Abstract
The clinical use of small interfering RNA (siRNA) is hampered by poor uptake by tissues and instability in circulation. In addition, off-target effects pose a significant additional problem for therapeutic use of siRNA. Chemical modifications of siRNA have been reported to increase stability and reduce off-target effects enabling possible therapeutic use of siRNA. Recently a large scale direct comparison of the impact of 21 different types of novel chemical modifications on siRNA efficiency and cell viability was published.1 It was found that several types of chemical modifications could enhance siRNA activity beyond that of an unmodified siRNA in vitro. In addition, a novel siRNA design, termed small internally segmented interfering RNA (sisiRNA), composed of an intact antisense strand and segmented guide strand stabilized using LNA was shown to be effective in cell based assays. In the present study we examined the in vivo efficacy of the LNA and UNA modified siRNA and sisiRNA in a mouse model bearing human tumor xenografts. We studied the biodistribution and efficacy of target knockdown in the mouse model. In addition we used whole genome profiling to assess the off-target effects in the liver of the mouse and the tumor xenografts. We report that LNA and UNA modified siRNA and sisiRNA improve the efficacy in target knockdown as compared with unmodified siRNA in the tumor xenografts without formulation. However, the level of off-target gene regulation in both the tumor and the liver correlated with the increase in efficacy in target knockdown, unless the seed region of the siRNA was modified.
Key words: LNA, unlocked nucleic acid, siRNA, sisiRNA, off-target effects
Introduction
Post-transcriptional gene silencing mediated by double stranded RNA represents an evolutionarily conserved cellular mechanism. Small dsRNA, such as microRNA (miRNA) are actually part of the main regulatory mechanisms of gene expression in cells. The possibilities of harnessing this intrinsic natural mechanism of gene silencing for therapeutic applications was opened up by the discovery by Tom Tuschl's team a few years ago that chemically synthesised small 21-mers of double stranded RNA (small interfering RNA, siRNA) could inhibit gene expression without induction of cellular antiviral-like responses.2 However, recent insights make it clear that siRNA faces some major hurdles before it can be used as a drug. Some of these problems are similar to those associated with classic antisense approaches, such as stability in circulations and lack of delivery to specific tissues (other than the liver), while other problems are more specific for siRNA, such as non sequence specific and off-target effects that can be caused for three reasons. First, non-specific effects can be caused by the unintended incorporation of the passenger or “sense” strand into RNA induced silencing complex (RISC). Second, (partial) homology to the seed region of a microRNA (miRNA) can cause the deregulation of hundreds of genes.3 Third, the innate mammalian immune system is very adapted in recognizing nucleic acid species as signatures of potential pathogens leading to inadvertent stimulation of immune responses through toll like receptors is a distinct possibility.4 Chemical modifications might provide solutions for these three problems. For instance, by the use of chemically modified nucleotides at specific positions, RISC can be forced to preferentially incorporate one strand by either making it thermodynamically more favorable to accept one of the strands5 or by physically disabling the incorporation of the incorrect strand.6 Furthermore it was reported that disruption of the seed region by chemical modifications minimizes off-targeting by the seed region of a siRNA.3 Recognition of oligonucleotides by toll like receptors can be prevented by incorporation of LNA or 2′OMe modified RNA.7–9
So there is a considerable amount of incidental knowledge about the use of chemical modifications in siRNA. However, just recently a concerted effort was made to characterize a wide range of chemical modifications. In a large scale direct comparison of the impact of 21 different types of novel chemical modifications on siRNA efficiency and cell viability it was found that several types of chemical modifications could enhance siRNA activity beyond that of an unmodified siRNA in vitro.1 Amongst the most reliable modifications were LNA and UNA. LNA and UNA have stark contrasting properties, and can almost be considered opposites. UNA is an acyclic 2′,3′-seco RNA nucleotide which has a high structural flexibility but provides a negative effect on duplex stability.10 In contrast LNA is a structurally rigid bicyclic nucleotide which confers a very high stability in a duplex.11 LNA has a well documented track record for use in siRNA. Modifications of the overhangs confer great stability to the siRNA, which is improved with further modifications of the guide (sense) strand. LNA is compatible with the intracellular siRNA machinery and can be used to reduce undesired, sequence-related off-target effects while LNA-modified siRNAs show improved efficiency over unmodified siRNA on certain RNA motifs.5,12 In addition the high duplex stability of LNA allows the design of small internally segmented interfering RNA (sisiRNA) composed of an intact antisense strand complemented with two shorter 10–12 nt sense (guide) strands.6 The sisiRNA design prevents sense strand induced off-target effects by physically disabling the incorporation of the incorrect strand into RISC.
The use of the acyclic UNA modification in siRNA is novel. Because UNA lowers the stability of the duplex it might have a beneficial property to force the correct strand into RISC. Interestingly, in the large screen of the 21 modifications a combination of UNA and LNA modifications within an siRNA proved to confer both highest levels in efficacy and lowest level of toxicity.1
We previously reported the effects of LNA modications in siRNA duplexes on the in vivo efficacy and non-specific off target effects in a tumor xenograft model.12 For the present study we used the same in vivo model to test the in vivo characteristics of LNA and UNA modified siRNA and compared it with sisiRNA and non-modified siRNA. We compared the in vivo efficacy and studied the differential gene regulation in the liver and the tumor xenograft after treatment with the LNA and UNA modified siRNA, sisiRNA and non-modified siRNA. We find that unformulated LNA and UNA modified siRNA and sisiLNA have superior efficacy in vivo as compared to non-modified siRNA. However, the amount of non-specific differential regulated genes correlated with the increase in efficacy of the siRNA/sisiRNA and could only be decreased when chemical modifications were introduced in the seed region of the siRNA.
Results
The efficacy of the selected siRNA designs in vitro.
The siRNA design and the chemical modifications used were selected based on the results of previous screening.1 We tested five types of modified double stranded RNA molecules i.e., a end LNA modified siRNA, a LNA modified siRNA with multiple LNA modifications within the passenger (sense) strand which we named heavy LNA modified siRNA. An UNA modified siRNA, an LNA/UNA modified siRNA and a sisiRNA. All the molecules were designed to target EGFP mRNA. The sequences were kept similar in all types of modifications, only the types of chemical modifications and their positions within the sequence were varied. The tested sequences and modifications and their nomenclature are depicted in Figure 1. First we confirmed the efficacy of the siRNA and sisiRNA designs in an EGFP expressing tumor cell line. All modified siRNAs and sisiRNA were very efficient in knockdown of EGFP in MiaPaca II cells (Fig. 2). This could be best observed on western blot, i.e., 72 hrs post transfection. The UNA modified II sequence proved to be the best able in EGFP knockdown while the seed region modified siRNAs were slightly less efficient.
Figure 1.
Above, Sequences of the siRNA molecules used in this study. The positions of the modifications are indicated by underscored letters. Below, chemical structures of LNA, UNA and CENA are depicted.
Figure 2.
EGFP knockdown efficacy in vitro in transfected MiaPaca II cells using lipofectamin 2000. The siRNA molucules and nomenclature used are as depicted in Figure 1. Three concentrations were used: 1 nM, 5 nM and 25 nM. Top, mRNA levels were measured 24 hrs post transfection, and were corrected for 28S ribosomal RNA levels. Botom, Protein levels were measured 72 hrs post transfection and are corrected for eEF2α expression. MM indicates the control mismatch siRNA.
In vivo properties of the siRNA and sisiRNA.
Having confirmed the efficacy in vitro we then proceeded towards a mouse model to test the properties and efficacy of the siRNA and sisiRNA compounds. We first tested the stability in fresh mouse serum. LNA-modified siRNAs gave superior serum stability as compared to non-modified siRNA. Also the UNA I-siRNA, showed increased serum stability as compared to the unmodified siRNA. However, the central modification with UNA II decreased serum stability at least partially (Fig. 3). This latter effect might be caused by a reduction in duplex stability due to lowering of the Tm by use of UNA modifications. The breakdown of the UNA II siRNA was partial, but interestingly this molecule retained a high efficacy in target knockdown in vivo as is demonstrated below in Figure 5.
Figure 3.
Stability of modified siRNAs in mouse serum at 37°C assayed using non denaturing PAGE. The numbers indicate the number of hours of incubation at 37°C.
Figure 5.
In vivo EGFP knockdown (mRNA) in the EGFP tumor xenografts after treatment with modified siRNAs and sisiRNA as indicated. mRNA was measured using northern blots and the mRNA levels were corrected for 28S levels.
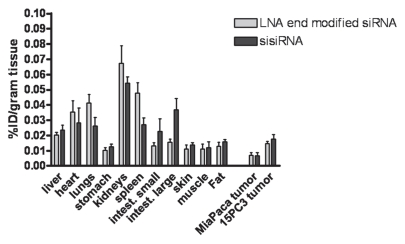
Since the sisiRNA is a novel and different type of tri-molecular design not yet tested in vivo we first wanted to know how the biodistribution compared with an LNA modified siRNA design. Nude mice bearing MiaPaca-II tumor xenografts were administered the tritiated siRNA and sisiRNA dissolved in PBS and the tissue distribution was measured after 30 minutes of circulation. LNA modified siRNA and sisiRNA displayed a very similar tissue distribution profile (Fig. 4). Kidneys were the principle site of uptake. The only small significant differences in uptake between LNA modified siRNA and sisiRNA were found in the small and large intestines. Uptake in both the tumors and liver was identical.
Figure 4.
Comparison of tissue distribution in female NMRI nu/nu mice of tritiated LNA end modified siRNA (siLNA) and sisiRNA. Data is expressed as percentage of the injected dose per gram tissue.
Having established that the sisiRNA design displayed a similar biodsitribution as a traditional siRNA design we compared the efficacy of EGFP knockdown in the tumor xenografts. Mice bearing MiaPaca II-EGFP xenografts received the different modified siRNAs via osmotic minipump dosed at 0.25 mg/kg/day for one week. The siRNAs were administered “naked” dissolved in PBS without formulation. EGFP knockdown was evaluated at the mRNA level. Mismatched siRNA was used as control. The different siRNA and sisiRNA treatments did not affect the growth rate of the tumors. EGFP mRNA levels were reduced by 50% after treatment with LNA modified siRNA, sisiRNA and UNA modified siRNA (Fig. 5). Unmodified siRNA was not successful in EGFP knockdown (Fig. 5). The high efficacy of UNA II modified siRNA was surprising because the UNA II modified siRNA was not as resistant against degradation in serum as compared to the other modified siRNA designs. Either, the efficacy of the UNA II siRNA is apparently high enough to cause target knockdown before breakdown occurs or the stable breakdown product (Fig. 3) is long enough to elicit knockdown of the target. Our data supports the latter option considering the fact that unmodified siRNA is stable for the first 8 hrs in mouse serum but shows little efficacy in target knockdown, while 50% of all the UNA II siRNA is clipped within 2 hrs resulting in a stable breakdown product, but still causes efficient knockdown of EGFP in the tumors. Aspartate aminotransferase (ASAT) and alanine aminotransferase (ALAT) levels in the blood were determined as a measurement of general liver toxicity. Changes in ASAT and ALAT levels were negligible in all siRNA/sisiRNA administrations when compared to mice treated with PBS alone (not shown).
Measurement of differential gene regulation in vivo after treatment.
We assessed the non-specific and off-target effects of siRNA treatment in both the tumors and the host (mouse liver). As a control we grafted a similar non-EGFP expressing tumor on the other flanks of the mice. Whole genome expression analysis was performed to measure the genome wide response in the tumor xenografts and the liver of mice treated with modified siRNA. In order to assay for effects in the host organism we also determined the expression pattern of the mouse liver as model tissue because this is an organ that is known to play an important role in the uptake and processing of (chemically modified) oligonucleotides.
A large number of genes were differentially regulated in liver, MiaPaca tumor without EGFP and MiaPaca tumor with EGFP after treatment with (modified) siRNA as compared to saline (control) treated animals (Table 1). It is immediately clear that a high number of genes respond to the treatment with those siRNAs that showed the highest efficacy in EGFP knockdown. In this study it is clear that chemical modifications per se do not limit the amount of differentially regulated genes. The siRNAs that were not able to knockdown EGFP did not cause much off-target effects. So there is a positive correlation between efficacy in target knockdown and differential regulation of genes.
Table 1.
The number of differentially regulated genes after treatment with modified siRNA and sisiRNA, as compared to saline treated animals
| Liver | FC2* | FC3* | MiaPaca | FC2* | FC3* | MiaPaca-GFP | FC2* | FC3* |
| Saline vs. siRNA | 17 ↑ | 8 ↑ | Saline vs. siRNA | 11 ↑ | 5 ↑ | Saline vs. siRNA | 0 ↑ | 0 |
| 12 ↓ | 7 ↓ | 12 ↓ | 5 ↓ | |||||
| Saline vs. end-mod | 347 ↑ | 115 ↑ | Saline vs. end-mod | 651 ↑ | 253 ↑ | Saline vs. end-mod | 12 ↑ | 3 ↑ |
| siLNA | 87 ↓ | 21 ↓ | siLNA | 115 ↓ | 23 ↓ | siLNA | 17 ↓ | 3 ↓ |
| Saline vs. heavy mod | 381 ↑ | 120 ↑ | Saline vs. heavy mod | 832 ↑ | 303 ↑ | Saline vs. heavy mod | 16 ↑ | 7 ↑ |
| siLNA | 45 ↓ | 16 ↓ | siLNA | 275 ↓ | 85 ↓ | siLNA | 46 ↓ | 10 ↓ |
| Saline vs. sisiRNA | 368 ↑ | 122 ↑ | Saline vs. sisiRNA | 667 ↑ | 259 ↑ | Saline vs. sisiRNA | 12 ↑ | 6 ↑ |
| 76 ↓ | 20 ↓ | 141 ↓ | 38 ↓ | 38 ↓ | 9 ↓ | |||
| Saline vs. UNA I | 417 ↑ | 134 ↑ | Saline vs. UNA I | 648 ↑ | 240 ↑ | Saline vs. UNA I | 5 ↑ | 0 ↑ |
| 73 ↓ | 24 ↓ | 286 ↓ | 128 ↓ | 29 ↓ | 10 ↓ | |||
| Saline vs. UNA II | 297 ↑ | 82 ↑ | Saline vs. UNA II | 1071↑ | 391 ↑ | Saline vs. UNA II | 21 ↑ | 7 ↑ |
| 63 ↓ | 15 ↓ | 266 ↓ | 82 ↓ | 9 ↓ | 6 ↓ | |||
| Saline vs. Seed | 104 ↑ | 27 ↑ | Saline vs. Seed | 25 ↑ | 6 ↑ | Saline vs. Seed | 25 ↑ | 11 ↑ |
| modified | 28 ↓ | 10 ↓ | modified | 18 ↓ | 9 ↓ | modified | 29 ↓ | 1 ↓ |
| Saline vs. MM | 257 ↑ | 67 ↑ | Saline vs. MM | 55 ↑ | 8 ↑ | Saline vs. MM | 9 ↑ | 3 ↑ |
| 64 ↓ | 27 ↓ | 13 ↓ | 7 ↓ | 1 ↓ | 0 ↓ |
Arrow up indicates upregulated genes, arrow down indicates downregulated genes. FC2, fold change 2; FC3, Fold chance 3; all p < 0.05 ANOVA with Benjamini-Hochberg false discovery rate correction for multiple testing.
The high number of differentially regulated genes is not an effect of EGFP knockdown because most differential gene regulation is actually observed in liver and tumors that do not express EGFP.
In general, we find that in the EGFP tumors the majority of the significantly regulated genes are actually knocked down. While in the liver and non EGFP tumors the majority of the significantly regulated genes are actually upregulated. There was no overlap in the differentially regulated genes between treatment groups in the EGFP tumors (Fig. 6). However, in the non-EGFP tumors we found a significant common regulation of 391 genes in the treatment groups with significant efficacy of EGFP knockdown: siLNA end modified, siLNA heavy modified, sisiRNA and UNA modified (Fig. 6 and Suppl. Table 1). However, Gene Ontology (GO) analysis of these 391 genes did not reveal significant GO classifications (i.e., a corrected p value below 0.05). We also compared the potential targets of the siRNA seed region sequence, predicted using a seed locator algorithm13 with the 391 common regulated genes in the Non-EGFP tumors. NONE of the 391 differentially regulated genes could be explained by direct “seed region effects”. Furthermore, all of these 391 genes were found to be upregulated.
Figure 6.
Venn diagrams showing the overlap of significant differential gene expression in the non-EGFP and the EGFP tumor xenografts in the mice treated with LNA modified siRNA, sisiRNA and UNA modified siRNA. Fold change 2 p < 0.05 ANOVA with Benjamini-Hochberg false discovery rate correction for multiple testing. The numbers within the Venn diagrams indicated the no. of genes found to be altered in expression level.
However, it is still possible that the large amount of differentially regulated genes is caused indirectly by minor pertubations in gene regulation as a consequence of seed region sequence complementarity. Because it is known that chemical modifications in the seed region of a siRNA can diminish seed region effects14 we designed another siRNA based on results obtained previously1 containing a single CENA (2′,4′-carbocyclic-ethylene-bridged-locked nucleic acid) at the 3rd position of the antisense strand (Fig. 1). The rationale to choose this CENA seed modified siRNA for in vivo testing was because in our previous studies1 this modification showed good properties in target efficacy and had low toxicity in vitro. Indeed, in vivo this seed modified siRNA was almost as effective in knocking down EGFP in the tumors as compared to LNA modified siRNA (Fig. 5), but importantly showed a stark reduction in the amount of differentially regulated genes in the tumors not expressing EGFP. The EGFP tumors showed similar numbers of deregulated genes as compared to the other effective siRNA's.
When comparing the gene expression in livers (where EGFP is not expressed) of the mice treated with LNA modified end modified, LNA heavy modified, UNA modified and sisiRNA (i.e., those molecules with highest efficacy) there is a relative large group of 51 genes with a fold change of 3, which is common in al four groups (Suppl. Table 2). All these genes were found to be upregulated and represent a general response of the liver towards treatment with siRNAs. Gene Ontology analysis revealed that no specific pathways were affected significantly. Instead the genes we found to represent a general stimulation of liver function due to treatment with siRNA or sisiRNA. In analogy with the results seen in the non-EGFP tumors we observed that the use of the seed modified siRNA resulted in a stark decrease in differentially regulated genes.
Discussion
A score of studies have tried to harness the possibilities of specific gene knockdown using siRNA in vivo, and several clinical studies are underway (www.clinicaltrials.gov). This very rapid progress of siRNA from the bench towards the clinic within a decade have made it clear that there are two major challenges that have to be overcome for succesfull therapeutic use. First, delivery and targeting of the siRNA molecules is essential for this type of drugs since the negative charge of siRNAs as well as their size makes it difficult for them to cross cell membranes. Although “naked” delivery as done in this study and in work by others,15,16 can work it will probably be suboptimal in effect. Despite this fact, currently most clinical trials only use naked siRNA which are delivered directly to the tissue of interest by direct injection or treating cells ex vivo as reviewed by rossi et al.17 The second and maybe more fundamental problem in application of siRNA as therapeutic which still claims target specificity is the non-specific effect on gene expression. This was first reported in 2003 by Jackson et al.18 Recent studies indicate that widespread perturbation of gene expression can be caused by interference with the endogenous gene regulation by microRNAs.19 Also stimulation of the immune system through toll like receptors can be the cause of the phenotype and not the knockdown of the primary target.20 The use of chemical modifications within the siRNA might be used to diminish these off-target effects. However, this report shows that the use chemical modified siRNA can still perturb the expression of genes on a large scale, unless the seed region is modified. When testing for differentially regulated genes in our model we conclude that systemic treatment with modified siRNA or sisiRNA can cause considerable de-regulation of gene expression in vivo. We found a clear correlation between differential effects on gene expression and the efficacy of the modified siRNA and sisiRNA. The chemical modifications tested in this study primarily increased the potency of the siRNAs and also the off-target effects. It is difficult to distinguish which genes are responding directly to the siRNA or which genes are secondary effects because of de-regulation of “primary off-targeted” genes. The reduced effect on off-target gene expression of the seed modified siRNA in the livers and the non-EGFP tumors suggests that for a large part seed sequence homology is responsible for a large number of differentially regulated genes. These results are in line with earlier reports that placing chemical modifications in the seed region is an important method to reduce off-target effects.14
It is tempting to hypothesize that the difference in responses between EGFP and non-EGFP MiaPaca II tumors can be caused by the absence of a perfect homologous target sequence (i.e., EGFP) in the non-EGFP tumor. This provides a greater possibility of efficacious siRNA to find other target sites slightly mismatched instead of the EGFP sequence in the non-EGFP tumors. It is possible that the high EGFP expression acts like a “sink” absorbing siRNA thus preventing de-regulation of genes.
The sets of differentially regulated genes in the non-EGFP tumors and the livers did not show any overlap. Apparently the cells in the different tissues react completely different towards treatment with the siRNA. A recent study showed that off-target transcript regulation by siRNAs is species specific.21 Since the tumor xenografts are human in origin this is a likely explanation. This study shows that chemical modifications are essential for in vivo use of siRNA's when no formulation is used. A wide variety of chemical modifications were tested in this study and it was shown that for most they increase the efficacy in vivo. Especially LNA and UNA modifications in siRNA and sisiRNA show great potential for future therapeutic development. However, as a consequence of this increase in potency the number of other genes affected increase as well. However, this study also shows that modifications in the seed region can help minimize these side effects. We conclude that by careful design and selection of the right chemistries therapeutic application of siRNA and sisiRNA will be possible in the near future.
Materials and Methods
siRNA.
All siRNA and modified siRNA and sisiRNA were synthesized by Ribotask ApS.
The phosphoramidites were incorporated into the siRNA sequence through solid-phase DNA/RNA synthesis on an automated synthesizer (for structures of chemically modified nucleotide monomers, see Fig. 1).22 For a standard RNA synthesis cycle (1–5 µmol scale), O2′-TBDMS protected RNA phosphoramidites and common reagents were used and the stepwise coupling yield of all monomers was >99%. For incorporation of modified nucleotides, a coupling time of 10 min was used. Following standard deprotection, purification and work-up, the composition and purity (>80%) of the resulting oligonucleotides was confirmed by MALDI-MS analysis and ion exchange HPLC. UNA (unlocked nucleic acid) and LNA (locked nucleic acid) modified oligonucleotides were obtained by using commercially available UNA (www.ribotask.com) and LNA (www.exiqon.com) phosphoramidites, respectively. The UNA phosphoramidites were synthesized as O2′-TBDMS derivatives by an optimized version of the procedure for synthesis of the thymine monomer.10,23 The chemical synthesis of CENA (2′,4′-carbocyclic-ethylene-bridged-locked nucleic acid) is previously described.24 CENA modified oligonucleotide were synthesized using O2′-TEM based RNA synthesis strategy.25,26
Cell lines.
The pancreatic cancer cell line MiaPaca-II and MiaPaca-II stably expressing EGFP were maintained at 37°C and 5% CO2 by serial passage in DMEM supplemented with 10% FCS, 2 mmol/L L-glutamine, 100 units/mL penicillin and 100 µg/mL streptomycin. The MiaPaca II cell line expressing EGFP was described previously.27
In vitro test.
The non-modified and modified siRNA were tested for their ability to repress EGFP expression in vitro. MiaPaca II-EGFP cells were transfected with the different siRNAs and EGFP knockdown was evaluated at the mRNA and protein level. Transfections were done in six-well culture plates with LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) as liposomal transfection agent. Protein samples were prepared in lysis buffer (PBS; 1% Triton X-100, 0.01% sodium azide) 72 h posttransfection. Total RNA was isolated with Trizol (Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions 24 h posttransfection. Northern blots were used to determine mRNA levels. RNA was denatured using glyoxal and separated on 1% agarose gels following standard protocols. RNA was subsequently transferred to Hybond-N+ membrane (Amersham, Piscataway, NJ) in 20x SSC. Following transfer, the RNA was UV crosslinked and baked at 80°C. BcgI/HindIII fragment of pEGFP-C1 was used as a probe. A 28S probe was used as loading control. Hybridisations and posthybridisation washes were according to Church and Gilbert.28 Western blots were used to determine protein levels. Cell extracts and tumor homogenates were subjected to 10% SDS-PAGE, and the resolved proteins were transferred electrophoretically to polyvinylidene difluoride membranes (Invitrogen). EGFP was detected with a rabbit anti- GFP polyclonal antibody (Molecular Probes, Eugene, OR). Elongation factor 2a (Cell Signaling, Beverly, MA) was used as loading control. Chemiluminescent detection was done on a LAS3000 in accordance with the manufacturer's instructions. EGFP signals were quantified using Aida software version 3.44 and normalized to those of elongation factor 2a.
Serum stability.
All animal experiments were conducted according to the law in the Netherlands and sanctioned by the local animal ethics committee. Modified siRNAs (10 µmol/L) were incubated at 37°C in fresh mouse serum (NMRI nu/nu). Aliquots of 4 µL were withdrawn after 2, 4, 6, 8, 24 and 48 h, of incubation. Samples were analysed on 16% nondenaturing polyacrylamide gels. Gels were stained with ethidium bromide, and siRNA was visualized on a LAS3000.
Biodistribution.
Modified siRNA and sisiRNA were tritiated using the heat-exchange method described by Graham et al.29 In brief: 1 mg of Annealed siRNA in PBS was lyophilized and subsequently dissolved in 100 µl tritiated water (185 GBq/ml; Amersham, little Chalfont, UK) with 4 µl β-mercaptoethanol. The siRNA was heated for 6 hrs at at 95°C under mineral oil. The siRNA was slowly cooled down to allow re-annealing. Then 500 µl PBS was added and applied to a 30 cm Sephadex G10 (Pharmacia) column using PBS as eluant. 500 µl fractions were collected and analyzed by liquid scintillation counting and UV absorption. Three more purification rounds with 30 cm G10 sephadex columns were done each time with the first 500 ul peak fraction to remove free tritium. After labeling the siRNA and sisiRNA were found to be still intact by using polyacrylamide gel electrophoresis. Tissue distribution studies of tritiated LNA modified siRNA and sisiRNA were done according to Bijsterbosch et al.30 For the tissue distribution NMRI nu/nu mice were used bearing tumor xenografts of MiaPaca II (pancreas tumor). Subcutaneous tumors were induced in 8- to 10-week-old female NMRI nu/nu mice (Charles River, Maastricht, the Netherlands) by injection of 106 tumor cells in Matrigel (BD Biosciences). Distribution was studied after 30 min of circulation of a bolus injection (200 µL) of tritiated siRNA (1.4 mol/L; 0.15 mg/kg). Distribution was calculated as disintegrations per minute per gram tissue present at the different organs at the time of sacrifice.
In vivo efficacy.
siRNA was tested for its ability to repress EGFP expression in vivo. Subcutaneous MiaPaca II tumors were induced in 8- to 10-week-old NMRI nu/nu mice (Charles River, Maastricht, the Netherlands) by injection of 106 tumor cells in Matrigel (BD Biosciences). One week after tumor cell injection, when tumor take was positive, administration of the siRNA started. Administration was via osmotic minipumps (model 1007D; Alzet Corp., Palo Alto, CA) placed subcutaneously. The siRNA administered using the Alzet pumps were dosed at 0.25 mg/kg/day. RNA and protein was isolated from the tumors and EGFP levels were determined in these samples using northern and western blots as indicated above.
Whole genome expression analysis.
Whole genome expression analysis was performed to measure the genome wide response in the tumor xenografts and the liver of mice treated with modified siRNA. Tumors and livers (per group of five mice three representative tumors/livers were selected) of mice that received saline, or (modified) siRNA via osmotic pumps dosed at 0.25 mg/kg/day were homogenized in Trizol using the Magnalyzer (Roche). RNA from the aqueous phase after chloroform extraction was further purified and DNase-treated on RNAeasy columns (Qiagen) using the Qiacube (Qiagen). RNA was eluted with RNase-free H2O. The quantity and quality of the RNA was assessed with a spectrophotometer (ND1000; NanoDrop Technologies, Rockland, DE) and a Bioanalyzer (model 2100; Agilent, Palo Alto, CA). RNA was labeled using Illumina TotalPrep RNA amplification kit (Ambion). The xenograft mRNA was profiled using Illumina Sentrix HumanWG-6 v3.0 expression beadchips, while the liver mRNA samples were profiled using Illumina Sentrix mouseWG-6 v.2.0 expression beadchips. The chip data was extracted using Beadstudio 3.0 (Illumina). Analysis was done with Rosetta Resolver version 7.2. Statistical analysis (ANOVA with Benjamini-Hochberg false discovery rate correction for multiple testing) was used to detect significant differences. Genes with an ANOVA p-value <0.05 and an absolute fold change greater than two were considered significantly regulated.
Acknowledgements
This work was supported by the EU-FP6 RIGHT project (no. LSHB-CT-2004-005276).
Abbreviations
- ALAT
alanine aminotransferase
- ASAT
aspartate aminotransferase
- CENA
2′,4′-carbocyclic-ethylene-bridged-locked nucleic acid
- EGFP
enhanced green fluorescent protein
- LNA
locked nucleic acid
- MALDI-MS
matrix-assisted laser desorption/ionization mass spectrometry
- miRNA
microRNA
- siRNA
small interfering RNA
- sisiRNA
small internally segmented interfering RNA
- RISC
RNA induced silencing complex
- TBDMS
tert-butyldimethylsilyl
- UNA
unlocked nucleic acid
- 2′-O TEM
2′-O-(4-tolylsulfonyl)ethoxymethyl
Footnotes
Previously published online: http://www.landesbioscience.com/journals/artificialdna/article/12204
Supplementary Material
References
- 1.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;3:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 5.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, et al. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFNα by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 8.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 10.Langkjaer N, Paternak A, Wengel J. UNA (unlocked nucleic acid): a flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg Med Chem. 2009;17:5420–5425. doi: 10.1016/j.bmc.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 11.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 12.Mook OR, Baas F, de Wissel MB, Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Mol Cancer Ther. 2007;6:833–843. doi: 10.1158/1535-7163.MCT-06-0195. [DOI] [PubMed] [Google Scholar]
- 13.Anderson EM, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, et al. Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 2008;14:853–861. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duxbury MS, Matros E, Ito H, Zinner MJ, Ashley SW, Whang EE. Systemic siRNA-mediated gene silencing: a new approach to targeted therapy of cancer. Ann Surg. 2004;240:667–674. doi: 10.1097/01.sla.0000140755.97224.9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- 17.Glud SZ, Bramsen JB, Dagnaes-Hansen F, Wengel J, Howard KA, Nyengaard JR, Kjems J. Naked siLNA-mediated gene silencing of lung bronchoepithelium EGFP expression after intravenous administration. Oligonucleotides. 2009;19:163–168. doi: 10.1089/oli.2008.0175. [DOI] [PubMed] [Google Scholar]
- 18.Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAse globally perturbs gene regulation by endogenous microRNAs. Nature Biotech. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burchard J, Jackson AL, Malkov V, Needham RH, Tan Y, Bartz SR, et al. MicroRNA-like off-target transcript regulation by siRNAs is species specific.RNA. RNA. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beaucage SL, Caruthers MH. In: Current Protocols in Nucleic Acid Chemistry. Beaucage SL, editor. New Jersey US: John Wiley & Sons, Inc.; 2001. Ch 3, Unit 3.3. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen P, Dreiøe LH, Wengel J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg Med Chem. 1995;3:19–28. doi: 10.1016/0968-0896(94)00143-q. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava P, Barman J, Pathmasiri W, Plashkevych O, Wenska M, Chattopadhyaya J. Five- and six-membered conformationally locked 2′,4′-carbocyclic ribo-thymidines: synthesis, structure and biochemical studies. J Am Chem Soc. 2007;129:8362–8379. doi: 10.1021/ja071106y. [DOI] [PubMed] [Google Scholar]
- 25.Zhou C, Honcharenko D, Chattopadhyaya J. 2-(4-Tolylsulfonyl)ethoxymethyl (TEM)-a new 2′-OH protecting group for solid-supported RNA synthesis. Org Biomol Chem. 2007;5:333–343. doi: 10.1039/b614210a. [DOI] [PubMed] [Google Scholar]
- 26.Zhou C, Pathmasiri W, Honcharenko D, Chatterjee S, Barman J, Chattopadhyaya J. High-quality oligo-RNA synthesis using the new 2′-O-TEM protecting group by selectively quenching the addition of p-tolyl vinyl sulphone to exocyclic amino functions. Can J Chem. 2007;85:293–301. [Google Scholar]
- 27.Fluiter K, ten Asbroek ALMA, van Groeningen M, Nooij M, Aalders MCG, Baas F. Tumor genotype-specific growth inhibition in vivo by antisense oligonucleotides against a polymorphic site of the large subunit of human RNA polymerase II. Cancer Res. 2002;62:2024–2028. [PubMed] [Google Scholar]
- 28.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham MJ, Freier SM, Crooke RM, Ecker DJ, Maslova RN, Lesnik EA. Tritium labeling of antisense oligonucleotides by exchange with tritiated water. Nucleic Acids Res. 1993;21:3737–3743. doi: 10.1093/nar/21.16.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bijsterbosch MK, Manoharan M, Rump ET, De Vrueh RL, van Veghel R, Tivel KL, et al. In vivo fate of phosphorothioate antisense oligodeoxynucleotides: predominant uptake by scavenger receptors on endothelial liver cells. Nucleic Acids Res. 1997;25:3290–3296. doi: 10.1093/nar/25.16.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.