base, which further interacts with neighboring phosphate groups. The cobalt hexamine complex not only stabilizes the flipped out base but also the packing arrangement through hydrogen bonding interactions between different chains.
Most cobalt hexamine complexes are located in-between the pseudo helical columns and provide a close and stable packing of the duplexes, either through direct hydrogen bonding or indirectly via the coordinated water molecules surrounding the cobalt hexamine complexes. Furthermore the cobalt hexamine complexes stabilize the CeNA ring puckering by binding to the phosphate group of the CeNA residue as well as to the adenine phosphate group in duplex 1.
Discussion
The heptamer duplex, which holds a single CeNA residue in the central region, crystallizes with two duplexes in the asymmetric unit in the space group P212121. Both helices were found to form an anti-parallel right-handed duplex, closely related to the classical B-type helix. The high resolution with data truncated to 1.17 Å allows observation of the fine structure of the DNA sugar-phosphate backbone and the sugar puckering of the CeNA incorporation. Standard Watson-Crick base pairs are observed for six bases, the terminal base pair being flipped out.
The conformation of the 2′-deoxyribofuranose sugar moieties, apart from the flipped out residues, varies from C4′-endo over C2′-endo to O4′-endo. The CeNA sugar ring is in the 3H2-halfchair conformation in duplex 2, and in the 2H3-halfchair conformation, slightly distorted towards the 2E conformation in duplex 1. This difference in sugar conformation shows the flexibility of the CeNA sugar moiety. Because of the high resolution it was also possible to view alternative conformations in the sugar-phosphate backbone, which may arise from the incorporation of the flexible CeNA residue. When comparing the helical regions of duplex 1 and 2 with the previously determined NMR structure,1 the overall structural features are similar, with low rms deviations, especially when comparing only the three central base pairs around the CeNA incorporation (0.96 Å for duplex 1 and 1.01 Å for duplex 2).
Cobalt hexamine complexes were found to stabilize the CeNA ring puckering, the flipped out bases and the overall crystal packing. The occurrence of cobalt hexamine in the crystal structure and in the previously analyzed crystal structure with incorporated CeNA residues2 might show the necessity of cobalt hexamine in stabilizing B-type helices with incorporated CeNA residues. The CeNA residue, which was designed as a flexible residue, is here in both half-chair conformations that mimic the classical C2′-endo (2H3-conformation) and C3′-endo (2H3-conformation) conformations. The cobalt hexamine residues play an important role in stabilizing the B-type (2H3-conformation) of the CeNA residue.
These results may be interpreted in function of the use of CeNA as potential synthetic aptamers, for which evolution of polymerases for the enzymatic synthesis of CeNA is needed.3 For the selection of CeNA dependent polymerases, information about the structure of the artificial polymer and its interaction with the enzyme may be very helpful to decide where to introduce mutations in the enzyme to generate potential successful polymerase libraries. Such a selection process starts with the introduction of only few modified nucleotides in a natural DNA, and study the recognition by natural polymerases. The observation that a cyclohexenyl nucleotide can adopt different conformations when introduced in the same DNA sequence is important, as the active site of a polymerase is dynamic and the polymerization process might need conformational changes in both the enzyme and the modified nucleotide. When triphosphates of unnatural nucleotides are used as substrate for natural DNA polymerases, chain termination is often observed after incorporation of two to six modified nucleotides. An explanation for this phenomenon could be the need for a conformational change of the nucleic acids during the elongation process, a few nucleotides away from the active site.8–10 CeNA might therefore be an excellent candidate to generate a polymerase that may copy DNA into CeNA and vice versa. Further investigations will demonstrate if it will be easier to evolve a polymerase for the synthesis of CeNA than for other six-membered modified nucleic acids.
Evolving a DNA polymerase so that it can be used for CeNA synthesis is a bottleneck in CeNA aptamer design. Aptamers are nucleic acids sequences, folded in such a way that they recognize another chemical substance in a very specific way and with high affinity. A certain degree of flexibility combined with a not too high tendency for self-hybridization might be beneficial to generate a diverse nucleic acids library adopting a variety of three dimensional nucleic acids structures. CeNA seems to fulfill these requirements.
Materials and Methods
Oligonucleotide synthesis.
The CeNA nucleoside,11 with a thymine base, as well as the protected phosphoramidite nucleoside, that was used in the oligonucleotide synthesis, were synthesized by the Laboratory of Medicinal Chemistry at the Rega Institute (Leuven).12 Assembly of the monomers into oligonucleotides was done as described by Maurinsh et al.13 for the nucleic acid strand d(GCG(xT)GCG) with (xT) the thymine base with CeNA sugar modification (Fig. 1). The unmodified strand d(CGCACGC) was purchased from RNAtec (Leuven).
Crystallization conditions.
The crystallization conditions for the heptamer were screened using a 24-matrix screen developed for oligonucleotides.14 The screening showed the necessity of cobalt hexamine as a crystallization agent. Crystals were obtained using a crystallization screen containing either 10% (v/v) 2-methyl-2,4-pentanediol (MPD), 20 mM cobalt hexamine, 40 mM potassium cacodylate buffered at pH = 5.5, and 40 mM LiCl, 20 mM MgCl2 or alternatively 80/12 mM KCl/NaCl as source of monovalent cations and no Mg2+ ions.
The oligonucleotide was crystallized using the ‘hanging-drop’ vapor diffusion technique. Droplet size was 2 µL (1 µL screen, and 1 µL of oligonucleotide at 1.2 mM) with an overall single-strand oligonucleotide concentration of 0.6 mM. The oligonucleotide sample was obtained by titration of a DNA solution d(CGCACGC) with the complementary modified sequence d(GCG(xT)GCG). The degree of complex formation was monitored with one-dimensional NMR spectra of non-exchangeable base protons and anomeric protons, after which the solution was lyophilized.
The crystallization experiment was set up at 16°C and produced crystals after only one day. Crystals have dimensions of about 0.15 × 0.15 × 0.44 mm and were stable over time.
Data collection.
A ‘native’ dataset was collected at the Swiss Light Source (SLS) synchrotron facility, beamline X06SA (λ = 0.7749 Å). 180 data images were collected under cryo-conditions (100 K) on a Pilatus detector with 1° increments. The crystals diffracted beyond 1 Å and were processed with ‘mosflm’15 and scaled with ‘Scala’.16 The resolution was truncated to 1.17 Å and data collection statistics are summarized in Table 1. A SAD dataset (λ = 1.5498 Å) was collected on a second crystal (same crystallization conditions as the native set) in order to exploit the anomalous signal from the phosphate atoms (600° at Δφ increments of 1.5°, 1.7 Å resolution).
Table 1.
Data collection statistics for the modified heptamer sequence d(GCG(xT)GCG)/d(CGCACGC) with (xT) a cyclohexene residue
| Space group | P 21 21 21 |
| Resolution Range (Å) | 18.64–1.17 (1.23–1.17)† |
| Measured reflections/Unique | 134041/24411 (20204/3448)† |
| Completeness (%) | 99.6 (98.8)† |
| Rmerge (%) | 9.4 (57.1)† |
| Mean I/σ | 21.7 (4.8)† |
| Multiplicity | 5.5 (5.9)† |
| Mosaicity (°) | 0.67 |
| B-value (Wilson) (Å2) | 6.3 |
Values in parentheses are for the outmost shell.
Structure determination.
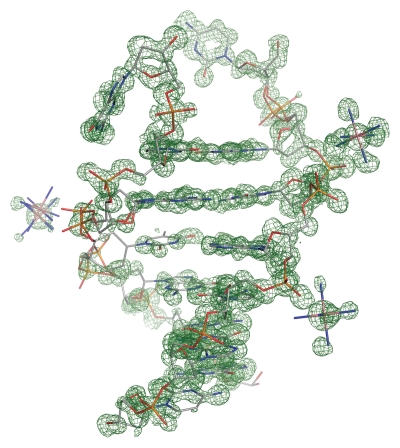
Molecular replacement using an NMR structure of the heptamer duplex failed, indicating that the crystal structure could show some atypical characteristics. The required files for SAD phasing with SHELXD/E17,18 were prepared using XPREP.19 The substructure was found with SHELXD, searching for 24 sites (two duplexes in the a.s.) followed by 400 cycles of density modifications using SHELXE with extrapolated data to 1 Å. The structure could be built in the phased electron density (Fig. 6) maps using COOT.20 Visualization of the initial maps immediately revealed that the phasing was performed on Co(NH3)6 residues (seven Co(NH3)63+ residues located) that were trapped in the crystal packing.
Figure 6.
Final atomic model of one d(GCG(xT)GCG)/d(CGCACGC) duplex superimposed onto the initial map after SAD phasing displayed in green. Electron density map contoured at the 1σ level.
The structure was refined with SHELXL using the ‘native’ dataset to 1.17 Å. For the unmodified residues standard dictionary restraints were used, for the CeNA residues restraints were used in analogy of previously solved crystal structures by our group, containing CeNA moieties.2
Because of the high 1.17 Å resolution it was clearly visible in both the 2|Fo| - |Fc| and |Fo| - |Fc| maps that the sugar-phosphate backbone showed alternative conformations. SHELXL was used to refine the occupancies of some regions of the backbone as well as some Co(NH3)63+ residues. Furthermore the two heptamer duplexes both show some distinct features such as bases flipped away, bases interacting with neighboring helices and conformational flexibility of the sugar-phosphate backbone (especially close to the CeNA residue).
Full least squares refinement for all 24372 unique reflections converged to an R1-value of 14.38%, ωR2-value of 34.92% and GooF (S) of 1.241 (restrained GooF 1.043). Refinement was performed using 9930 restraints for 6402 parameters. All atoms, except for the water molecules, were refined anisotropically. The Rfree-value (5% of the data) was used to cross-validate the (anisotropic) refinement (Rfree drop of 3.5% after anisotropic refinement), the above R-values being calculated for the whole dataset. Further refinement details are listed in Table 2.
Table 2.
Refinement statistics for the modified heptamer sequence d(GCG(xT)GCG)/d(CGCACGC) with (xT) a cyclohexene residue
| Resolution range (Å) | 19.86-1.17 |
| Number of reflections | 24372 |
| Number of atoms: | |
| Nucleic acid | 600 |
| Waters (treated as O) | 130 |
| Co(NH3)63+ | 49 |
| Final R-value (all data) (%) | 15.80 |
| RMS deviation from restraint target value: | |
| Bond lengths (Å) | 0.009 |
| Angle distances (Å) | 0.036 |
| Distances from restraint planes (Å) | 0.003 |
| Anti-bumping distance restraints (Å) | 21 |
| Mean B-values (Å2) | |
| DNA atoms | 9.0 |
| Solvent Atoms (H2O) | 18.2 |
The automated water divining procedure SHELXwat was used to locate the majority of the 131 water molecules, the remaining water molecules were found after inspection of the difference maps. Waters were set to half occupancy when isotropic B-values exceeded 40 Å2, and monitored in further refinement cycles. In total 42 out of 131 water molecules were given occupancy less than 1 (Some water molecules form hydrogen bonds with the flexible regions of the sugar-phosphate backbone, in which case the water molecules were given the occupancy of the non-solvent atoms they interact with).
All molecular figures were created using the program PyMol [DeLano, W.L. The PyMOL Molecular Graphics System (2002) on World Wide Web http://www.pymol.org].
Acknowledgements
Koen Robeyns thanks the K.U. Leuven Research Fund for a postdoctoral position. We thank the staff of the Swiss Light Source in Villigen (beamline X06SA) for help with the synchrotron experiments. BioMacS, the K.U. Leuven Interfacultary Centre for Biomacromolecular Structure, is supported by the Impulse Project of the K.U. Leuven.
Accession codes. Final coordinates and structure factor amplitudes have been deposited with the Protein Data Bank (3FL6) and Nucleic Acid Data Bank (BD0108).
Footnotes
Previously published online: http://www.landesbioscience.com/journals/artificialdna/article/10952
References
- 1.Nauwelaerts K, Lescrinier E, Sclep G, Herdewijn P. Cyclohexenyl nucleic acids: conformationally flexible oligonucleotides. Nucleic Acids Research. 2005;33:2452–2463. doi: 10.1093/nar/gki538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robeyns K, Herdewijn P, Van Meervelt L. Influence of the incorporation of a cyclohexenyl nucleic acid (CeNA) residue onto the sequence d(CGCGAATTCGCG) Nucleic Acids Research. 2008;36:1407–1414. doi: 10.1093/nar/gkm1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herdewijn P, Marliere P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem Biodivers. 2009;6:791–808. doi: 10.1002/cbdv.200900083. [DOI] [PubMed] [Google Scholar]
- 4.Robeyns K, Herdewijn P, Van Meervelt L. Structure of the fully modified left-handed cyclohexene nucleic acid sequence GTGTACAC. J Am Chem Soc. 2008;130:1979–1984. doi: 10.1021/ja077313f. [DOI] [PubMed] [Google Scholar]
- 5.Lu XJ, Olson WK. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Research. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider B, Cohen D, Berman HM. Hydration of DNA bases—analysis of crystallographic data. Biopolymers. 1992;32:725–750. doi: 10.1002/bip.360320703. [DOI] [PubMed] [Google Scholar]
- 7.Nunn CM, Neidle S. Structure of the DNA decamer d(GGCAATTGCG) contains both major- and minor-groove binding G.(G.C) base triplets. Acta Crystallographica Section D-Biological Crystallography. 1998;54:577–583. doi: 10.1107/s090744499701559x. [DOI] [PubMed] [Google Scholar]
- 8.Vastmans K, Pochet S, Peys A, Kerremans L, Van Aerschot A, Hendrix C, et al. Enzymatic incorporation in DNA of 1,5-anhydrohexitol nucleotides. Biochemistry. 2000;39:12757–12765. doi: 10.1021/bi001297g. [DOI] [PubMed] [Google Scholar]
- 9.Renders M, Abramov M, Froeyen M, Herdewijn P. Polymerase-catalysed incorporation of glucose nucleotides into a DNA duplex. Chem-Eur J. 2009;15:5463–5470. doi: 10.1002/chem.200801951. [DOI] [PubMed] [Google Scholar]
- 10.Marx A, MacWilliams MP, Bickle TA, Schwitter U, Giese B. 4′-acylated thymidines: A new class of DNA chain terminators and photocleavable DNA building blocks. J Am Chem Soc. 1997;119:1131–1132. [Google Scholar]
- 11.Wang J, Herdewijn P. Enantioselective synthesis and conformational study of cyclohexene carbocyclic nucleosides. J Org Chem. 1999;64:7820–7827. [Google Scholar]
- 12.Gu P, Griebel C, Van Aerschot A, Rozenski J, Busson R, Gais HJ, Herdewijn P. Synthesis of enantiomeric-pure cyclohexenyl nucleoside building blocks for oligonucleotide synthesis. Tetrahedron. 2004;60:2111–2123. [Google Scholar]
- 13.Maurinsh Y, Rosemeyer H, Esnouf R, Medvedovici A, Wang J, Ceulemans G, et al. Synthesis and pairing properties of oligonucleotides containing 3-hydroxy-4-hydroxymethyl-1-cyclohexanyl nucleosides. Chem-Eur J. 1999;5:2139–2150. [Google Scholar]
- 14.Berger I, Kang CH, Sinha N, Wolters M, Rich A. A highly efficient 24-condition matrix for the crystallization of nucleic acid fragments. Acta Crystallogr D Biol Crystallogr. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- 15.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992:26. [Google Scholar]
- 16.Evans PR. Scala. Joint CCP4 + ESF-EACBM Newsletter on Protein Crystallography. 1997:33. [Google Scholar]
- 17.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 18.Sheldrick GM. Macromolecular phasing with SHELXE. Zeitschrift Fur Kristallographie. 2002;217:644–650. (Ger). [Google Scholar]
- 19.Bruker, XPREP, Bruker AXS Inc., author Madison WI, USA: 1997. [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]